Abstract
Merkel cell-neurite complexes are somatosensory receptors that initiate the perception of gentle touch. The role of epidermal Merkel cells within these complexes is disputed. To ask whether Merkel cells are genetically programmed to be excitable cells that may participate in touch reception, we purified Merkel cells from touch domes and used DNA microarrays to compare gene expression in Merkel cells and other epidermal cells. We identified 362 Merkel-cell-enriched transcripts, including neuronal transcription factors, presynaptic molecules, and ion-channel subunits. Antibody staining of skin sections showed that Merkel cells are immunoreactive for presynaptic proteins, including piccolo, Rab3C, vesicular glutamate transporter 2, and cholecystokinin 26-33. These data indicate that Merkel cells are poised to release glutamate and neuropeptides. Finally, by using Ca2+ imaging, we discovered that Merkel cells have L- and P/Q-type voltage-gated Ca2+ channels, which have been shown to trigger vesicle release at synapses. These results demonstrate that Merkel cells are excitable cells and suggest that they release neurotransmitters to shape touch sensitivity.
The somatic senses of touch, proprioception, and pain are mediated by mechanosensory cells that transduce pressure or stretch into electrical signals. In vertebrates, the discovery of molecules that underlie mechanosensory signaling has been difficult because somatosensory neurons are diverse and their afferents are dispersed throughout skin and other target tissues.
The Merkel cell-neurite complex, which is among the most sensitive vertebrate touch receptors, may serve as a model for studying mechanosensory signaling. These complexes, comprising sensory afferents and epidermal Merkel cells, are one of a few somatosensory receptors whose morphology and response properties have been correlated (1). They mediate slowly adapting type I responses, which are important for the perception of shapes and textures (2). Additionally, they may be studied in living tissue because Merkel cells and sensory afferents can be fluorescently labeled in vivo (3, 4).
Whether the Merkel cell, the afferent, or both are sites of mechanotransduction is a controversial issue raised more than a century ago (5). Because somatosensory terminals often contact them, Merkel cells have been proposed to be mechanosensory cells that activate sensory afferents. This role would be analogous to that of hair cells, specialized epithelial cells that mediate transduction in the acousticolateralis system.
Parallels between Merkel cells and hair cells have fueled the idea that Merkel cells are mechanosensory cells (6). For example, Merkel cells have microvilli that are reminiscent of stereocilia, the sites of mechanotransduction in hair cells. Also, both cell types express the transcription factors Math1 and Gfi1 (7-9).
If Merkel cells are sensory receptor cells, then they must transmit signals through synaptic contacts with somatosensory neurons. Consistent with this notion, Merkel cells contain dense-core vesicles that resemble neurosecretory vesicles (10). Moreover, Merkel cell-neurite complexes have membrane densities like those at synaptic active zones (11); however, some have argued that these are merely sites of adhesion (12).
Studies that asked whether Merkel cells are required for touch sensitivity have produced conflicting results (13). For example, removing Merkel cells by enzymatic treatment, photoablation, or genetic modification abolished the responses of slowly adapting afferents in some studies (14, 15), but not in others (16). Reports of the involvement of synaptic transmission in slowly adapting type I responses are likewise contradictory (12, 17). Recent evidence for excitatory neurotransmission is the finding that an inhibitor of ionotropic glutamate receptors reduces slowly adapting type I responsiveness (18). Additionally, sinus hair follicles, which are rich in Merkel cell-neurite complexes, show immunoreactivity for vesicular glutamate transporters (VGLUTs), which fill synaptic vesicles with glutamate (19).
Because the question of whether Merkel cells are sensory cells is unresolved, other functions have been proposed. For example, Merkel cells may play a passive role in touch by efficiently transmitting force to mechanosensitive afferents (12). Alternatively, they may release neuromodulators to regulate the sensitivity of mechanoreceptive neurons (20). Merkel cells have also been proposed to influence the development or innervation of epithelia (ref. 21, but see ref. 22).
To ask whether Merkel cells express genes that are indicative of excitable cells that play a direct role in touch, we used DNA microarrays to profile gene expression in Merkel cells. Our results show that Merkel cells express the molecular tools to send both excitatory and modulatory signals to sensory neurons.
Methods
Animals. Math1/nGFP mice [postnatal day (P)1-P19] were used for all experiments (4). Animal use was approved by the University of California, San Francisco Committee on Animal Research.
Cell Isolation. Epidermal cells were dissociated by using procedures modified from Morris (23). Hairy skin was dissected from neonatal mice (P1-P6) and placed in cold PBS. Subcutaneous tissue was removed and remaining tissue was cut into ≈1-cm2 pieces, which were attached to a Petri dish with Super Glue and then submerged in 0.25% trypsin (Invitrogen) for 1.3 h at 32°C. After peeling away the dermis, epidermal tissue was stirred with a magnetic stir bar at 60 rpm for 30 min at 4°C in S-MEM (Invitrogen) plus 10% FBS (HyClone) and 50 units/ml DNase I (Worthington). Cells were filtered through cell strainers, collected by centrifugation (300 × g for 10 min), and washed by trituration. Final cell pellets were resuspended in S-MEM/10% FBS (107 cells per ml) for flow cytometry. This procedure yielded 3.0 ± 0.4 × 107 epidermal cells per mouse (SEM; n = 141 mice), and cell viability, as determined by Trypan blue exclusion, was 60-90%.
Fluorescence-Activated Cell Sorting (facs). Cells were purified with a multiparameter cell sorter. We excluded dead cells by setting gates on forward-versus side-scatter plots. Next, we used plots of GFP fluorescence (530/30 nm) versus red autofluorescence (580/30 nm) to set gates around GFP-positive (GFP+) and GFP-negative (GFP-) cells. Equal numbers of GFP+ and GFP- epidermal cells were collected with each sort so that cells from the same animals could be compared directly. Cells were sorted into lysis buffer for RNA isolation or into S-MEM/50% FBS for culture. The isolation procedure (from skin harvesting to lysis buffer) typically lasted ≈4 h.
RNA Amplification. Total RNA was harvested from epidermal cells with a Mini RNA isolation kit (Zymo Research, Orange, CA), and DNA was removed by using RNase-free DNase (Promega). RNA was amplified from matched numbers of GFP+ and GFP- cells (P2-P5; 1-2 × 104 cells per reaction). To prepare samples for screening cDNA microarrays, we used published linear amplification methods (24). Two rounds of reverse transcription and in vitro transcription produced 7-28 μg of amplified RNA per reaction. For screening Affymetrix arrays, amplification through the second round of cDNA synthesis was performed as described (24), then in vitro transcription was accomplished with a BioArray HighYield kit (Enzo Life Sciences, Farming-dale, NY). These methods yielded 5-40 μg of biotinylated RNA per reaction. Biotinylated RNA was pooled from three amplification reactions for each Affymetrix array.
Microarray Analysis. Glass-slide cDNA microarrays were generated by the University of California, San Francisco Mouse Microarray Consortium. Fluorescent cDNAs were produced from amplified RNA (2 μg) and microarrays were screened as described (ref. 25 and Supporting Text, which is published as supporting information on the PNAS web site). Affymetrix GeneChips (Murine Genome Array U74v2) were hybridized and analyzed as described in Supporting Text. Enrichment thresholds were chosen to yield a manageable number (<300) of elements for further analysis.
RT-PCR. Total RNA from 1-2 × 104 sorted cells served as the template for reverse transcription. First-strand cDNA was synthesized with oligo(dT)12-18 primers at 42°C for 2 h using SuperScriptII (Invitrogen). PCR products were amplified with touchdown PCR; 1/100-1/20 of a reverse transcription reaction was used for each PCR. In all experiments, control PCRs lacking cDNA template were performed to confirm that products were not due to contamination. CaV2.2/α1B and keratin (KRT) primers were designed to span introns to demonstrate that products were amplified from cDNA and not genomic DNA.
Immunohistochemistry. Hairy skin from P15-P19 mice was fixed, embedded, and stained as described (ref. 4 and Supporting Text). Primary antibodies were mouse anti-KRT1-18 (RGE 53, MP Biomedicals, Aurora, OH), mouse neurofilament 200 (N52, Sigma), rabbit anti-Rab3C (Calbiochem), rabbit anti-cholecystokinin (CCK) 26-33 (CCK8, Phoenix Pharmaceuticals, Belmont, CA), rabbit anti-VGLUT2 (gift from R. Edwards, University of California, San Francisco), and rabbit anti-Piccolo (Synaptic Systems, Göttingen, Germany).
Live-Cell Imaging. FACS-purified Merkel cells were plated in Lab-Tek II CC2 chamber slides (Nalge Nunc, Rochester, NY) at ≈104 cells per well and cultured for 2 d at 37°C in keratinocyte media/10% FBS (Invitrogen). Merkel cells were loaded for 45 min with 5 μM fura-2 acetoxymethyl ester and 0.02% pluronic F-127 in Ringer's solution containing 140 mM NaCl, 5 mM KCl, 10 mM Hepes (pH 7.4), 10 mM glucose, 2 mM MgCl2, and 2 mM CaCl2. Cells were rinsed and imaged in Ringer's solution. Merkel cells were depolarized approximately once per hour with high-K+ Ringer's solution containing 73 mM NaCl, 73 mM KCl, 10 mM Hepes (pH 7.4), 10 mM glucose, 2 mM MgCl2, and 2 mM CaCl2. Ca2+-channel antagonists included 10 μM nimodipine (Tocris Cookson, Ellisville, MO), 1 μM ω-conotoxin GVIA (Tocris Cookson), and 1 μM ω-agatoxin IVA (Bachem). Antagonists and high-K+ solutions were removed with at least five washes with Ringer's solution. Cells were imaged as described in Supporting Text with a ×10, 0.3 numerical aperture Plan Fluor objective lens (Nikon). Data are expressed as means ± SEM. Statistical significance was assessed with two-tailed Student's t tests.
Results
Isolating Merkel Cells. Merkel cells represent a miniscule fraction of cells in the skin; we therefore developed a strategy to genetically label these rare cells and purify them by FACS. To obtain labeled Merkel cells, we used a transgenic mouse strain (Math1/nGFP) in which math1 enhancer sequences drive expression of GFP. In these animals, Merkel cells are the only skin cells with detectable GFP fluorescence (4).
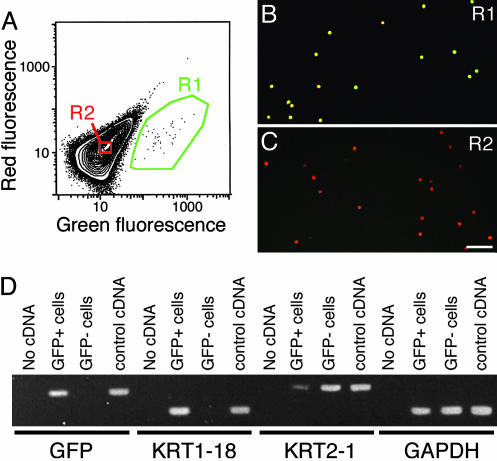
We dissociated epidermal cells from neonatal Math1/nGFP mice for FACS. To identify GFP-expressing cells, we plotted the red versus green fluorescence of viable epidermal cells (Fig. 1A). Whereas signals of autofluorescent epidermal cells fell near a line of slope unity (R2), the signals of GFP+ cells were displaced along the abscissa (R1). GFP+ cells constituted 0.08% of viable epidermal cells (n = 141 animals).
Fig. 1.
Purifying Merkel cells. (A) A plot of red versus green fluorescence of 5 × 105 epidermal cells from Math1/nGFP mice was used to set regions around GFP+ cells (R1) and GFP- cells (R2). (B and C) Confocal micrographs show epidermal cells collected from R1 (B) or R2 (C). Nuclei were stained with 4′,6-diamidino-2-phenylindole (shown in red). GFP+ cells appear yellow. (Scale bar, 50 μm.) (D) PCR products were amplified from sorted cells. Templates were dH2O, GFP+ cell cDNA, GFP- cell cDNA, and whole-skin cDNA (control cDNA). Cell-type-specific markers were amplified with the primers indicated. GAPDH product confirmed that comparable amounts of template were used from sorted cells.
Three lines of evidence confirmed that sorted GFP+ cells were highly enriched. First, by using flow cytometry to analyze FACS-purified GFP+ cells, we found that 85-95% of the cells expressed GFP, which corresponds to an enrichment of >1,000-fold (n = 3 experiments). Second, epifluorescence microscopy showed that most cells in the sorted GFP+ population displayed GFP fluorescence (Fig. 1B), whereas GFP- cells did not (Fig. 1C). Third, we used PCR to amplify transcripts that are known to be expressed specifically in either Merkel cells or keratinocytes (Fig. 1D). We amplified robust PCR products for GFP and the Merkel-cell marker KRT1-18 only from GFP+ cells. By contrast, the keratinocyte marker KRT2-1 (26) was more abundant in GFP- cells than in GFP+ cells.
Profiling Gene Expression in Merkel Cells. To identify molecules that define the specialized role of the Merkel cell in the epidermis, we compared the gene-expression profile of GFP+ Merkel cells with that of an equivalent number of GFP- epidermal cells. This comparison may identify transcripts that are specifically up-regulated in Merkel cells or those that are specifically down-regulated in other epidermal cells. The latter population primarily consisted of keratinocytes, as evidenced by the expression of KRT2-1, KRT1-14, and integrin β 1 (ref. 27 and data not shown).
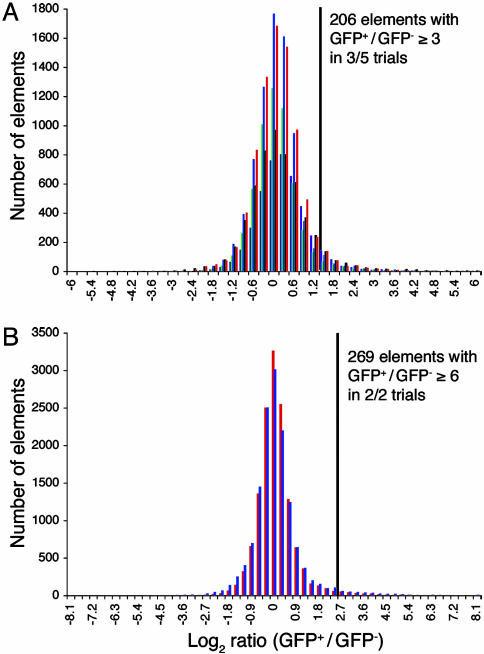
We screened cDNA microarrays containing ≈20,000 murine clones, including the riken fantom 1.1 set (28). GFP+ Merkel cells and GFP- epidermal cells were collected by FACS in two independent experiments. Total RNA was harvested and linearly amplified. One FACS collection (1.9 × 104 cells) yielded sufficient fluorescent cDNA probe to screen microarrays in triplicate. A second FACS collection (1.1 × 104 cells) produced sufficient cDNA to screen two additional microarrays. For each replicate, we determined a transcript's enrichment in Merkel cells over GFP- epidermal cells by dividing the hybridization signal from the Merkel cell probe by that of the GFP- cell probe (Fig. 2A and Table 2, which is published as supporting information on the PNAS web site). For further analysis, we chose 206 clones that exceeded an enrichment of 3-fold in at least three trials.
We also screened Affymetrix arrays representing ≈36,000 probe sets (Fig. 2B and Table 3, which is published as supporting information on the PNAS web site). Two replicates were performed from sorts and amplification reactions independent of each other and those used for screening cDNA arrays. For further analysis, we chose 269 probe sets that were enriched at least 6-fold in Merkel cells in both experimental trials.
Some transcripts were represented by multiple array elements in the two types of microarrays; we therefore compared the datasets of Merkel cell-enriched genes from the RIKEN and Affymetrix array screens to identify 362 unique genes whose mean fold enrichment in Merkel cells ranged from 3 to 1,748 (Table 4, which is published as supporting information on the PNAS web site). These genes included 225 named genes and 137 transcripts of unknown function. Eighty-five transcripts were identified with at least two independent elements on the arrays.
Nine of the Merkel cell-enriched transcripts in the dataset encoded proteins that have been previously shown by immunostaining to be expressed in Merkel cells (Table 1 and refs. 7-9, 21, and 29-34). These transcripts included transcription factors (Math1 and Gfi1), intermediate filament subunits (e.g., KRT1-18), and a dense-core vesicle protein (7B2/Sgne1). These results demonstrate that our approach is suitable for identifying transcripts that are enriched in Merkel cells in vivo.
Table 1. Selected list of Merkel cell-enriched transcripts.
| Name | Gene | Affymetrix* | RIKEN* |
|---|---|---|---|
| Previously characterized Merkel cell markers | |||
| 7B2 protein | Sgne1 | 25 | 15 |
| G protein, α-o | Gnao | 7 | |
| Gfi1 | Gfi1 | 95 | |
| KRT1-18 | Krt1-18 | 18 | |
| KRT1-20 | Krt20 | 175 | 46 |
| KRT2-8 | Krt2-8 | 443 | |
| Math1 | Atoh1 | 1,748 | |
| Neurotrophin 3 | Ntf3 | 16 | |
| PGP9.5 | Uchl1 | 20 | 7 |
| Neuronal transcription factors | |||
| AP-2β | Tcfap2b | 6 | |
| Dach1 | Dach1 | 14 | |
| Hes6 | Hes6 | 7 | |
| Insm1 | Insm1 | 43 | |
| Islet1 | Isl1 | 51 | |
| Lhx3 | Lhx3 | 19 | |
| Mash1 | Ascl1 | 68 | |
| Myt1 | Myt1 | 86 | |
| Nhlh1/NSCL1 | Nhlh1 | 19 | |
| Brn3B | Pou4f2 | 13 | |
| Sox2 | Sox2 | 87 | |
| Presynaptic molecules | |||
| Complexin 1 | Cplx1 | 15 | |
| Munc13-1 | Unc13h1 | 11 | |
| Mint1 | Apba1 | 8 | |
| Neurexin I | Nrxn1 | 13 | |
| Piccolo | Pclo | 65 | |
| Rab3C | Rab3c | 12 | 18 |
| RIM2 | Rims2 | 24 | |
| SNAP25 | Snap25 | 39 | |
| Synaptotagmin 1 | Syt1 | 31 | |
| Synaptotagmin 7 | Syt7 | 99 | |
| Synapsin II | Syn2 | 57 | 17 |
| VGLUT2 | Slc17a6 | 24 | 13 |
| CADPS | Cadps | 47 | |
| Carboxypeptidase E | Cpe | 7 | 39 |
| CCK | Cck | 631 | |
| Chromogranin B | Chgb | 136 | |
| PC 2 | Pcsk2 | 21 | 13 |
| Ion-channel subunits | |||
| α2δ1 | Cacna2d1 | 16 | |
| HCN2 | Hcn2 | 25 | |
| BK β-1 | Kcnmb1 | 18 | |
| Kv4.2 | Kcnd2 | 117 | |
| Kv1.4 | Kcna4 | 16 | |
| Kv8.1 | Kcnv1 | 71 | 40 |
Mean fold enrichment from all trials.
Merkel Cells Express Neuronal Transcription Factors. A cell's identity is determined largely by its complement of cell-type-specific transcription factors. Our microarray analysis identified 14 transcription factors that are enriched in Merkel cells. Thirteen of these transcription factors, including Math1 and Gfi1, act in neuronal development (Table 1). These data support the idea that Merkel cells function as excitable cells.
Merkel Cells Express Synaptic Proteins. Consistent with a neuron-like fate for Merkel cells, our dataset of Merkel cell-enriched transcripts included a number of molecules that regulate cell adhesion, synaptic transmission, and electrical excitability (Table 1). For example, cadherin 10, which we found to be 25-fold enriched in Merkel cells, is an adhesion molecule that is thought to regulate the formation of specific neuronal connections (35).
Fig. 2.
Histograms of the log2 ratio of signals from GFP+ cells to GFP- cells (GFP+/GFP-). For each array type, data from each trial are plotted in a different color. Gray bars indicate the enrichment values exceeded by clones that were analyzed further. (A) Results from five trials with glass-slide cDNA microarrays. Microarray elements shown had sums of median Cy3 and Cy5 signals ≥500 fluorescence units. (B) Results from two trials with Affymetrix oligonucleotide microarrays. Elements shown were scored as “present” by the absolute-call algorithm.
Moreover, we identified 17 Merkel cell-enriched transcripts encoding presynaptic and neurosecretory molecules, in addition to 7B2/Snge1 (Table 1). These transcripts included active-zone molecules such as Piccolo, and molecules required for Ca2+-triggered vesicle release, such as synaptotagmin I and synaptosomal-associated protein of 25 kDa (SNAP25). We also found that Merkel cells express molecules that modulate release, such as Rab3C and synapsin II. Moreover, our array data showed that the neuropeptide-precursor CCK and the transporter VGLUT2 are highly enriched in Merkel cells.
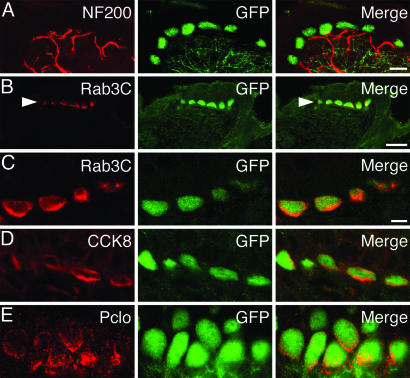
To test whether the enrichment we observed at the transcript level translates into differences in protein abundance, we labeled skin cryosections with antibodies against presynaptic proteins (Fig. 3). An antibody against the vesicle protein Rab3C showed immunoreactivity only in Merkel cells in the skin (Fig. 3B). This immunoreactivity was concentrated on the lower half of the Merkel cell (Fig. 3C), which is where afferent fibers make contact (Fig. 3A). Antibodies against the neuropeptide CCK8 (Fig. 3D) and the active-zone-matrix protein Piccolo also stained Merkel cells specifically (Fig. 3E). Similar staining patterns were seen with antibodies against SNAP25, RIM2, and synaptotagmin 13 (data not shown). The latter is an unconventional synaptotagmin (36) that we found to be enriched in Merkel cells (42-fold-enriched, Affymetrix; 64-fold-enriched, RIKEN).
Fig. 3.
Merkel cells express presynaptic proteins in vivo. Confocal micrographs show immunohistochemical staining of touch domes in Math1/nGFP skin cryosections. Each row includes antibody staining (red, Left), GFP fluorescence (green, Center) and a merged image (Right). (A) An anti-NF200 antibody labeled sensory afferents that contacted Merkel cells. Dermal fluorescence reflects autofluorescence that is independent of GFP expression. (Scale bar, 10 μm.) (B) Low-magnification micrographs demonstrate that, in the skin, Rab3C staining was detectable only in Merkel cells (arrowhead). (Scale bar, 25 μm.) (C-E) High-magnification images show immunoreactivity of Rab3C (C), CCK8 (D), and Piccolo (Pclo, E) in Merkel cells. (Scale bar in C, 5 μm, applies to C-E.)
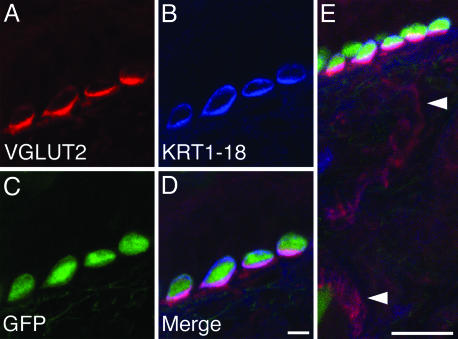
We additionally used an antibody against VGLUT2 to determine whether in vivo Merkel cells express this glutamate transporter (37). Like other presynaptic proteins (Fig. 3), VGLUT2 immunoreactivity in the skin was most intense in Merkel cells and was strongest on the side of the cell that abuts sensory nerve terminals (Fig. 4 A-D). We also observed weak VGLUT2 staining in dorsal-root-ganglion fibers, including those that contacted Merkel cells (Fig. 4E). Notably, our array data revealed that Merkel cells express receptors that monitor glutamate release, including the ionotropic receptor GluR2 (29-fold enriched). Furthermore, we found that Merkel cells express Homer2 (7-fold-enriched, Affymetrix; 15-fold-enriched, RIKEN), which regulates metabotropic glutamate receptors. The expression of such receptors has recently been detected in Merkel cells (38).
Fig. 4.
Merkel cells express VGLUT2 protein. An antibody against VGLUT2 (red in A, D, and E) labeled KRT1-18-positive (blue in B, D, and E), GFP-expressing Merkel cells (green in C-E) in a touch dome. Dorsal root ganglion fibers that contacted Merkel cells and those that formed palisade endings around hair shafts displayed weak VGLUT2 staining (arrowheads in E). The image in E is a projection of a confocal z series collected with 2-μm axial steps. (Scale bars, 5 μm in D; 20 μm in E.)
Merkel Cells Have Voltage-Gated Ion Channels. Our array data revealed that Merkel cells preferentially express six ion-channel subunits that may control signaling between Merkel cells and sensory afferent terminals (Table 1). These transcripts include three voltage-gated K+-channel subunits.
We also observed that Merkel cells preferentially express the α2δ1-subunit of voltage-gated Ca2+ channels. To determine which pore-forming subunits are expressed in Merkel cells, we performed PCRs with subtype specific primers (Fig. 5A; n = 6-9 experiments). We consistently amplified products for the P/Q-type Ca2+ channel CaV2.1/α1A, the N-type channel CaV2.2/α1B, and the L-type channel CaV1.2/α1C. Products from other α1-subunits were detected only sporadically or not at all (data not shown).
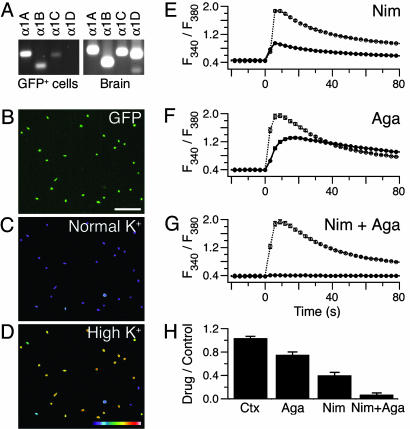
Fig. 5.
Merkel cells have functional P/Q- and L-type Ca2+ channels. (A) PCR products were amplified from sorted Merkel cells with primers specific for the indicated voltage-gated Ca2+-channel α1-subunits. (B) An epifluorescence micrograph shows sorted GFP+ cells after 2 d in culture. (Scale bar, 100 μm.) (C and D) Pseudocolor images of fura-2 fluorescence ratios (F340/F380) of the cells in B just before (C) and 6 s after (D) perfusion with high-K+ Ringer's solution. Pseudocolor scale denotes F340/F380 from 0.1 (black) to 3 (white). (E-G) Plots of mean fura-2 ratios versus time in the absence (dashed line) or presence (solid line) of Ca2+-channel antagonists. Cells were exposed to drugs for 15-20 min before depolarization. Application of high-K+ solution began at t = 0 and lasted throughout the recording. Each trace represents the average fura-2 ratio of 57-119 cells. Error bars indicate SEM. Antagonists used were 10 μM nimodipine (Nim) (E), 1 μM ω-agatoxin IVA (Aga) (F), and 10 μM nimodipine plus 1 μM ω-agatoxin IVA (G). (H) Quantification of the effects of Ca2+-channel antagonists (n = 4-5 experiments per group). Responses of Merkel cells exposed to antagonists were normalized to those measured from control cells. The effect of ω-agatoxin IVA or nimodipine was significantly different from that of ω-conotoxin GVIA (Ctx) (P ≤ 0.002). Inhibition by ω-agatoxin IVA plus nimodipine was significantly greater than that achieved with either alone (P ≤ 8 × 10-4).
To determine whether voltage-gated Ca2+ channels are functional in Merkel cells, we used the ratiometric Ca2+ indicator fura-2 acetoxymethyl ester to monitor the cytoplasmic free Ca2+ concentration in FACS-purified Merkel cells (Fig. 5 B-G). In normal Ringer's solution, Merkel cells exhibited a low ratio of fura-2 fluorescence when excited at 340 and 380 nm (Fig. 5C). When depolarized with high-K+ Ringer's solution after 2 d in culture, ≈90% of Merkel cells exhibited robust increases in fura-2 ratio (Fig. 5D). On average, the peak fura-2 response was 4-fold that of baseline signals (n = 30 experiments).
To delineate the voltage-gated Ca2+ channels that mediate depolarization-evoked Ca2+ influx in Merkel cells, we used specific antagonists (39) of L-type (nimodipine), P/Q-type (ω-agatoxin IVA), and N-type Ca2+ channels (ω-conotoxin GVIA). We observed that the peak fura-2 ratios of Merkel cells were reduced by 10 μM nimodipine (Fig. 5 E and H) and by 1 μM ω-agatoxin IVA (Fig. 5 F and H). Together, nimodipine and ω-agatoxin IVA blocked almost all of the response to high-K+ solution in cultured (93 ± 3%; Fig. 5 G and H) and acutely dissociated Merkel cells (93 ± 1%; Fig. 6, which is published as supporting information on the PNAS web site). By contrast, 1 μM ω-conotoxin GVIA had no effect on Ca2+ signals in Merkel cells (Fig. 5H).
Discussion
Our principal finding is that Merkel cells express presynaptic active-zone constituents, synaptic vesicle proteins, and molecules required for neuropeptide production and glutamate release. Moreover, our live-cell imaging experiments revealed that Merkel cells have functional voltage-gated Ca2+ channels; such channels are essential for synaptic transmission. Together, these data demonstrate that Merkel cells are excitable cells and designate glutamate and CCK8 as candidate neurotransmitters at synapses between Merkel cells and sensory afferents in vivo. Our conclusion that Merkel cells function as excitable cells is strengthened by the abundance of neuronal transcription factors that we found to be enriched in Merkel cells (Table 1).
The discovery of molecules that are necessary for touch reception has been hindered by the paucity of somatosensory mechanoreceptors and by the fact that their mechanosensitive structures are scattered throughout target tissues. In this study, we have surmounted these obstacles by combining genetic labeling, in vitro methods, and microarray techniques to identify 362 transcripts that are enriched in Merkel cells. A similar strategy has been used to discover genes expressed in worm touch receptors (40). To our knowledge, this report represents the first extensive molecular profiling of Merkel cells, and it provides a rich data set of molecules that help to define the Merkel cell's function in the epidermis.
These data afford an assessment of the molecules expressed by Merkel cells at the message level. For 16 of the named genes, we and others have used antibodies to demonstrate protein enrichment in vivo. Such verification is important because the correlation between transcript abundance and protein levels is imperfect. Furthermore, technical limitations may have led to the inclusion of false positives in our data set. For example, it is conceivable that Merkel cells copurified with fragments of somatosensory afferents that contained neuronal transcripts. By directly demonstrating that Merkel cells express presynaptic proteins in vivo, we have ruled out the possibility that synaptic molecules are found only in somatosensory afferents.
Along with describing molecular components of Merkel-cell synapses, our expression data offer a means for discovering targets of transcription factors. Interestingly, three of the Merkel cell-enriched transcription factors we found have been implicated in mechanosensory cell development. For example, Math1 and Gfi1, which are expressed in Merkel cells at the protein level (7, 9), are essential for proper hair-cell differentiation (9, 41). Atonal, the Drosophila ortholog of Math1, is a proneural gene for chordotonal organs, which mediate hearing and proprioception (42). Additionally, expression of Brn3B has been shown in lateral-line hair cells (43). The closest homolog to Brn3B in Caenorhabditis elegans, UNC-86, is needed for differentiation of neurons that respond to gentle body touch (44).
Our data also support the idea that Merkel cells or their precursors give rise to Merkel-cell carcinoma, a skin tumor whose origin is controversial (45). Comparison of our expression data with those from Merkel-cell carcinomas (46) identifies five transcripts that are enriched in both cell types: SNAP25, carboxypeptidase E, proprotein convertase 2, 7B2/Sgne1, and protein phosphatase 2A B56β.
As well as molecular profiles, we have developed in vitro methods for purifying and imaging the activity of living Merkel cells. These methods represent a significant advance because they allow signal transduction in Merkel cells to be characterized with high-resolution techniques. Such dissociated cell preparations have been essential for discovering mechanisms of sensory signaling in hair cells and thermosensitive nociceptors (47, 48). In this study, we used these methods to ascertain which voltage-gated Ca2+ channels are active in murine Merkel cells. Our results extend a previous report of Ca2+ currents in Merkel cells (49). We found that almost all of the depolarization-induced Ca2+ influx in Merkel cells is through two types of channels. These are L-type channels, which trigger neurotransmission in hair cells and retinal bipolar cells (50, 51), and P/Q-type channels, which are found at central synapses (52). Although we found that Merkel cells expressed transcripts encoding CaV2.2/α1B, these channels did not significantly contribute to Ca2+ entry. This finding suggests that, under our experimental conditions, either such channels are not activated or significant protein is not expressed.
Our finding that Merkel cells express presynaptic molecules indicates that the sites of Merkel cell-afferent contact observed ultrastructurally are most likely synaptic active zones. The presence of such active zones is consistent both with the idea that Merkel cells are sensory receptor cells that signal afferents through neurotransmission and with the hypothesis that Merkel cells release neuromodulators to influence the sensitivity of mechanoreceptive afferents.
How might neurotransmitter release be stimulated from Merkel cells? Merkel cells may be mechanoreceptive cells that are directly activated by touch. Alternatively, Merkel cells may receive input from active afferent terminals. The latter conjecture is bolstered by reports of reciprocal connections in Merkel cell-neurite complexes (11).
By identifying molecular components of Merkel-cell synapses, our results strongly suggest that Merkel cells are active participants in somatosensory signaling. Moreover, this study provides tools for interfering with synaptic transmission so its role in touch reception can be defined.
Supplementary Material
Acknowledgments
We thank Jane Johnson (University of Texas Southwestern Medical Center, Dallas) for providing mice; Rich Locksley, Dan Stetson, and Cliff McArthur for assistance with flow cytometry; Caroline Mrejen for cDNA arrays; Anita Sil and Ansgar Klebes for assistance with array experiments; Robert Fremeau and Robert Edwards (University of California, San Francisco), and Thomas Südhof (University of Texas Southwestern Medical Center) for antibodies; David Julius and his colleagues for discussions; and Diana Bautista, David Julius, Helmut Krämer, Dan Minor, and Anita Sil for providing invaluable comments on the manuscript. This work was supported by the Sandler Program in Basic Sciences (to E.A.L.) and by National Institute of Arthritis and Musculoskeletal and Skin Diseases Grant AR051219 (to E.A.L.). H.H. is a National Science Foundation Predoctoral Fellow.
Abbreviations: CCK, cholecystokinin; FACS, fluorescence-activated cell sorting; VGLUT, vesicular glutamate transporter; Pn, postnatal day n; KRT, keratin; SNAP25, synaptosomal-associated protein of 25 kDa.
References
- 1.Iggo, A. & Muir, A. R. (1969) J. Physiol. (London) 200, 763-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson, K. O. (2001) Curr. Opin. Neurobiol. 11, 455-461. [DOI] [PubMed] [Google Scholar]
- 3.Meyers, J. R., MacDonald, R. B., Duggan, A., Lenzi, D., Standaert, D. G., Corwin, J. T. & Corey, D. P. (2003) J. Neurosci. 23, 4054-4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lumpkin, E. A., Collisson, T., Parab, P., Omer-Abdalla, A., Haeberle, H., Chen, P., Doetzlhofer, A., White, P., Groves, A., Segil, N., et al. (2003) Gene Expr. Patterns 3, 389-395. [DOI] [PubMed] [Google Scholar]
- 5.Merkel, F. (1875) Arch. Mikrosk. Anat. 11, 636-652. [Google Scholar]
- 6.Iggo, A. & Findlater, G. S. (1984) in Sensory Receptor Mechanisms, eds. Hamann, W. & Iggo, A. (World Scientific, Singapore), pp. 117-131.
- 7.Helms, A. W., Abney, A. L., Ben-Arie, N., Zoghbi, H. Y. & Johnson, J. E. (2000) Development (Cambridge, U.K.) 127, 1185-1196. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Arie, N., Hassan, B. A., Bermingham, N. A., Malicki, D. M., Armstrong, D., Matzuk, M., Bellen, H. J. & Zoghbi, H. Y. (2000) Development (Cambridge, U.K.) 127, 1039-1048. [DOI] [PubMed] [Google Scholar]
- 9.Wallis, D., Hamblen, M., Zhou, Y., Venken, K. J., Schumacher, A., Grimes, H. L., Zoghbi, H. Y., Orkin, S. H. & Bellen, H. J. (2003) Development (Cambridge, U.K.) 130, 221-232. [DOI] [PubMed] [Google Scholar]
- 10.Hartschuh, W., Weihe, E. & Egner, U. (1990) Neurosci. Lett. 116, 245-249. [DOI] [PubMed] [Google Scholar]
- 11.Mihara, M., Hashimoto, K., Ueda, K. & Kumakiri, M. (1979) J. Invest. Dermatol. 73, 325-334. [DOI] [PubMed] [Google Scholar]
- 12.Gottschaldt, K. M. & Vahle-Hinz, C. (1981) Science 214, 183-186. [DOI] [PubMed] [Google Scholar]
- 13.Halata, Z., Grim, M. & Bauman, K. I. (2003) Anat. Rec. 271, 225-239. [DOI] [PubMed] [Google Scholar]
- 14.Mills, L. R. & Diamond, J. (1995) J. Neurocytol. 24, 117-134. [DOI] [PubMed] [Google Scholar]
- 15.Kinkelin, I., Stucky, C. L. & Koltzenburg, M. (1999) Eur. J. Neurosci. 11, 3963-3969. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda, I., Yamashita, Y., Ono, T. & Ogawa, H. (1994) J. Physiol. (London) 479, 247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pacitti, E. G. & Findlater, G. S. (1988) Prog. Brain Res. 74, 37-42. [DOI] [PubMed] [Google Scholar]
- 18.Fagan, B. M. & Cahusac, P. M. (2001) NeuroReport 12, 341-347. [DOI] [PubMed] [Google Scholar]
- 19.Hitchcock, I. S., Genever, P. G. & Cahusac, P. M. (2004) Neurosci. Lett. 362, 196-199. [DOI] [PubMed] [Google Scholar]
- 20.Tachibana, T. & Nawa, T. (2002) Anat. Sci. Int. 77, 26-33. [DOI] [PubMed] [Google Scholar]
- 21.Pasche, F., Merot, Y., Carraux, P. & Saurat, J. H. (1990) J. Invest. Dermatol. 95, 247-251. [DOI] [PubMed] [Google Scholar]
- 22.Cronk, K. M., Wilkinson, G. A., Grimes, R., Wheeler, E. F., Jhaveri, S., Fundin, B. T., Silos-Santiago, I., Tessarollo, L., Reichardt, L. F. & Rice, F. L. (2002) Development (Cambridge, U.K.) 129, 3739-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris, R. J. (1994) in High Calcium Defined Medium: Keratinocyte Methods, eds. Leigh, I. & Watt, F. (Cambridge Univ. Press, Cambridge, U.K.), pp. 25-31.
- 24.Klebes, A., Biehs, B., Cifuentes, F. & Kornberg, T. B. (2002) Genome Biol. 3, RESEARCH0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeRisi, J. L., Iyer, V. R. & Brown, P. O. (1997) Science 278, 680-686. [DOI] [PubMed] [Google Scholar]
- 26.Chu, P. G. & Weiss, L. M. (2002) Histopathology 40, 403-439. [DOI] [PubMed] [Google Scholar]
- 27.Brakebusch, C., Grose, R., Quondamatteo, F., Ramirez, A., Jorcano, J. L., Pirro, A., Svensson, M., Herken, R., Sasaki, T., Timpl, R., et al. (2000) EMBO J. 19, 3990-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawai, J., Shinagawa, A., Shibata, K., Yoshino, M., Itoh, M., Ishii, Y., Arakawa, T., Hara, A., Fukunishi, Y., Konno, H., et al. (2001) Nature 409, 685-690. [DOI] [PubMed] [Google Scholar]
- 29.Tachibana, T., Endoh, M. & Nawa, T. (2001) Histochem. Cell Biol. 116, 205-213. [DOI] [PubMed] [Google Scholar]
- 30.Vielkind, U., Sebzda, M. K., Gibson, I. R. & Hardy, M. H. (1995) Acta Anat. (Basel) 152, 93-109. [DOI] [PubMed] [Google Scholar]
- 31.Moll, I., Kuhn, C. & Moll, R. (1995) J. Invest. Dermatol. 104, 910-915. [DOI] [PubMed] [Google Scholar]
- 32.Dalsgaard, C. J., Rydh, M. & Haegerstrand, A. (1989) Histochemistry 92, 385-390. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Caballero, A., Gallego, R., Garcia-Caballero, T., Fraga, M., Blanco, M., Fernandez-Redondo, V. & Beiras, A. (1997) Anat. Rec. 248, 159-163. [DOI] [PubMed] [Google Scholar]
- 34.Szeder, V., Grim, M., Kucera, J. & Sieber-Blum, M. (2003) Dev. Dyn. 228, 623-629. [DOI] [PubMed] [Google Scholar]
- 35.Bekirov, I. H., Needleman, L. A., Zhang, W. & Benson, D. L. (2002) Neuroscience 115, 213-227. [DOI] [PubMed] [Google Scholar]
- 36.von Poser, C. & Südhof, T. C. (2001) Eur. J. Cell Biol. 80, 41-47. [DOI] [PubMed] [Google Scholar]
- 37.Fremeau, R. T., Jr., Troyer, M. D., Pahner, I., Nygaard, G. O., Tran, C. H., Reimer, R. J., Bellocchio, E. E., Fortin, D., Storm-Mathisen, J. & Edwards, R. H. (2001) Neuron 31, 247-260. [DOI] [PubMed] [Google Scholar]
- 38.Tachibana, T., Endoh, M., Kumakami, R. & Nawa, T. (2003) Histochem. Cell Biol. 120, 13-21. [DOI] [PubMed] [Google Scholar]
- 39.Catterall, W. A. (2000) Annu. Rev. Cell Dev. Biol. 16, 521-555. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, Y., Ma, C., Delohery, T., Nasipak, B., Foat, B. C., Bounoutas, A., Bussemaker, H. J., Kim, S. K. & Chalfie, M. (2002) Nature 418, 331-335. [DOI] [PubMed] [Google Scholar]
- 41.Bermingham, N. A., Hassan, B. A., Price, S. D., Vollrath, M. A., Ben-Arie, N., Eatock, R. A., Bellen, H. J., Lysakowski, A. & Zoghbi, H. Y. (1999) Science 284, 1837-1841. [DOI] [PubMed] [Google Scholar]
- 42.Jarman, A. P. (2002) Hum. Mol. Genet. 11, 1215-1218. [DOI] [PubMed] [Google Scholar]
- 43.DeCarvalho, A. C., Cappendijk, S. L. & Fadool, J. M. (2004) Dev. Dyn. 229, 869-876. [DOI] [PubMed] [Google Scholar]
- 44.Duggan, A., Ma, C. & Chalfie, M. (1998) Development (Cambridge, U.K.) 125, 4107-4119. [DOI] [PubMed] [Google Scholar]
- 45.Haag, M. L., Glass, L. F. & Fenske, N. A. (1995) Dermatol. Surg. 21, 669-683. [DOI] [PubMed] [Google Scholar]
- 46.Van Gele, M., Boyle, G. M., Cook, A. L., Vandesompele, J., Boonefaes, T., Rottiers, P., Van Roy, N., De Paepe, A., Parsons, P. G., Leonard, J. H., et al. (2004) Oncogene 23, 2732-2742. [DOI] [PubMed] [Google Scholar]
- 47.Lewis, R. S. & Hudspeth, A. J. (1983) Nature 304, 538-541. [DOI] [PubMed] [Google Scholar]
- 48.Caterina, M. J. & Julius, D. (2001) Annu. Rev. Neurosci. 24, 487-517. [DOI] [PubMed] [Google Scholar]
- 49.Yamashita, Y., Akaike, N., Wakamori, M., Ikeda, I. & Ogawa, H. (1992) J. Physiol. (London) 450, 143-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts, W. M., Jacobs, R. A. & Hudspeth, A. J. (1990) J. Neurosci. 10, 3664-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tachibana, M., Okada, T., Arimura, T., Kobayashi, K. & Piccolino, M. (1993) J. Neurosci. 13, 2898-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reuter, H. (1996) Curr. Opin. Neurobiol. 6, 331-337. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.