Abstract
Cajal-Retzius (CR) cells are early-generated transient neurons and are important in the regulation of cortical neuronal migration and cortical laminar formation. Molecular entities characterizing the CR cell identity, however, remain largely elusive. We purified mouse cortical CR cells expressing GFP to homogeneity by fluorescence-activated cell sorting and examined a genomewide expression profile of cortical CR cells at embryonic and postnatal periods. We identified 49 genes that exceeded hybridization signals by >10-fold in CR cells compared with non-CR cells at embryonic day 13.5, postnatal day 2, or both. Among these CR cell-specific genes, 25 genes, including the CR cell marker genes such as the reelin and calretinin genes, are selectively and highly expressed in both embryonic and postnatal CR cells. These genes, which encode generic properties of CR cell specificity, are eminently characterized as modulatory composites of voltage-dependent calcium channels and sets of functionally related cellular components involved in cell migration, adhesion, and neurite extension. Five genes are highly expressed in CR cells at the early embryonic period and are rapidly down-regulated thereafter. Furthermore, some of these genes have been shown to mark two distinctly different focal regions corresponding to the CR cell origins. At the late prenatal and postnatal periods, 19 genes are selectively up-regulated in CR cells. These genes include functional molecules implicated in synaptic transmission and modulation. CR cells thus strikingly change their cellular phenotypes during cortical development and play a pivotal role in both corticogenesis and cortical circuit maturation.
In the developing neocortex, the generation of distinct classes of cortical neurons is controlled by a hierarchical series of developmental events (1). In this process, postmitotic cortical neurons migrate along radial glial cell fibers from the ventricular zone. These cells form the cortical plate and subdivide the preexisting preplate into the superficial marginal zone (MZ) and the subplate (2). Cajal-Retzius (CR) cells represent the key neuronal subtype that regulates radial migration of cortical neurons and the laminar formation of the neocortex (2, 3). In mice, CR cells appear at embryonic days (E) 10 and 11, occupy a major cell population in the MZ throughout the entire cortex, and gradually decrease during the postnatal period. CR cells produce an extracellular matrix protein called reelin (4). Reelin is defective in the reeler mouse mutant and human congenital lissencephaly, both of which show altered cortical cell migration and an abnormal cortical layer formation (4, 5). CR cells also form synaptic connections with migrating cortical neurons, thus serving as a physiological scaffolding of cortical synaptic circuits during neocortical development (3, 6, 7). Despite great advances in the characterization of CR cells in neocortical development, neither the molecular entities that govern the CR cell identity nor the molecular mechanisms in which CR cells participate in the control of cortical organization were well understood.
In our previous studies, we reported that the membrane-anchored GFP transgene, when driven by the promoter function of metabotropic glutamate receptor subtype 2 (8), is specifically expressed by CR cells both in the MZ during the embryonic stage and in layer 1 during the postnatal period (7, 9). This specific expression of GFP in CR cells provided a unique opportunity to purify CR cells to homogeneity by fluorescence-activated cell sorting (FACS) and to investigate a genomewide expression profile of CR cells during neocortical development with microarray techniques. Here we report that CR cells not only acquire the generic properties of CR cell specificity from early development but also change their gene expression profiles markedly during cortical development. Furthermore, the identification of many functional molecules specific for CR cells suggests that CR cells play a pivotal role in the control of corticogenesis and cortical circuit maturation.
Methods
Cell Purification and FACS. Cerebral cortices from 10-24 embryos or newborns of the IG17 line of homozygous transgenic mice (8) were cut into small pieces in ice-cold L15 medium (Invitrogen). These pieces were treated with 10-20 units/ml papain (Nacalai Tesque, Kyoto) and 0.01-0.02% DNase I (Sigma) in a solution containing 0.02% BSA, 0.02% l-cysteine, and 0.5% glucose for 20 min at 37°C. Single cells were prepared by passing them through a plastic pipette 40 times. Dissociated cells were stained with propidium iodide (PI) (1 μg/ml, Sigma), and two-color cell sorting based on GFP and PI fluorescences was performed with a FACSVantage flow cytometer (BD Biosciences). Approximately 5 × 104 purified GFP-positive, PI-negative cells and GFP-negative, PI-negative cells were isolated from cortices at E13.5 and postnatal day (P) 2 and used for microarray analysis.
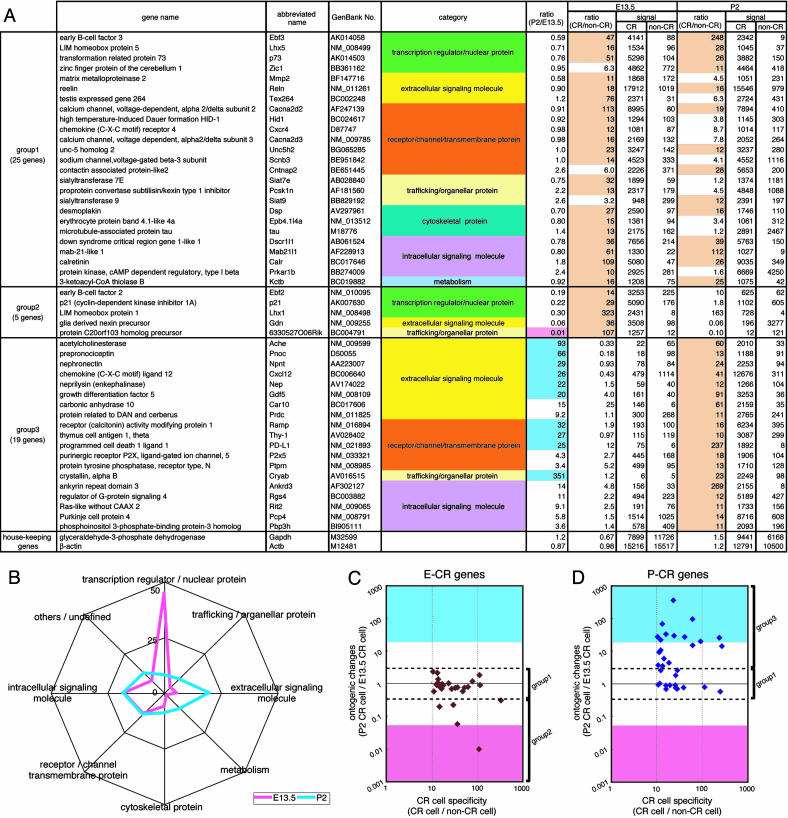
Microarray Analysis. An Affymetrix (Santa Clara, CA) MOE430A mouse expression microarray consisting of 22,690 probe sets was used in all microarray experiments. cRNA probes were synthesized according to a two-cycle in vitro transcription labeling protocol (Affymetrix), and four sets of biotinylated cRNA probes were prepared from total RNAs isolated from CR and non-CR cells at E13.5 and P2. Hybridization and scanning were performed according to the manufacturer's instructions (Affymetrix). Hybridization signals were calculated by analyzing raw data with microarray suite 5.0 (Affymetrix) and were further analyzed with genespring 6.1 (Silicon Genetics). The data were normalized to the 50th percentile for per-chip normalization. EST sequences were searched from the UniGene and GenBank databases and with the blast search engine at the National Center for Biotechnology Information (NCBI). The information on functions of candidate genes was obtained from the PubMed database (National Library of Medicine) and literature. The functions of candidate genes were classified in eight categories (Fig. 2B).
Fig. 2.
Gene-expression profile of CR cells at E13.5 and P2. (A) The 49 genes that exceeded hybridization signals by >10-fold (indicated with pale orange) between CR and non-CR cells at E13.5, P2, or both are listed. Relative ratios of hybridization signals of E13.5 CR cells to P2 CR cells [ratio (P2/E13.5)] and those of CR to non-CR cells at E13.5 and P2 [ratio (CR/non-CR)] were calculated from microarray-analysis data. The CR genes were classified into three groups according to ontogenic changes in expression levels between E13.5 and P2 CR cells. The functional category of each gene is indicated with different colors. The 11 genes that showed a >20-fold increase or decrease in hybridization signals in P2 CR cells compared with E13.5 CR cells are indicated with blue and pink in the ratio column (P2/E13.5), respectively. At the bottom, the data of two housekeeping genes are indicated for comparison. The full and abbreviated names of genes are taken from the GenBank database. (B) The functions of 131 genes at E13.5 (pink) and 111 genes at P2 (blue) in which hybridization signals were more than three times different between E13.5 CR cells and P2 CR cells were annotated on the basis of available databases (Tables 1 and 2), and the proportion of eight different categories is expressed as a percentage for each category. (C and D) The E-CR genes (27 genes) and the P-CR genes (33 genes), which had hybridization signals >10 times higher in CR cells compared with non-CR cells at E13.5 (C) and P2 (D), respectively, were separately analyzed. Relative ratios of the expression levels of P2 to E13.5 CR cells (ontogenic changes) were plotted against relative ratios of the expression levels of CR to non-CR cells (CR cell specificity). Group 1-3 CR genes are marked on the right side. Expression levels exceeding a 20-fold increase or decrease in P2 CR cells compared with E13.5 CR cells are indicated with blue and pink, respectively.
Quantitative PCR and in Situ Hybridization. Total RNA was isolated from purified CR cells and subjected to quantitative PCR as described in ref. 10. Specific primers were designed to generate 150- to 200-bp PCR products corresponding to the 3′ region of each gene. All reactions were performed in triplicate, and the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was used as an internal control for mRNA quantification. In situ hybridization with 33P- and digoxigenin-labeled RNA probes was carried out as described in ref. 11.
Results
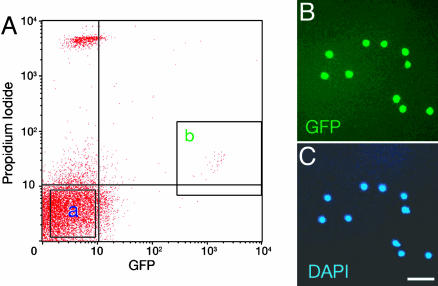
Purification of CR Cells by FACS. We focused on CR cells at E13.5 and P2, because the expression of the GFP transgene is not only specific to CR cells within the developing neocortex but also sufficiently appears at both stages (7, 9). The neocortices were prepared from the brains of E13.5 and P2 transgenic mice, and cells were dissociated into single cells by papain treatment. Trypan blue staining, a marker of dead cells, showed that almost all and approximately four-fifths of purified GFP-positive cells were viable in the E13.5 and P2 cell populations, respectively. To remove nonviable cells, dissociated cells were stained with PI, a fluorescent marker of dead cells, and the GFP-positive and PI-negative viable CR cells were purified by FACS with GFP and PI fluorometry (Fig. 1A). In this sorting, GFP-negative, PI-negative cells were also pooled and used as a cell population of non-CR cells (Fig. 1 A). Under microscopic observation, GFP-positive CR cells were almost 100% homogenous in the cell preparations analyzed (Fig. 1 B and C), ensuring that the microarray analysis reflects a gene-expression profile of CR cells.
Fig. 1.
Purification of CR cells by FACS. (A) GFP-positive, PI-negative cells (fraction b) and GFP-negative, PI-negative cells (fraction a) at P2 were sorted by FACS as CR cells and non-CR cells, respectively. The PI fluorescence of viable CR cells was slightly higher than that of viable non-CR cells because of a slight overlap of GFP fluorescence in PI fluorescence measurement. (B and C) Purified GFP-positive CR cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and detected with microscopy. (Scale bar, 50 μm.)
Gene-Expression Profiles of Embryonic and Postnatal CR Cells. We used an Affymetrix MOE430A expression microarray consisting of 22,690 probe sets. Four sets of biotinylated cRNA probes were prepared from E13.5 and P2 CR cells and the corresponding non-CR cells. Each set of cRNA probes was subjected to microarray hybridization analysis. As a criterion to select candidate genes, we used hybridization signals of >1,000 in E13.5 and/or P2 CR cells, a level sufficiently higher than that of the background signal. On the basis of this criterion, we identified 3,657 and 3,275 probe sets from data analysis of E13.5 and P2 CR cells, respectively. Hybridization signals of the well known CR cell marker reelin, transformation related protein 73 (p73), and calretinin genes (2, 12) were prominently higher in CR cells compared with non-CR cells at both E13.5 and P2 (Fig. 2A), and this expression profile was in marked contrast to the intense hybridization signals of the housekeeping GAPDH and β-actin transcripts in both CR and non-CR cells (Fig. 2 A). This examination validated that our microarray data are reliable for further characterization of a gene-expression profile characteristic of CR cells.
We first examined developmental changes of gene expression in CR cells per se by comparing the 3,657 probe sets at E13.5 and the 3,275 probe sets at P2 with the corresponding probe sets at P2 and E13.5, respectively. Upon this comparison, we identified 150 and 121 genes that increased by more than three times in E13.5 and P2 CR cells, respectively. The functions of 131/150 genes at E13.5 and 111/121 genes at P2 were annotated on the basis of available databases and literature and classified into eight categories (Tables 1 and 2, which are published as supporting information on the PNAS web site). For convenience, the 131 genes characteristic of E13.5 CR cells and the 111 genes of P2 CR cells are termed “E13.5 genes” and “P2 genes” hereafter.
When the functions of the 131 E13.5 genes and the 111 P2 genes are inspected, transcription regulators/nuclear proteins are strikingly prominent in E13.5 CR cells, accounting for almost half (45%) of the E13.5 genes (Fig. 2B). By contrast, extracellular signaling molecules, cytoskeletal proteins, and metabolic enzymes are up-regulated at P2 CR cells, as compared with E13.5 cells (Fig. 2B). Furthermore, the E13.5 gene products are more closely related to cell proliferation, cell differentiation, neurogenesis, and cell morphogenesis, whereas the P2 gene products are implicated in synaptic organization, synaptic transmission, and transmission-linked signal transduction (see Discussion). CR cells thus dramatically change their cellular phenotypes from the embryonic period to the postnatal period.
CR Cell-Specific Gene Expression in the Developing Neocortex. We next analyzed a gene-expression profile specific for CR cells by comparing the above 3,657 and 3,275 probe sets of CR cells with the corresponding sets of non-CR cells. We identified 69 and 96 probe sets that were more than five times higher in hybridization signals of CR cells at E13.5 and P2 than those of non-CR cells at the corresponding periods, respectively. The functions of 63/69 genes at E13.5 and 87/96 genes at P2 were annotated on the basis of available databases and literature (Tables 3 and 4, which are published as supporting information on the PNAS web site). Among these 150 genes, 49 had a signal >10 times higher in CR cells compared with non-CR cells at E13.5, P2, or both. These 49 genes, hereafter termed “CR genes,” can be classified into three groups according to the difference in expression levels between E13.5 and P2 (Fig. 2 A). Group 1 CR genes showed high levels of expression in both E13.5 and P2 CR cells, with the relative ratios of P2 to E13.5 CR cells within the range from 0.33 to 3.0. This group includes not only the CR cell marker genes (reelin, calretinin, and p73) but also 22 other genes. The reelin gene showed the highest hybridization signals, but hybridization signals of several other genes (Zic1, Cacna2d2, and Scnb3) were as high as those of the p73 and calretinin genes at both E13.5 and P2. The functions of the group 1 genes are diverse, suggesting that diverse functional molecules are necessary for inducing and maintaining cellular phenotypes characteristic of CR cells. Group 2 of the CR genes comprises five genes that exhibited a marked reduction (>3.0-fold) in expression levels from E13.5 to P2. Group 3 CR genes showed a prominent elevation (>3.0-fold) in expression levels from E13.5 to P2.
The relationship between the CR cell specificity and the ontogenic expression profile was further examined by analyzing ontogenic changes in expression levels of the CR genes listed in Fig. 2 A. In this comparison, the CR genes exhibiting a >10-fold increase in expression levels at either E13.5 or P2 CR cells, termed “E-CR genes” and “P-CR genes,” respectively, were separately analyzed. Relative ratios of expression levels between E13.5 and P2 CR cells (ontogenic changes) were then inspected against the extent of difference in expression levels between CR and non-CR cells (CR cell specificity) (Fig. 2 C and D). More than 80% (22/27) of the E-CR genes also were highly and comparably expressed in CR cells at P2 (Fig. 2C), indicating that the generic properties of CR cell specificity are acquired from the early period of CR cell development. When this relationship was analyzed on the P-CR genes, ≈40% (14/33) of the P-CR genes also were highly expressed at E13.5 CR cells (Fig. 2D). Importantly, the remaining ≈60% (19/33) of the P-CR genes showed a dramatic increase in expression levels at P2 (Fig. 2D). Moreover, the majority of these up-regulated genes encode extracellular signaling molecules, membrane-integral receptor proteins, and intracellular signaling molecules. These findings demonstrate not only that many genes other than the well known CR cell marker genes are selectively and highly expressed in both embryonic and postnatal CR cells but also that postnatal CR cells acquire distinct properties that can be characterized by the CR genes up-regulated during the postnatal period.
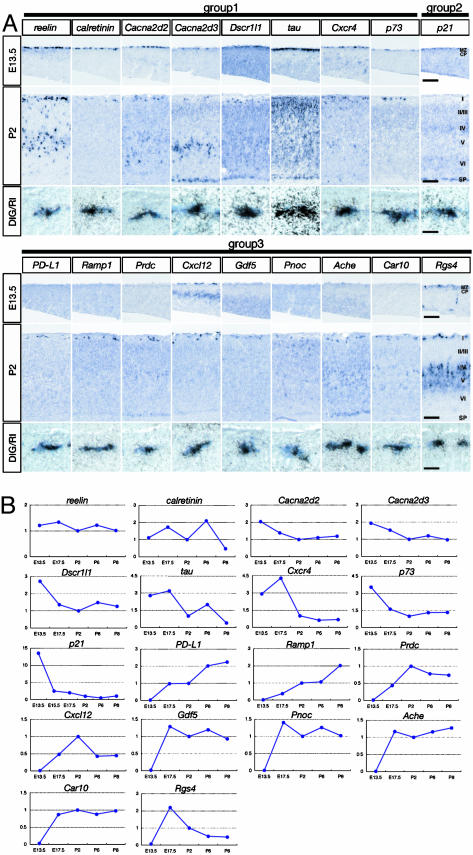
Spatial and Temporal Expression Patterns of CR Genes. To confirm the spatially restricted expressions of the CR genes, we performed in situ hybridization analysis of 30 CR genes, as well as double in situ hybridization analysis of 18 CR genes, by using a digoxigenin-labeled GFP probe and radioisotope (RI)-labeled probes for CR genes (Fig. 3A). Hybridization signals of the group 1 genes (those shown in Fig. 3A and Ebf3, Lhx5, Zic1, Siat7e, Unc5h2, and Mab21l1) were detected both in the MZ at E13.5 and in layer 1 at P2. The signals of these transcripts were mostly confined to the MZ at E13.5 but in some cases were distributed not only in layer 1 but also in other cortical layers at P2 (Fig. 3A) (for example, see reelin, Cxcr4, and tau), and these data are consistent with other reports (13-15).
Fig. 3.
Analysis of in situ hybridization and temporal expression patterns of CR genes. (A) In situ hybridization analysis of CR genes was conducted in neocortical sections at E13.5 and P2, and the results are displayed in Top and Middle, respectively. CP, cortical plate; I-VI, layers 1-6; SP, subplate; DIG, digoxigenin. In Bottom, double in situ hybridization analysis in neocortical sections at E13.5 (Cacna2d3, tau, and p21) and at P2 (others) is indicated. (Scale bars: Top and Middle, 100 μm; Bottom, 10 μm.) (B) Temporal changes in expression levels of indicated CR genes were analyzed at five to six time points from E13.5 to P8. Expression levels at P2 were taken as 1 in all analyses.
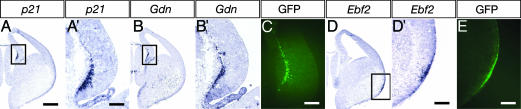
Most of the group 1 CR genes also were detected at the MZ of the caudomedial wall of the telencephalic vesicle (CMWT), including at the cortical hem and at the cortical surface of the pallium (data not shown), where newly generated CR cells from origins are supposed to be transiently accumulated (9, 16). Importantly, signals of p21 and Gdn at E13.5 were abundantly seen at the CMWT possessing massive GFP-positive CR cells, whereas the signal of Ebf2 was considerably detected at the MZ adjacent to the ventral pallium (Fig. 4). These group 2 CR genes are thus distinctly expressed at the regions corresponding to the plausible CR cell origins at the early stage of corticogenesis (9, 16-18).
Fig. 4.
Expression of group 2 CR genes at the CMWT or the ventral pallium. In situ hybridization was conducted at sections of the right hemisphere at E13.5 (A, B, and D). The boxed areas comprising the CMWT (A and B) and the ventral pallium (D) are expanded in A′, B′, and D′. GFP-positive CR cells at the corresponding regions were detected at an adjacent section of in situ hybridization analysis (C and E). (Scale bars: A, B, and D, 400 μm; A′, B′, and C, 100 μm; D′ and E, 133 μm.)
Upon hybridization analysis of group 3 CR genes, signals of Rgs4, Thy-1, and Pcp4 were observed at layer 1, as well as at other cortical layers (Fig. 3A and data not shown). These distributed expression patterns of group 3 CR genes, however, were rather rare, and signals of other group 3 genes were restrictedly seen at layer 1 of the neocortex at P2 (Fig. 3A). Furthermore, in all 18 CR genes analyzed, the expression of these genes at CR cells was verified by double in situ hybridization analysis (Fig. 3A).
When temporal expression patterns were analyzed with quantitative RT-PCR techniques (Fig. 3B), expression levels of the reelin, calretinin, Cacna2d2, and Cacna2d3 transcripts of group 1 genes showed no appreciable change from E13.5 to P8, but other group 1 gene transcripts (Dscr1l1, tau, Cxcr4, and p73) gradually decreased from the embryonic stage to the postnatal stage (Fig. 3B). Interestingly, the p21 transcript of group 2 genes rapidly decreased at the very early embryonic stage (Fig. 3B). In contrast, all transcripts of group 3 genes considerably but differentially increased during development. The PD-L1 and Ramp1 transcripts increased up to at least P8. The Gdf5, Pnoc, Ache, Car10, and Rgs4 transcripts and the Prdc and Cxcl12 transcripts displayed a maximal level at the prenatal and postnatal periods, respectively. This analysis explicitly demonstrates that group 3 CR genes are up-regulated in CR cells during development but in distinct temporal fashions.
Discussion
In this investigation, a large number of CR genes were newly identified, and their functions appear to be diversified. However, sets of the CR genes are functionally closely related, and several possible functional roles of the CR genes can be envisaged in relation to the characteristic features of CR cells.
Origins of CR Cells. With respect to the CR cell origins, many sources have been proposed, including the pallium (17), the subpallium (18), the retrobulbar area (16), and the cortical hem region (9). The Efb2 gene and the Gdn and p21 genes of the group 2 CR genes are preferentially expressed at the plausible CR cell origins (Fig. 4). Ebf2 is a member of the helix-loop-helix transcription factors (19) and serves as an upstream transcription factor in the helix-loop-helix transcriptional cascade (see below), whereas p21 is a downstream signaling molecule of the transforming growth factor β (TGF-β) signaling pathway (20). The TGF-β signaling enhances the formation of the Smad-Foxo3 signaling complex (20). Interestingly, Foxo3 (Table 3, No. 12) is selectively expressed in E13.5 CR cells, whereas Tgfbr1 and AcvrII, which are the members of TGF-β receptor subtypes I and II, respectively, also are highly expressed in E13.5 CR cells and then down-regulated at P2. The selective expressions of Ebf2, Gdn, and p21 at two distinct regions corresponding to the plausible CR cell origins suggest that prospective CR cells are heterogeneous and are distributed through the MZ by tangential migration.
Transcriptional Regulation of CR Cells. We identified seven transcription regulators/nuclear proteins as the group 1 or group 2 CR genes (Fig. 2 A). In the Xenopus nervous system, Ebf2, NeuroD, and Ebf3 serve as a transcriptional cascade in this sequential order and regulate neural cell development and differentiation (19). Ebf2 is expressed at the very early stage of CR cells and is down-regulated thereafter. NeuroD (Table 1, No. 4) is highly expressed in both CR and non-CR cells at E13.5 and also is down-regulated at P2. In contrast, Ebf3 is prominently expressed in CR cells at both E13.5 and P2. The Ebf2-NeuroD-Ebf3 transcriptional cascade may thus participate in the regulation of CR cell development and differentiation. Lhx1 and Lhx5, another family of the CR genes, belong to the LIM family of the homeobox genes (21). The expression of Lhx1 is restricted at the MZ at E13.5. Lhx2 (Table 1, No. 27) and Lhx9 (Table 1, No. 11) also are highly expressed in CR cells at E13.5 and down-regulated at P2. In contrast, Lhx5 is persistently high in CR cells throughout development. Gene targeting of Lhx5 results in the disappearance of hippocampal CR cells and hippocampal malformation (21). Zic1 also was identified as a group 1 CR gene, and its family, Zic3 (Table 1, No. 43) and Zic5 (Table 3, No. 9), are highly and relatively selectively expressed in CR cells at E13.5 and down-regulated at P2. Zic1 was originally isolated as a specific transcription factor of embryonic cerebellar granule cells that secrete reelin during cerebellar development (4, 22). These findings suggest that several distinct transcriptional cascades coordinately regulate the fate and cellular phenotypes of CR cells.
Calcium Signaling of CR Cells. Cacna2d2 and Cacna2d3 of group 1 CR genes encode subunit composites of voltage-dependent calcium channels (VDCCs) (23). In addition, Cacng4 (Table 3, No. 39), Cacng5 (Table 2, No. 38), and Cacna2d1 (Table 4, No. 45), other VDCC composites, are relatively selectively expressed in CR cells. These VDCC composites per se have no channel activity and modulate the VDCC activity (23). Scnb3, identified as a group 1 CR gene, also encodes a modulatory composite of sodium channels (24). The mutant of Cacna2d2 called ducky shows a reduction of a current density of VDCCs in Purkinje cells and impairs the dendritic formation of these cells (23). In addition, several intracellular calcium signaling molecules, including Dscr1l1, calretinin, Rit2, and Pcp4 (25-27), are selectively expressed in CR cells (Fig. 2 A). The CR cell-specific Ca2+ signaling components could thus play an important role in development and functions that are characteristic of CR cells.
Cell Migration, Adhesion, and Neurite Extension. CR cells migrate along the most apical layer and dramatically change their morphology. This layer also is filled with growing dendrites, axons, and extracellular matrix proteins during development. Many functional molecules involved in cell migration, cell adhesion, neurite/axon extension, and extracellular matrix maturation are specifically expressed in CR cells. These include Cxcr4, Dsp, Mmp2, Siat7e, and Siat9 of the group1 CR genes and Cxcl12, Npnt, and Thy-1 of the group 3 CR genes (28-31) (Fig. 2 A). Particularly interesting is the CR cell-specific expression of chemokine signaling molecules. Cxcr4 is the receptor for the Cxcl12 chemokine and is persistently up-regulated in embryonic and postnatal CR cells (Fig. 2 A). In the developing neocortex, Cxcl12 is continuously produced at adjacent meningeal cells (15). Furthermore, this chemokine is strikingly up-regulated in CR cells per se at the postnatal period (Fig. 3B). In addition, chemokine orphan receptor 1 (Cmkor1; Table 3, No. 37) also is selectively and highly expressed in CR cells at E13.5. Although gene targeting of either Cxcr4 or Cxcl12 has been shown to keep CR cells at the MZ, it is possible that the chemokine signaling may regulate the migration of immature CR cells and/or axonal responsiveness to a variety of guidance cues (15, 32). Another interesting feature is the selective expression of Siat7e and Siat9. In the MZ/layer 1, the neural cell adhesion molecule is preferentially polysialylated, and this polysialylation attenuates the adhesive property of the neural cell adhesion molecule, thereby facilitating neural cell migration (2, 33). Polysialylation requires the preceding α2,3- or α2,6-linked sialylation reaction (34). Siat7e and Siat9 are α2,6- and α2,3-sialyltransferases, respectively (35). The CR cell-specific expression of these sialyltransferases could be important for sialylation-mediated regulation of cell-cell interactions and neurite extension.
Induction of CR Cell-Specific Genes During Cortical Circuit Maturation. Many functional molecules involved in synaptic transmission and modulation are selectively up-regulated at the late developmental stage (Figs. 3A and 4B). P2x5 is a purinergic receptor and is capable of exciting neuronal cells by means of cation permeation (36). Acetylcholinesterase (Ache) may also regulate the cholinergic transmission to layer 1 γ-aminobutyric acid (GABA)-ergic interneurons, because these interneurons receive cholinergic inputs by means of muscarinic cholinergic receptors (6). The selective expression of prepronociceptin (Pnoc) is also interesting because the nociceptin receptor is expressed in cortical pyramidal neurons from early development to the adult stage (37). Furthermore, Pcsk1n is an inhibitor of proteolytic cleavage of many peptide precursors, whereas Nep, also called enkephalinase, inactivates biologically active peptides such as enkephalins and substance P (38, 39). Another interesting biological relevance is the relationship between Prdc and Gdf5. Gdf5 is a member of the bone morphogenetic protein (BMP) family (40), whereas Prdc is an inhibitory protein for binding of BMPs to their receptors (41). The interference of BMP signaling with the BMP antagonist has been reported as a key mechanism of many neural cell developments (40). Interestingly, Gdf5 is rapidly up-regulated and thus could be controlled by the slowly up-regulated Prdc at the late stage (Fig. 3B). In conclusion, this investigation strongly indicates that CR cells are involved more actively than previously envisioned in corticogenesis and cortical circuit maturation and contribute as a key signaling center to regulating dynamic cortical development.
Supplementary Material
Acknowledgments
We thank Kohei Kometani for FACS and Kumlesh Dev for careful reading of the manuscript. This work was supported in part by research grants from the Ministry of Education, Science, and Culture of Japan and Precursory Research for Embryonic Science and Technology of the Japan Science and Technology Agency. H.Y. is a fellow of the 21st Century Center of Excellence Program of the Ministry of Education, Science, and Culture of Japan.
Abbreviations: CMWT, caudomedial wall of the telencephalic vesicle; CR, Cajal-Retzius; En, embryonic day n; FACS, fluorescence-activated cell sorting; MZ, marginal zone; Pn, postnatal day n; PI, propidium iodide; VDCC, voltage-dependent calcium channel.
References
- 1.O'Leary, D. D. & Nakagawa, Y. (2002) Curr. Opin. Neurobiol. 12, 14-25. [DOI] [PubMed] [Google Scholar]
- 2.Supèr, H., Soriano, E. & Uylings, H. B. (1998) Brain Res. Brain Res. Rev. 27, 40-64. [DOI] [PubMed] [Google Scholar]
- 3.Marín-Padilla, M. (1998) Trends Neurosci. 21, 64-71. [DOI] [PubMed] [Google Scholar]
- 4.Rice, D. S. & Curran, T. (2001) Annu. Rev. Neurosci. 24, 1005-1039. [DOI] [PubMed] [Google Scholar]
- 5.Feng, Y. & Walsh, C. A. (2001) Nat. Rev. Neurosci. 2, 408-416. [DOI] [PubMed] [Google Scholar]
- 6.Schwartz, T. H., Rabinowitz, D., Unni, V., Kumar, V. S., Smetters, D. K., Tsiola, A. & Yuste, R. (1998) Neuron 20, 541-552. [DOI] [PubMed] [Google Scholar]
- 7.Soda, T., Nakashima, R., Watanabe, D., Nakajima, K., Pastan, I. & Nakanishi, S. (2003) J. Neurosci. 23, 6272-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Watanabe, D., Inokawa, H., Hashimoto, K., Suzuki, N., Kano, M., Shigemoto, R., Hirano, T., Toyama, K., Kaneko, S., Yokoi, M., et al. (1998) Cell 95, 17-27. [DOI] [PubMed] [Google Scholar]
- 9.Takiguchi-Hayashi, K., Sekiguchi, M., Ashigaki, S., Takamatsu, M., Hasegawa, H., Suzuki-Migishima, R., Yokoyama, M., Nakanishi, S. & Tanabe, Y. (2004) J. Neurosci. 24, 2286-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Gelder, R. N., von Zastrow, M. E., Yool, A., Dement, W. C., Barchas, J. D. & Eberwine, J. H. (1990) Proc. Natl. Acad. Sci. USA 87, 1663-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tomita, K., Moriyoshi, K., Nakanishi, S., Guillemot, F. & Kageyama, R. (2000) EMBO J. 19, 5460-5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer, G., Perez-Garcia, C. G., Abraham, H. & Caput, D. (2002) J. Neurosci. 22, 4973-4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alcántara, S., Ruiz, M., D'Arcangelo, G., Ezan, F., de Lecea, L., Curran, T., Sotelo, C. & Soriano, E. (1998) J. Neurosci. 18, 7779-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brion, J. P., Octave, J. N. & Couck, A. M. (1994) Neuroscience 63, 895-909. [DOI] [PubMed] [Google Scholar]
- 15.Stumm, R. K., Zhou, C., Ara, T., Lazarini, F., Dubois-Dalcq, M., Nagasawa, T., Höllt, V. & Schulz, S. (2003) J. Neurosci. 23, 5123-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer, G., Soria, J. M., Martínez-Galán, J. R., Martín-Clemente, B. & Fairén, A. (1998) J. Comp. Neurol. 397, 493-518. [PubMed] [Google Scholar]
- 17.Hevner, R. F., Neogi, T., Englund, C., Daza, R. A. & Fink, A. (2003) Brain Res. Dev. Brain Res. 141, 39-53. [DOI] [PubMed] [Google Scholar]
- 18.Lavdas, A. A., Grigoriou, M., Pachnis, V. & Parnavelas, J. G. (1999) J. Neurosci. 19, 7881-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pozzoli, O., Bosetti, A., Croci, L., Consalez, G. G. & Vetter, M. L. (2001) Dev. Biol. 233, 495-512. [DOI] [PubMed] [Google Scholar]
- 20.Seoane, J., Le, H. V., Shen, L., Anderson, S. A. & Massague, J. (2004) Cell 117, 211-223. [DOI] [PubMed] [Google Scholar]
- 21.Zhao, Y., Sheng, H. Z., Amini, R., Grinberg, A., Lee, E., Huang, S., Taira, M. & Westphal, H. (1999) Science 284, 1155-1158. [DOI] [PubMed] [Google Scholar]
- 22.Aruga, J., Yokota, N., Hashimoto, M., Furuichi, T., Fukuda, M. & Mikoshiba, K. (1994) J. Neurochem. 63, 1880-1890. [DOI] [PubMed] [Google Scholar]
- 23.Brodbeck, J., Davies, A., Courtney, J. M., Meir, A., Balaguero, N., Canti, C., Moss, F. J., Page, K. M., Pratt, W. S., Hunt, S. P., et al. (2002) J. Biol. Chem. 277, 7684-7693. [DOI] [PubMed] [Google Scholar]
- 24.Morgan, K., Stevens, E. B., Shah, B., Cox, P. J., Dixon, A. K., Lee, K., Pinnock, R. D., Hughes, J., Richardson, P. J., Mizuguchi, K., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao, X., Kambe, F., Miyazaki, T., Sarkar, D., Ohmori, S. & Seo, H. (2002) Biochem. J. 367, 459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johanson, R. A., Sarau, H. M., Foley, J. J. & Slemmon, J. R. (2000) J. Neurosci. 20, 2860-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, C. H., Della, N. G., Chew, C. E. & Zack, D. J. (1996) J. Neurosci. 16, 6784-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kowalczyk, A. P., Bornslaeger, E. A., Borgwardt, J. E., Palka, H. L., Dhaliwal, A. S., Corcoran, C. M., Denning, M. F. & Green, K. J. (1997) J. Cell Biol. 139, 773-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagase, H. & Woessner, J. F., Jr. (1999) J. Biol. Chem. 274, 21491-21494. [DOI] [PubMed] [Google Scholar]
- 30.Brandenberger, R., Schmidt, A., Linton, J., Wang, D., Backus, C., Denda, S., Müller, U. & Reichardt, L. F. (2001) J. Cell Biol. 154, 447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leyton, L., Schneider, P., Labra, C. V., Ruegg, C., Hetz, C. A., Quest, A. F. & Bron, C. (2001) Curr. Biol. 11, 1028-1038. [DOI] [PubMed] [Google Scholar]
- 32.Chalasani, S. H., Sabelko, K. A., Sunshine, M. J., Littman, D. R. & Raper, J. A. (2003) J. Neurosci. 23, 1360-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brusés, J. L. & Rutishauser, U. (2001) Biochimie 83, 635-643. [DOI] [PubMed] [Google Scholar]
- 34.Angata, K., Suzuki, M., McAuliffe, J., Ding, Y., Hindsgaul, O. & Fukuda, M. (2000) J. Biol. Chem. 275, 18594-18601. [DOI] [PubMed] [Google Scholar]
- 35.Harduin-Lepers, A., Vallejo-Ruiz, V., Krzewinski-Recchi, M. A., Samyn-Petit, B., Julien, S. & Delannoy, P. (2001) Biochimie 83, 727-737. [DOI] [PubMed] [Google Scholar]
- 36.North, R. A. (2002) Physiol. Rev. 82, 1013-1067. [DOI] [PubMed] [Google Scholar]
- 37.Neal, C. R., Jr., Akil, H. & Watson, S. J., Jr. (2001) J. Chem. Neuroanat. 22, 219-249. [DOI] [PubMed] [Google Scholar]
- 38.Wei, S., Feng, Y., Che, F. Y., Pan, H., Mzhavia, N., Devi, L. A., McKinzie, A. A., Levin, N., Richards, W. G. & Fricker, L. D. (2004) J. Endocrinol. 180, 357-368. [DOI] [PubMed] [Google Scholar]
- 39.Carson, J. A. & Turner, A. J. (2002) J. Neurochem. 81, 1-8. [DOI] [PubMed] [Google Scholar]
- 40.Balemans, W. & Van Hul, W. (2002) Dev. Biol. 250, 231-250. [PubMed] [Google Scholar]
- 41.Sudo, S., Avsian-Kretchmer, O., Wang, L. S. & Hsueh, A. J. (2004) J. Biol. Chem. 279, 23134-23141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.