Abstract
Ribosomal translocation catalyzed by EF-G hydrolyzing GTP entails multiple conformational changes of ribosome and positional changes of tRNAs and mRNA in the ribosome. However, the detailed dynamic relations among these changes and EF-G sampling are not clear. Here, based on our proposed pathway of ribosomal translocation, we study theoretically the dynamic relations among these changes exhibited in the single molecule data and those exhibited in the ensemble kinetic data. It is shown that the timing of these changes in the single molecule data and that in the ensemble kinetic data show very different. The theoretical results are in agreement with both the available ensemble kinetic experimental data and the single molecule experimental data.
Keywords: Biophysics, Mathematical Bioscience
Introduction
Ribosomal translocation is one of crucial steps in the elongation cycle of protein synthesis by the ribosome. It involves the precise movement of mRNA by one codon in the 3′ direction, which is coupled with two tRNAs that moves from the ribosomal aminoacyl (A) site and peptidyl (P) site to the P site and exit (E) site, respectively. In bacteria, the translocation is catalyzed by elongation factor G (EF-G) hydrolyzing GTP. The translocation also entails multiple conformational changes of the ribosome and other positional or conformational changes of tRNAs in the ribosome such as the forward (counterclockwise) and backward (clockwise) rotations of the small 30S subunit relative to the large 50S subunit (viewed from the 30S subunit) [1, 2, 3, 4, 5, 6], forward (counterclockwise) and reverse (clockwise) rotations of the 30S head relative to the 30S body (viewed from the 30S head) [7, 8, 9, 10, 11, 12], movements of tRNAs in the 50S subunit [4, 5, 6, 13, 14], compaction of the two tRNAs [9, 15], deacylated tRNA dissociation from the E/E site [16, 17, 18, 19, 20], etc.
To understand molecular mechanism of ribosomal translocation, it is important to know in more detail the dynamic relations among these changes or state transitions and EF-G binding and dissociation during the translocation. To this end, both single molecule fluorescence resonance energy transfer (smFRET) and ensemble kinetic (or ensemble FRET) methods were recently employed to record simultaneously the FRET data between different pairs of multiple reporters placed on both ribosomal subunits, tRNA and EF-G [15, 21]. The smFRET data revealed direct evidence of structurally distinct late intermediates during translocation such as exaggerated, reversible fluctuations of the 30S head [15]. The ensemble kinetic data revealed an early forward rotation or swiveling of the 30S head taking place while the 30S body rotating in the opposite, clockwise direction, and the backward swiveling of the 30S head starting upon tRNA translocation and continuing until the posttranslocation state is reached [21]. However, what similarities and differences are between the dynamic relations among different state transitions or processes in the translocation pathway exhibited in the smFRET data and those exhibited in the ensemble kinetic data are not clear. Do the timing and sequence of state transitions or processes exhibited in the ensemble kinetic data reflects directly the real situations in the translocation pathway? Moreover, it is imperative to have a translocation pathway that can explain various available single molecule and ensemble kinetic data.
In this work, with our proposed pathway of ribosomal translocation, the dynamic relations among different state transitions or processes in the translocation pathway exhibited in the smFRET data and those exhibited in the ensemble kinetic data are theoretically studied in detail, addressing the unclear issues mentioned above. The theoretical results are in agreement with both the available ensemble kinetic data and smFRET data.
2. Model
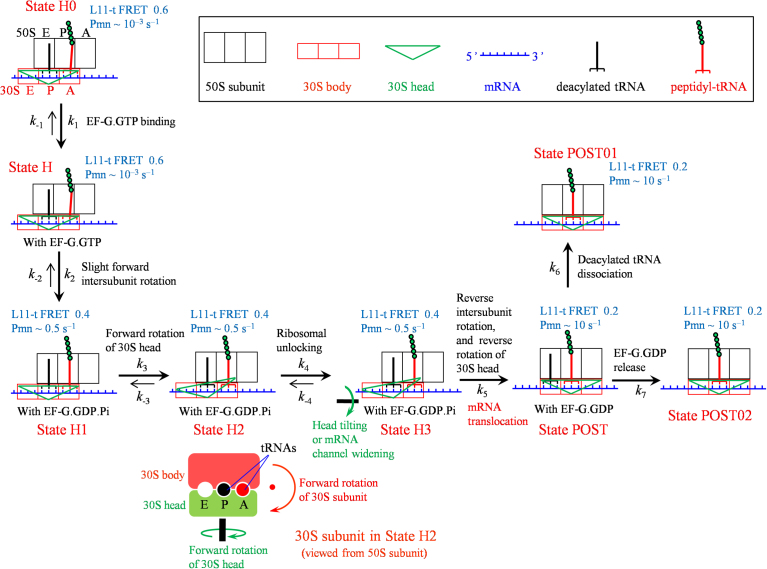
The minimal pathway of the ribosomal translocation is schematically shown in Fig. 1 [22]. Since before EF-G binding the pretranslocation ribosomal complexes are mainly in rotated hybrid state (State H0) [3], for simplicity, we consider here that before EF-G.GTP binding the ribosomal complex is in State H0 with nearly 100% probability (for the case that the ribosomal complex is not in State H0 with nearly 100% probability, the pathway is shown in Fig. S1 [22]). After EF-G.GTP binding (State H), the subsequent GTP hydrolysis causes a further slight forward 30S subunit rotation relative to the 50S subunit. This results in State H transiting to another hybrid state (State H1), which causes the peptidyl-tRNA to change the position in 50S subunit, with FRET between Cy3-labeled ribosomal protein L11 and Cy5-labeled peptidyl-tRNA (L11-t FRET) decreasing from 0.6 to 0.4, with the rate of reactivity of peptidyl-tRNA toward puromycin (denoted by Pmn) increasing from the order of to the order of 0.1–1 and with the peptidyl-tRNA becoming closer to deacylated tRNA. In State H1 with EF-G in GDP.Pi form, small conformational change in EF-G or small global rotational motion of EF-G relative to the ribosome causes the tip (loops I and II) of domain IV to move towards and interact with the decoding center in the 30S subunit, facilitating the forward rotation of the 30S head relative to the 30S body (State H2) [7, 23]. The forward 30S head rotation then leads to widening of the mRNA channel or tilting of the 30S head (State H3) [7, 8, 9, 10, 11, 12, 24], which is termed as ribosomal unlocking. Facilitated by the ribosomal unlocking, the reverse 30S subunit rotation, which is accompanied or followed immediately by the reverse rotation of the 30S head via the change of the conformation (particularly the orientation of domain IV) of EF-G, causes the translocation of the mRNA-tRNA complex in the 30S subunit, with State H3 transiting to posttranslocation state (State POST). The ribosomal unlocking also facilitates rapid Pi release, which is independent of the reverse intersubunit rotation [25]. After transition to the non-rotated conformation (State POST), as the 30S head is non-rotated the mRNA channel becomes tight again [1, 26], fixing the mRNA-tRNA complex in the 30S subunit, and the peptidyl-tRNA in the canonical P/P site induces the ribosome to be non-labile (termed as ribosomal locking in the literature) [2], inhibiting the reverse transition of State POST to State H3. Then the deacylated tRNA dissociates from the E/E site and EF-G.GDP releases independently, with State POST transiting irreversibly to State POST01 and State POST02, respectively (for clarity, we denote here by State POST01 the state where the deacylated tRNA dissociates from the posttranslocation state and by State POST02 the state where EF-G.GDP releases from the posttranslocation state, and when both the deacylated tRNA and EF-G.GDP are dissociated the two states become the same).
Fig. 1.
The minimal pathway of the ribosomal translocation catalyzed by EF-G hydrolyzing GTP (see text for detailed description). The panel below State H2 shows the interface view showing 30S subunit and head rotation in State H2. L11-t FRET denotes the FRET between Cy3-labeled ribosomal protein L11 and Cy5-labeled peptidyl-tRNA. The different values of L11-t FRET in different states are consistent with the available smFRET data [30]. Note that the rates of reactivity of peptidyl-tRNA toward puromycin (denoted by Pmn) are in the order of in State H0 and State H, in the order of 0.1–1 in State H1, State H2 and State H3, and in the order of 10 in State POST, State POST01 and State POST02, which are consistent with the available biochemical data [41].
Note that in the translocation pathway (Fig. 1), the last three transitions—State H3 to State POST, State POST to State POST01 and State POST to State POST02—are irreversible, while the other transitions may be reversible. However, catalyzed by EF-G hydrolyzing GTP, the rate constants of forward transitions, k2, k3 and k4, of the reversible transitions are much larger than the corresponding rate constants of backward transitions, k−2, k−3 and k−4 [25, 27]. Thus, for a good approximation, we take k−2 = 0, k−3 = 0 and k−4 = 0 throughout our analysis. Additionally, for simplicity, we take k−1 = 0 in our analysis. As done in the previous work [22], we take values of rate constants k2, k3, k4 and k5 as given in Table 1, which are consistent with the available experimental data [25]. By fitting to the single molecule data [20], it was shown that the rate constant of deacylated tRNA dissociation from the posttranslocation state is about 5.5 [28]. Thus, we take k6 = 5.5 (see Table 1). To be consistent with the biochemical data [25], we take rate constant of EF-G.GDP dissociation from the posttranslocation state to be k7 = 20 (see Table 1). In addition, we take k1 = 100 for the calculation (see Table 1).
Table 1.
Values of rate constants used in the calculation.
| Rate constant | Value () |
|---|---|
| k1 | 100 |
| k2 | 250 |
| k3 | 120 |
| k4 | 30 |
| k5 | 100 |
| k6 | 5.5 |
| k7 | 20 |
3. Results
As done in Belardinelli et al. [21], we consider here that the 30S head rotations are monitored by S13-L33 FRET (30S head protein S13 labeled with the nonfluorescent acceptor and large 50S subunit protein L33 labeled with the fluorescence donor), the 30S subunit rotations are monitored by S6-L9 FRET (30S body protein S9 labeled with the nonfluorescent acceptor and 50S subunit protein L9 labeled with the fluorescence donor), the movement of mRNA is monitored with reporter Alx405 or Alx488 attached to the 3′ end of the mRNA, the deacylated tRNA dissociation is monitored by tRNA-L33 FRET, and the binding and dissociation of EF-G are monitored by L12-EF-G FRET.
Based on the pathway (Fig. 1), we take following FRET values in our calculations. In State H0 and State H, the ribosomal complex is in the rotated/hybrid state with no forward 30S head rotation, and we take value of S13-L33 FRET to be A1 = 0.5 and that of S6-L9 FRET to be B1 = 0.5. The concrete values of A1 and B1 are not important because values of S13-L33 FRET and S6-L9 FRET in states after State H1 are chosen relative to A1 and B1. Since the transition from State H to State H1 causes a slight further forward intersubunit rotation, implying that S6 deviates slightly further away from L9, we take value of S6-L9 FRET in State H1 (B2 = 0.4) to be slightly smaller than that in State H (B1 = 0.5), i.e., taking B2 to be decreased by 20% relative to B1. After transition of State H1 to State H2, the 30S head makes a large forward rotation, with the distance between S13 and L13 becoming far away so that S13-L33 FRET becomes nearly zero, and thus we take value of S13-L33 FRET changing from A1 = 0.5 to A2 = 0. The transition from State H3 to State POST causes large reverse 30S subunit rotation and large reverse 30S head rotation, implying that S12 becomes much closer to L33 and S9 becomes much closer to L9 than in State H0 and State H1. Thus, we take value of S13-L33 FRET changing from A2 = 0 to A3 = 1 (i.e., taking A3 to be increased by 2-fold relative to A1 = 0.5) and value of S6-L9 FRET changing from B2 = 0.4 to B3 = 1 (i.e., taking B3 to be increased by 2-fold relative to B1 = 0.5). As will be seen below, taking these values of B2, B3, A2 and A3 relative to those of B1 and A1, the calculated changes in relative amplitudes of S13-L33 FRET and S6-L9 FRET versus time are in agreement with the ensemble biochemical data. For the mRNA movement and deacylated tRNA dissociation, we take value of Alx405-fluorescence changing from C1 = 1 (before mRNA movement) to C2 = 0 (after mRNA movement) and that of tRNA-L33 changing from D1 = 1 (before deacylated tRNA dissociation) to D2 = 0 (after tRNA dissociation). For clarity, these FRET values are summarized in Table 2.
Table 2.
Values of FRET in different states in the pathway of Fig. 1.
| State | S13-L33 FRET |
S6-L9 FRET | Alx405-Flu amplitude | tRNA-L33 FRET | L12-EF-G FRET |
|---|---|---|---|---|---|
| H0 | A1 = 0.5 | B1 = 0.5 | C1 = 1 | D1 = 1 | E1 = 1 |
| H | A1 = 0.5 | B1 = 0.5 | C1 = 1 | D1 = 1 | E2 = 0 |
| H1 | A1 = 0.5 | B2 = 0.4 | C1 = 1 | D1 = 1 | E2 = 0 |
| H2 | A2 = 0 | B2 = 0.4 | C1 = 1 | D1 = 1 | E2 = 0 |
| H3 | A2 = 0 | B2 = 0.4 | C1 = 1 | D1 = 1 | E2 = 0 |
| POST | A3 = 1 | B3 = 1 | C2 = 0 | D1 = 1 | E2 = 0 |
| POST01 | A3 = 1 | B3 = 1 | C2 = 0 | D2 = 0 | – |
| POST02 | – | – | – | – | E1 = 1 |
Symbol “–” represents that the value is not required in the calculation.
3.1. smFRET data
To simulate time courses of single-molecule S13-L33 FRET data, S6-L9 FRET data, Alx405-fluorescence data, tRNA-L33 FRET data and L12–EF-G FRET data, we use Monte Carlo algorithm, as used elsewhere [29]. In our simulations, during each time step ( s in our simulations), a random number x is generated with uniform probability between 0 and 1. If x Pi, the transition from one state to another state with the rate constant ki (i = 1, …, 7) in the pathway of Fig. 1 occurs; if x > Pi, the state transition with the rate constant ki does not occur, where is the probability of state transition. The FRET values in each state are given in Table 2.
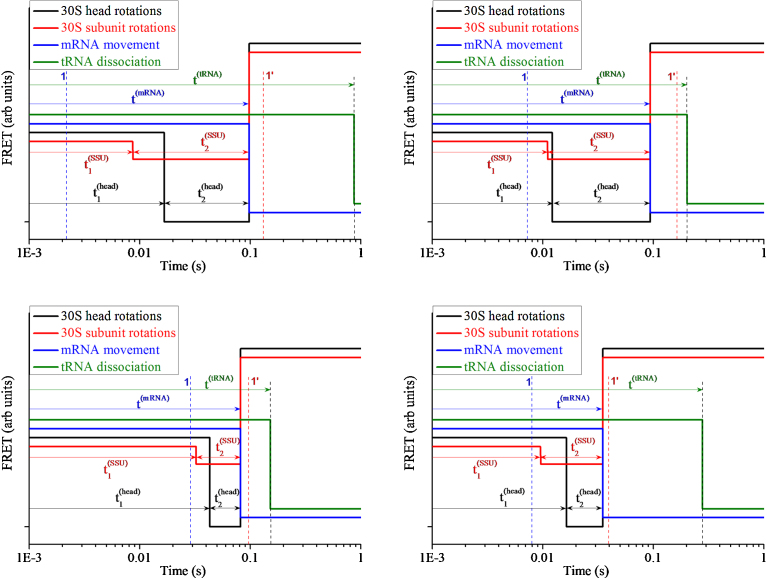
In Fig. 2, we show four examples of our simulated results for time courses of single-molecule S13-L33 FRET data, S6-L9 FRET data, Alx405-fluorescence data and tRNA-L33 FRET data, where line 1 (blue broken line) represents the moment when EF-G.GTP binds to the ribosome and line 1′ (red broken line) represents the moment when EF-G.GDP releases from the ribosome. In any of the four examples, the four smFRET data and the moments of EF-G sampling are observed simultaneously in the same ribosomal complex. As it is noted from the pathway (Fig. 1), the simulations show that the further slight forward 30S subunit rotation, forward 30S head rotation, reverse 30S head rotation, reverse 30S subunit rotation and mRNA movement occur after EF-G binding; the further slight forward 30S subunit rotation is followed by the large forward 30S head rotation; the tRNA dissociation occurs after the mRNA movement; and more importantly, the reverse 30S subunit rotation, reverse 30S head rotation and mRNA movement occur almost simultaneously.
Fig. 2.
Four examples of time courses of smFRET data characterizing 30S head rotations (S13-L33 FRET), 30S subunit rotations (S6-L9 FRET), mRNA movement (Alx405 fluorescence) and tRNA dissociation (tRNA-L33 FRET). The four examples correspond to the smFRET data observed simultaneously in four different ribosomal complexes. Line 1 (blue broken line) represents the moment when EF-G.GTP binds to the ribosome and line 1′ (red broken line) represents the moment when EF-G.GDP releases from the ribosome. For clarity, throughout the paper the four smFRET data are shifted in the vertical axis relatively with each other.
We denote by , , , , , , and the average values of , , , , , , and , respectively, i.e., , , etc, where , , etc., are defined in Fig. 2. From the pathway (Fig. 1), it is evident that when N , these average values can be calculated by
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
| (8) |
It is noted that , , , , and satisfy the relation
| (9) |
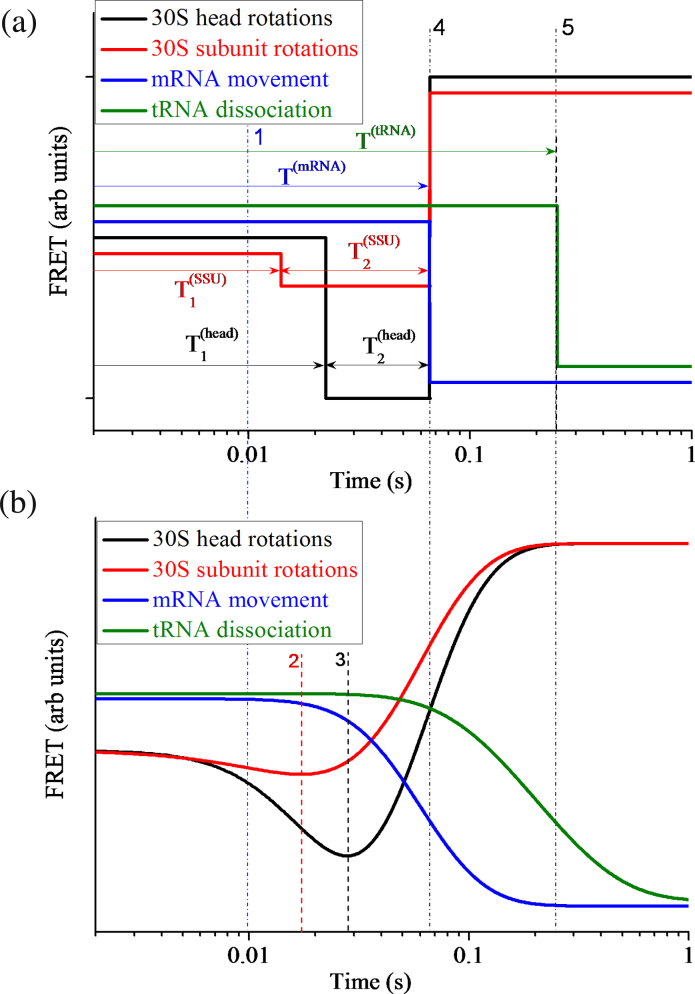
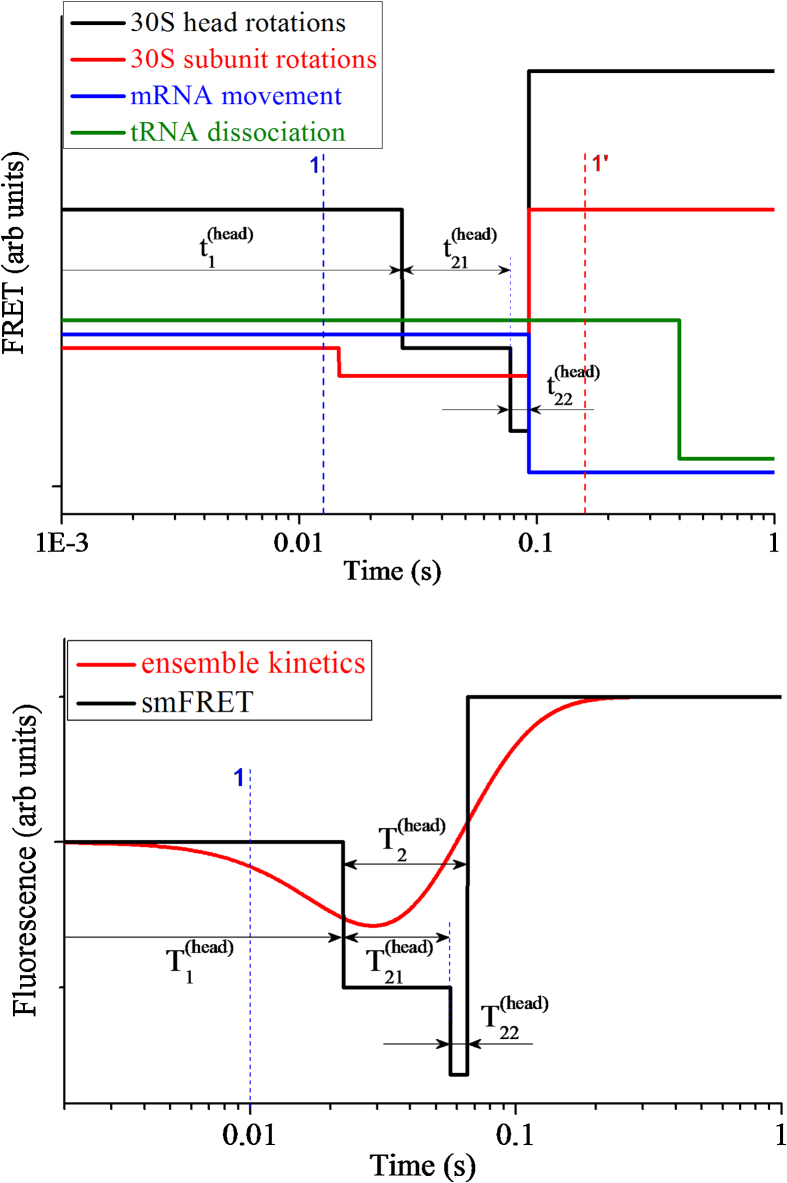
In Fig. 3a we show time courses of S13-L33 smFRET data, S6-L9 smFRET data, Alx405-fluorescence data and tRNA-L33 smFRET data, but with times , , , , and being replaced with their average values, , , , , and , respectively. Here, line 1 represents the average moment () when EF-G.GTP binds to the ribosome, line 4 represents the average moment () when the reverse 30S subunit, reverse 30S head and mRNA movement take place, and line 5 represents the average moment () when the deacylated tRNA dissociates from the E/E site of the ribosome.
Fig. 3.
Average smFRET data characterizing 30S head rotations (S13-L33 FRET), 30S subunit rotations (S6-L9 FRET), mRNA movement (Alx405 fluorescence) and tRNA dissociation (tRNA-L33 FRET). (a) Time courses of single smFRET data with the time when the smFRET changes value being replaced with the average time. (b) Time courses of ensemble smFRET data (with N 10000) or ensemble kinetic data calculated with differential equations (11)–(21). Line 1 (blue broken line) represents the average time when EF-G.GTP binds to the ribosome, line 4 represents the average time when the reverse 30S subunit, the reverse 30S head and the mRNA movement take place, and line 5 represents the average time when the deacylated tRNA dissociates from the E/E site of the ribosome. Line 2 in (b) represents the time when the ensemble FRET data characterizing 30S head rotations begin to increase, with the decrease of FRET data corresponding to the forward 30S head rotation while the increase of FRET data corresponding to the reverse 30S head rotation. Line 3 in (b) represents the time when the ensemble FRET data characterizing 30S subunit rotations begin to increase, with the decrease of FRET data corresponding to the forward 30S subunit rotation while the increase of FRET data corresponding to the reverse 30S subunit rotation.
3.2. Average smFRET data and ensemble kinetic data
In this section, we focus on theoretical studies of the ensemble kinetic data, which correspond to the case observed in bulky assays [21]. In the bulky assays, the solution contains a lot of ribosome molecules, with the number of molecules N . Thus, the experimentally observed FRET data corresponds to the average value of N smFRET data. The time course of the average value of N smFRET data can be calculated by
| (10) |
where Fi(t) is the time course of an smFRET data, which can be calculated with the Monte Carlo algorithm mentioned in the above section.
Based on the pathway of Fig. 1, the ensemble kinetic data characterizing 30S head rotations, 30S subunit rotations, mRNA movement and tRNA dissociation, which correspond to Faverage(t) calculated with N in Eq. (10), can also be calculated by following differential equations
| (11) |
| (12) |
| (13) |
| (14) |
| (15) |
| (16) |
| (17) |
where PH0, PH, PH1, PH2, PH3, PPOST and PPOST01 denote probabilities of State H0, State H, State H1, State H2, State H3, State POST and State POST01, respectively, in the pathway of Fig. 1. The ensemble S13-L33 FRET, which characterizes the 30S head rotations, can be calculated by
| (18) |
where A1 and A3 are constants (Table 2). The ensemble S6-L9 FRET, which characterizes the 30S subunit rotations, can be calculated by
| (19) |
where B1, B2 and B3 are constants (Table 2). The ensemble fluorescence of Alx405 attached to the 3′ end of the mRNA, which characterizes the mRNA movement, can be calculated by
| (20) |
The ensemble tRNA-L33 FRET, which characterizes tRNA dissociation, can be calculated by
| (21) |
Similarly, the ensemble kinetic data characterizing EF-G binding and dissociation can be calculated by Eqs. (11)–(15) supplemented by following differential equations
| (22) |
| (23) |
where PPOST02 denotes probability of State POST02 in the pathway of Fig. 1. The ensemble L12–EF-G FRET can be calculated by
| (24) |
To see at least how many smFRET data can give their average value to be nearly identical to the corresponding ensemble kinetic data calculated with Eqs. (11)–(18), we make calculations of the time course of the average value of N S13-L33 smFRET data, Faverage(t), with Eq. (10) and compare with the ensemble kinetic data of S13-L33 FRET, Fhead(t), calculated with Eqs. (11)–(18) (Fig. S2). It is seen that when N 10000, Faverage(t) becomes almost identical to Fhead(t) (Fig. S2). Similarly, the average values of S6-L9 smFRET data, Alx405-fluorescence data, tRNA-L33 smFRET data and L12-EF-G smFRET data with N 10000 are almost identical to Fsubunit(t), FmRNA(t), FtRNA(t) and FEF-G(t), respectively. Thus, we can use either Eq. (10) with N 10000 or the differential equations (11)–(24) to obtain the ensemble kinetic data monitored in the bulky assays that contain N molecules in the solution [21].
In Fig. 3b we show the ensemble kinetic data characterizing 30S head rotations, Fhead(t), 30S subunit rotations, Fsubunit(t), mRNA movement, FmRNA(t), and tRNA dissociation, FtRNA(t). Here, the decrease and increase of Fhead(t) correspond to the forward and reverse 30S head rotations, respectively; the decrease and increase of Fsubunit(t) correspond to the forward and reverse 30S subunit rotations, respectively; the decrease of FmRNA(t) corresponds to the mRNA movement; and the decrease of FtRNA(t) corresponds to the tRNA dissociation. To see the effect of slight variations of the parameter values given in Table 2 on the ensemble kinetic data, we take value of B2 to be decreased by 10–30% relative to that of B1, and take value of B3 to be increased by 1.9–2.1-fold relative to that of B1. Our calculations show that the variations of B2 and B3 only have slight effects on Fsubunit(t) (Fig. S3). Similarly, our calculations show that the slight variations of other parameters such as A3 also have only slight effects on Fhead(t) (Fig. S4).
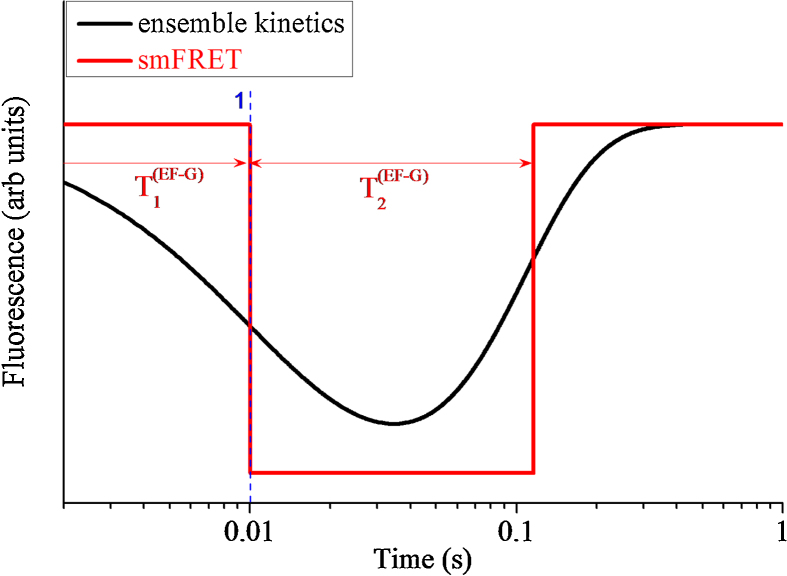
Similar to Fig. 3 showing the dynamic relations among 30S head rotations, 30S subunit rotations, mRNA movement and tRNA dissociation during ribosomal translocation catalyzed by EF-G hydrolyzing GTP, in Fig. 4 we show the dynamics and kinetics of EF-G binding and dissociation. Here, the decrease and increase of L12–EF-G data correspond to the EF-G binding and dissociation, respectively.
Fig. 4.
Average smFRET data characterizing EF-G dissociation (L12–EF-G FRET). Red line represents the time course of single smFRET data with the time when the smFRET changes value being replaced with the average time. Black line represents time course of ensemble smFRET data (with N 10000) or ensemble kinetic data calculated by using differential equations (11)–(15) and equations (22)–(24).
From our calculated data (Fig. 2, Fig. 3, Fig. 4) we have following conclusions. (i) Although the smFRET data (Fig. 2 and Fig. 3a) show that the reverse 30S head rotation, reverse 30S subunit rotation and mRNA movement occur almost simultaneously, the ensemble kinetic data (Fig. 3b) show that these processes starts to take place at very different moments: the process of reverse 30S subunit rotation starts before that of reverse 30S head rotation, and the process of mRNA movement starts before that of 30S head rotation and even before that of reverse 30S subunit rotation. In other words, the ensemble kinetic data show that before the 30S head has reached the largest angle of the forward rotation, the 30S subunit begins to make the reverse rotation and the mRNA begins to move; or, an early forward rotation of the 30S head takes place while the 30S subunit rotates in the reverse direction. (ii) The ensemble kinetic data (Fig. 3b) show that the process of tRNA dissociation starts to take place later than the beginning of the process of reverse 30S subunit rotation and that of mRNA movement. (iii) The ensemble kinetic data (Fig. 3b) show that the reverse rotation of the 30S head starts nearly upon tRNA dissociation and continues until the average time when the posttranslocation state is reached. (iv) The ensemble kinetic data (Fig. 4) show that the process of EF-G binding starts to take place before the average time of EF-G binding and the process of EF-G dissociation starts to take place later than the average time of EF-G dissociation. (v) The ensemble kinetic data (Fig. 3b) show that the process of slight forward 30S subunit rotation and that of forward 30S head rotation also start to take place before the average time of EF-G binding. (vi) The ensemble kinetic data (Fig. 3b and Fig. 4) show that after the average time of EF-G dissociation the process of reverse 30S subunit rotation and that of reverse 30S head rotation still continue. All these ensemble kinetic results are in good agreement with those of Belardinelli et al. [21].
4. Discussion
4.1. The pathway of ribosomal translocation is consistent with various experimental data
In the previous work [22], with the pathway (Fig. S1) that is similar to that of Fig. 1, we provided quantitative explanations of diverse single molecule and ensemble kinetic data obtained by different research groups in the literature, such as (i) the smFRET data on dynamic fluctuations between different tRNA states during translocation interfered with various EF-G mutants, the antibiotic fusidic acid and non-hydrolysable GTP analogue [30], (ii) the ensemble kinetic data on 30S head rotation catalyzed by EF-G hydrolyzing GTP [27], (iii) the ensemble kinetic data on tRNA movement in 50S subunit and mRNA movement in 30S subunit catalyzed by wild-type EF-G, different EF-G mutants and antibiotics [31], (iv) the ensemble kinetic data on biphasic characteristics of mRNA translocation in wild-type ribosome and in mutant ribosomes C2394A, C2394 G and C2394U, where nucleotide C2394 of 23S rRNA in the 50S subunit was mutated to A, G and U, respectively, so that the affinity of deacylated tRNA for the 50S E site was reduced greatly [32], (v) the single molecule data on the dynamics of EF-G sampling to the ribosome during ribosomal translocation [33], etc. Various ensemble kinetic data on sparsomycin-catalyzed ribosomal translocation [34, 35] were also explained well with the similar pathway [36]. As discussed in detail elsewhere [22, 26], the pathway is also consistent with various available structural data.
In this work, with the pathway of Fig. 1 we simulate the time courses of single smFRET data associated with the 30S subunit rotations, 30S head rotations, mRNA movement, tRNA dissociation, and EF-G binding and dissociation, and compare with the time courses of the corresponding ensemble kinetic data. The theoretical results are also in good agreement with the recent experimental data on the ensemble kinetic relations among 30S subunit rotations, 30S head rotations, mRNA movement, tRNA dissociation, and EF-G binding and dissociation during ribosomal translocation [21].
More interestingly, based on the pathway of Fig. 1 it is noted that at saturating concentration of EF-G.GTP, the slight decrease of the S6-L9 FRET (corresponding to the transition from State H to State H1) proceeds with a rate constant of kdecrease = k2 = 250 , which is followed by a large increase of S6-L9 FRET (corresponding to the transition from State H1 through State POST) with a rate constant of kincrease = = 21 . These results are also consistent with the ensemble kinetic data of Sharma et al. [37] showing that for the case of EF-G-induced translocation at saturating EF-G.GTP the time course of S6-L9 FRET has a small downward phase, with a rate constant of kdown = 200 or 210 , which is followed by an upward phase, with a rate constant of kup = 15 or 11 . Thus, our results imply that the experimental data of kdown = 200 or 210 in Sharma et al. [37] does not necessarily reflect the rate constant of the transition from the classical non-rotated pretranslocation to rotated hybrid state. Consider that a small fraction (denoted by Q) of the pretranslocation ribosomal complexes are in the classical non-rotated state. Then, the pathway of Fig. 1 is changed to Fig. S1, which is the same as that presented before [22]. Based on Fig. S1, at saturating concentration of EF-G.GTP the slight decrease of the S6-L9 FRET (corresponding to the transition from State H to State H1 combined with the transition from State C to State H1) proceeds with a rate constant of kdecrease having a value between those of k2 and kC, where kC is the rate constant of transition from State C to State H1 (Fig. S1). For example, in Fig. S5, we show that the results calculated based on the pathway of Fig. S1 for the time course of S6-L9 FRET (with Q = 0.2, kC = 10 and values of other rate constants given in Table 1) are almost coincident with those calculated based on the pathway of Fig. 1 (with values of rate constants k2 = 202 , k4 = 25 and other rate constants as given in Table 1); in Fig. S6, we show that the results calculated based on the pathway of Fig. S1 (with Q = 0.2, kC = 50 and values of other rate constants given in Table 1) are almost coincident with those calculated based on the pathway of Fig. 1 (with values of rate constants k2 = 180 , k4 = 26 and other rate constants as given in Table 1). Consequently, our theoretical results are consistent with both the smFRET data of Cornish et al. [3] showing that EF-G binding only mildly facilitates the transition from the classical non-rotated pretranslocation to rotated hybrid state and the recent ensemble kinetic data of Sharma et al. [37] showing a very fast (about 200 ) decrease of S6-L9 FRET data upon EF-G.GTP binding.
Additionally, the pathway of Fig. 1 is also consistent with the recent smFRET data. For example, the transition from State H to State H1 (see Fig. 1) results in the peptidyl-tRNA becoming closer to deacylated tRNA (i.e., the compaction of the two tRNAs) until the deacylated tRNA is dissociated in State POST01, causing FRET between the two tRNAs to increase from a low value to a high value and then become zero, which is consistent with the smFRET data of Wasserman et al. [15]. The positional change of the peptidyl-tRNA arising from the transition from State H to State H1 also causes the decrease of FRET between Cy3-labeled ribosomal protein L11 and Cy5-labeled peptidyl-tRNA (L11-t FRET) (see Fig. 1), which is consistent with the smFRET data of Adio et al. [30]. The characteristic of 30S head rotations in Fig. 1 is also consistent with the smFRET data of Wasserman et al. [15], as discussed as follows.
In the experiments of Belardinelli et al. [21], the 30S head rotations were monitored by S13-L33 FRET. In our above analysis (see Results), it is considered that the large forward 30S head rotation results in the distance between S13 and L33 becoming far away so that S13-L33 FRET becomes nearly zero in State H2. In State H3, the tilting of 30S head can cause a further increase in the distance between S13 and L13. Thus, S13-L33 FRET is also zero in State H3. Now, consider that the 30S head rotations are monitored by S13-L5 FRET, as done in the experiments of Wasserman et al. [15]. The large forward 30S head rotation in State H2 results in S13-L5 FRET being still larger than zero. Thus, it is expected that the tilting of 30S head in State H3 can cause a further decrease of S13-L5 FRET relative to that in State H2. To quantitatively simulate the time course of S13-L5 smFRET data, we take S13-L5 FRET to be 1 in State H0, State H and State H1, 0.5 in State H2, 0.2 in State H3, and 1.5 in State POST and State POST0. In Fig. 5a we show one example of our simulated results for the time course of S13-L5 smFRET data, where for comparison, the time courses of smFRET data characterizing 30S subunit rotations (S6-L9 FRET), mRNA movement (Alx405 fluorescence) and tRNA dissociation (tRNA-L33 FRET) are also shown. The time course of single S13-L5 smFRET data with times , and being replaced with their average values, , and , respectively, are shown in Fig. 5b (black line), where , , (with + = ). It is seen that the results for the time course of S13-L5 smFRET data are in good agreement with the recent smFRET data of Wasserman et al. [15], which show an exaggerated head rotation preceding ribosomal relocking.
Fig. 5.
Time courses of smFRET data and ensemble kinetic data characterizing 30S head rotations (S13-L5 FRET). (a) An example of the time course of the smFRET data characterizing 30S head rotations (S13-L5 FRET). For comparison, time courses of smFRET data characterizing 30S subunit rotations (S6-L9 FRET), mRNA movement (Alx405 fluorescence) and tRNA dissociation (tRNA-L33 FRET) are also shown. Line 1 (blue broken line) represents the time when EF-G.GTP binds to the ribosome and line 1′ (red broken line) represents the time when EF-G.GDP releases from the ribosome. (b) Average smFRET data characterizing 30S head rotations (S13-L5 FRET). Black line represents the time course of single smFRET data with the time when the smFRET changes value being replaced with the average time. Red line represents the time course of ensemble smFRET data (with N 10000) or ensemble kinetic data calculated with differential equations (11)–(17) and equation (25). Line 1 (blue broken line) represents the average time when EF-G.GTP binds to the ribosome.
The time course of the ensemble kinetic data of S13-L5 FRET can still be calculated by Eqs. (11)–(17) but with Eq. (18) being replaced with the following equation
| (25) |
The calculated results for the time course of ensemble kinetic data of S13-L5 FRET are shown in Fig. 5b (red line). It is seen that the time course of ensemble kinetic data of S13-L5 FRET (Fig. 5b) is much similar to that of S13-L33 FRET (Fig. 3b), with the position of the lowest value of S13-L5 FRET in Fig. 5b being almost identical to that of S13-L33 FRET in Fig. 3b.
4.2. Role of EF-G in ribosomal translocation
In the translocation pathway (Fig. 1), the small conformational change in EF-G or small global rotational motion of EF-G relative to the ribosome in State H1 causes the tip of domain IV to move towards and interact with the decoding center in the 30S subunit, facilitating the large forward rotation of the 30S head relative to the 30S body; the forward 30S head rotation widens the mRNA channel (termed as ribosomal unlocking); the unlocking facilitates the reverse 30S subunit rotation, driving the movement of mRNA coupled with tRNAs in the 30S subunit; and after transition to the non-rotated posttranslocation state, the mRNA channel becomes tight again, fixing the mRNA-tRNA complex in the 30S subunit and thus inhibiting the reverse movement of the mRNA-tRNA complex. In other words, the binding of EF-G hydrolyzing GTP is only purposed to facilitate the ribosomal unlocking, allowing the movement of mRNA-tRNA complex in the 30S subunit, and it does not provide the forward force to facilitate tRNA movement or act as a pawl to block the reverse movement of tRNA after the translocation. These are consistent with the recent single molecule data on conformational changes of EF-G in the ribosome [23], as discussed as follows.
The single molecule data showed that EF-G.GTP binding to the pretranslocation complex bound with two tRNAs is followed by a small (∼10°) global rotational motion of EF-G domains I, IV and V relative to the ribosome and by contrast, domain III consistently showed very large and variable rotational motions, whose angular changes were evenly distributed from 0° to 90° without a clear peak [23]. The small global rotational motion of EF-G domains I, IV and V facilitates the ribosomal unlocking via 30S head rotation. The even distribution of rotational angles (from 0° to 90°) of domain III implies that domain III is very flexible, so that it cannot provide even a small forward force to facilitate tRNA movement or cannot act as a pawl to block the reverse movement of tRNA after the translocation. The flexible rotations of domain III to the large extent, which occur after the A-site tRNA movement to the P site, are an intrinsic property of EF-G in GDP form [38], which can be noted from the single molecule evidence that when EF-G.GTP binds to the ribosomal complex in the absence of A-site tRNA, where no translocation can occur [39] (see, Refs. [40] for detailed discussion), the distributions of rotation angles for EF-G domains were quite similar to those when EF-G-GTP binds to pretranslocation complex with two tRNAs [23]. This can also be noted from the single molecule evidence that the antibiotics viomycin (Vio) and spectinomycin (Spc) considerably reduced the extent of rotations in EF-G domain III but had little or even no effect on rotations of domains I, IV and V in pretranslocation complex with two tRNAs [23], because the A-site tRNA restricts flexible rotations of domain III to the large extent. By contrast, when the A site was empty, neither Vio nor Spc had perceptible effects on the motion of EF-G domain III [23], because no A-site tRNA restricts the motion although no translocation occurs.
4.3. Conclusion
In summary, our proposed pathway of ribosomal translocation is consistent with diverse single molecule and ensemble kinetic data presented in the literature [15, 21, 23, 27, 30, 31, 32, 33, 34, 35, 37]. With the pathway we study the dynamic relations among different transitions or events in the translocation pathway exhibited in the smFRET data and those exhibited in the ensemble kinetic data. We show that the timing of these events in the smFRET data and that in the ensemble kinetic data show very different: for example, although the reverse 30S head rotation, reverse 30S subunit rotation and mRNA movement occur almost simultaneously in the smFRET data, these processes start to take place at very different moments in the ensemble kinetic data. Thus, one must be cautious to deduce the precise timing relationship among different conformational changes of ribosome and positional changes of tRNAs and mRNA in the ribosome from the ensemble kinetic data. In the future, it is hoped to test the timing relationship presented in Fig. 2 and Fig. 3a by either using high time-resolution smFRET techniques or by slowing down the translocation reaction via lowering the temperature or using mutant ribosomes.
Declarations
Author contribution statement
Ping Xie: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Competing interest statement
The author declares no conflict of interest.
Funding statement
This work was supported by the National Natural Science Foundation of China (Grant No. 11374352).
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
References
- 1.Frank J., Agrawal R.K. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature. 2000;406:318–322. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 2.Valle M., Zavialov A., Sengupta J., Rawat U., Ehrenberg M., Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 3.Cornish P.V., Ermolenko D.N., Noller H.F., Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol. Cell. 2008;30:578–588. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agirrezabala X., Lei J., Brunelle J.L., Ortiz-Meoz Rodrigo F., Green R., Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol. Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Julián P., Konevega A.L., Scheres S.H.W., Lázaro M., Gil D., Wintermeyer W., Rodnina M.V., Valle M. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16924–16927. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer N., Konevega A.L., Wintermeyer W., Rodnina M.V., Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 7.Taylor D.J., Nilsson J., Merrill A.D., Andersen G.R., Nissen P., Frank J. Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation. EMBO J. 2007;26:2421–2431. doi: 10.1038/sj.emboj.7601677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratje A.H., Loerke J., Mikolajka A. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature. 2010;468:713–716. doi: 10.1038/nature09547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramrath D.J., Lancaster L., Sprink T., Mielke T., Loerke J., Noller H.F., Spahn C.M.T. Proc. Natl. Acad. Sci. U. S. A. 2013;110:20964–20969. doi: 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tourigny D.S., Fernández I.S., Kelley A.C., Ramakrishnan V. Elongation Factor G Bound to the Ribosome in an Intermediate State of Translocation. Science. 2013;340:1235490. doi: 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J., Lancaster L., Donohue J.P., Noller H.F. Crystal structures of EF-G-ribosome complexes trapped in intermediate states of translocation. Science. 2013;340:1236086. doi: 10.1126/science.1236086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pulk A., Cate J.H. Control of Ribosomal Subunit Rotation by Elongation Factor G. Science. 2013;340:1235970. doi: 10.1126/science.1235970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J., Lancaster L., Donohue J.P., Noller H.F. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science. 2014;345:1188–1191. doi: 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agirrezabala X., Valle M. Structural insights into tRNA dynamics on the ribosome. Int. J. Mol. Sci. 2015;16:9866–9895. doi: 10.3390/ijms16059866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wasserman M.R., Alejo J.L., Altman R.B., Blanchard S.C. Multiperspective smFRET reveals rate-determining late intermediates of ribosomal translocation. Nat. Struct. Mol. Biol. 2016;23:333–341. doi: 10.1038/nsmb.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinos G., Wilson D.N., Teraoka Y., Szaflarski W., Fucini P., Kalpaxis D., Nierhaus K.H. Dissecting the ribosomal inhibition mechanisms of edeine and pactamycin: the universally conserved residues G693 and C795 regulate P-site tRNA binding. Mol. Cell. 2004;13:113–124. doi: 10.1016/s1097-2765(04)00002-4. [DOI] [PubMed] [Google Scholar]
- 17.Dinos G., Kalpaxis D.L., Wilson D.N., Nierhaus K.H. Deacylated tRNA is released from the E site upon A site occupation but before GTP is hydrolyzed by EF-Tu. Nucleic Acids Res. 2005;33:5291–5296. doi: 10.1093/nar/gki833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zavialov A.V., Hauryliuk V.V., Ehrenberg M. Guaninenucleotide exchange on ribosome-bound elongation factor G initiates the translocation of tRNAs. J. Biol. 2005;4:9. doi: 10.1186/jbiol24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C., Stevens B., Kaur J., Smilansky Z., Cooperman B.S., Goldman Y.E. Allosteric vs. spontaneous exit-site (E-site) tRNA dissociation early in protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16980–16985. doi: 10.1073/pnas.1106999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uemura S., Aitken C.E., Korlach J., Flusberg B.A., Turner S.W., Puglisi J.D. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–1017. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belardinelli R., Sharma H., Caliskan N., Cunha C.E., Peske F., Wintermeyer W., Rodnina M.V. Choreography of molecular movements during ribosome progression along mRNA. Nat. Struct. Mol. Biol. 2016;23:342–348. doi: 10.1038/nsmb.3193. [DOI] [PubMed] [Google Scholar]
- 22.Xie P. On the pathway of ribosomal translocation. Int. J. Biol. Macromol. 2016;92:401–415. doi: 10.1016/j.ijbiomac.2016.07.048. [DOI] [PubMed] [Google Scholar]
- 23.Chen C., Cui X., Beausang J.F., Zhang H., Farrell I., Barry S., Cooperman B.S., Goldman Y.E. Elongation factor G initiates translocation through a power stroke. Proc. Natl. Acad. Sci. U. S. A. 2016;113:7515–7520. doi: 10.1073/pnas.1602668113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen K., Whitford P.C. Steric interactions lead to collective tilting motion in the ribosome during mRNA–tRNA translocation. Nat. Commun. 2016;7:10586. doi: 10.1038/ncomms10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Savelsbergh A., Katunin V.I., Mohr D., Peske F., Rodnina M.V., Wintermeyer W. An elongation factor G-induced ribosome rearrangement precedes tRNA-mRNA translocation. Mol. Cell. 2003;11:1517–1523. doi: 10.1016/s1097-2765(03)00230-2. [DOI] [PubMed] [Google Scholar]
- 26.Xie P. Model of ribosomal translocation coupled with intra- and inter-subunit rotations. Biochem. Biophys. Rep. 2015;2:87–93. doi: 10.1016/j.bbrep.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Z., Noller H.F. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20391–20394. doi: 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie P. Dynamics of tRNA occupancy and dissociation during translation by the ribosome. J. Theor. Biol. 2013;316:49–60. doi: 10.1016/j.jtbi.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 29.Xie P. Origin of multiple intersubunit rotations before EF-G-catalyzed ribosomal translocation through the mRNA with a downstream secondary structure. BMC Biophys. 2014;7:12. [Google Scholar]
- 30.Adio S., Senyushkina T., Peske F., Fischer N., Wintermeyer W., Rodnina M.V. Fluctuations between multiple EF-G-induced chimeric tRNA states during translocation on the ribosome. Nat. Commun. 2015;6:7442. doi: 10.1038/ncomms8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holtkamp W., Cunha C.E., Peske F., Konevega A.L., Wintermeyer W., Rodnina M.V. GTP hydrolysis by EF-G synchronizes tRNA movement on small and large ribosomal subunits. EMBO J. 2014;33:1073–1085. doi: 10.1002/embj.201387465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker S.E., Shoji S., Pan D., Cooperman B.S., Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9192–9197. doi: 10.1073/pnas.0710146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Petrov A., Tsai A., O′Leary S.E., Puglisi J.D. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat. Struct. Mol. Biol. 2013;20:718–727. doi: 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fredrick K., Noller H.F. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–1162. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 35.Takyar S., Hickerson R.P., Noller H.F. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 36.Xie P. Modeling ribosomal translocation facilitated by peptidyl transferase antibiotics. Cell. Mol. Bioeng. 2016;9:289–302. [Google Scholar]
- 37.Sharma H., Adio S., Senyushkina T., Belardinelli R., Peske F., Rodnina M.V. Kinetics of spontaneous and EF-G-accelerated rotation of ribosomal subunits. Cell Rep. 2016;16:2187–2196. doi: 10.1016/j.celrep.2016.07.051. [DOI] [PubMed] [Google Scholar]
- 38.Czworkowski J., Wang J., Steitz T.A., Moore P.B. The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J. 1994;13:3661–3668. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joseph S., Noller H.F. EF-G–catalyzed translocation of anticodon stem–loop analogs of transfer RNA in the ribosome. EMBO J. 1998;17:3478–3483. doi: 10.1093/emboj/17.12.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie P. Model of the pathway of −1 frameshifting: long pausing. Biochem. Biophys. Rep. 2016;5:408–424. doi: 10.1016/j.bbrep.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan D., Kirillov S.V., Cooperman B.S. Kinetically competent intermediates in the translocation step of protein synthesis. Mol. Cell. 2007;25:519–529. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.