Abstract
This review examines the current state of electrophysiological endophenotype research and recommends best practices that are based on knowledge gleaned from the last decade of molecular genetic research with complex traits. Endophenotype research is being oversold for its potential to help discover psychopathology relevant genes using the types of small samples feasible for electrophysiological research. This is largely because the genetic architecture of endophenotypes appears to be very much like that of behavioral traits and disorders: they are complex, influenced by many variants (e.g., tens of thousands) within many genes, each contributing a very small effect. Out of over 40 electrophysiological endophenotypes covered by our review, only resting heart, a measure that has received scant advocacy as an endophenotype, emerges as an electrophysiological variable with verified associations with molecular genetic variants. To move the field forward, investigations designed to discover novel variants associated with endophenotypes will need extremely large samples best obtained by forming consortia and sharing data obtained from genome wide arrays. In addition, endophenotype research can benefit from successful molecular genetic studies of psychopathology by examining the degree to which these verified psychopathology-relevant variants are also associated with an endophenotype, and by using knowledge about the functional significance of these variants to generate new endophenotypes. Even without molecular genetic associations, endophenotypes still have value in studying the development of disorders in unaffected individuals at high genetic risk, constructing animal models, and gaining insight into neural mechanisms that are relevant to clinical disorder.
Keywords: endophenotype, biomarker, heritability, genes, GWAS, GREML, candidate gene, data sharing
1.0 Introduction
In a paper investigating the genetic basis for schizophrenia published in Nature in 1988, Sherrington et al. (1988) published what many thought was a breakthrough paper, wherein they concluded that they had found the “first strong evidence for the involvement of a single gene in the causation of schizophrenia.” Few would have thought then that in the ensuing three decades, and despite witnessing the human genome project completed, we still would know relatively little about how specific genes influence the development of schizophrenia and other psychiatric disorders. Moreover, what little we do know has no obvious public health significance. A major concern in the psychiatric literature has been that identifying genes is hamstrung by the inadequacy of the American Psychiatric Association's Diagnostic and Statistical Manuals (DSM) to carve nature at its joints with enough precision to facilitate success. The definition of DSM disorders depends little on biology; instead, they remain defined largely by consensus expert opinion, are obviously heterogeneous, and show substantial overlap. Against this backdrop, interest in strategies for gene finding that do not depend on the DSM has been high.
One such strategy involves identifying endophenotypes, genetically influenced quantifiable traits that have the potential to carve nature at its joints. Endophenotypes identify those at risk for psychopathology prior to its becoming manifest and can be used to identify etiologically relevant genetic variants. Assumed to be less genetically complex and more proximal to the effects of genes, endophenotypes offer a potentially refined and powerful approach to uncover genetic variants associated with psychopathology. Many thoughtful endophenotype articles populate the literature (for recent examples, see Anokhin, 2014; Beauchaine, 2009; Burton et al., 2015; Campanella, Pogarell, & Boutros, 2014; Euser et al., 2012; Faraone, Bonvicini, & Scassellati, 2014; Ferrarelli, 2013; Glahn et al., 2014; Goldstein & Klein, 2014; W. G. Iacono & Malone, 2011; Lenzenweger, 2013; Loo, Lenartowicz, & Makeig, 2015; Manoach & Agam, 2013; Miller & Rockstroh, 2013; Moses-Kolko, Horner, Phillips, Hipwell, & Swain, 2014; Owens, Bachman, Glahn, & Bearden, 2016; Pearlson, 2015; Rosen, Spellman, & Gordon, 2015; Rubenstein, Wiggins, & Lee, 2015; Salvatore, Gottesman, & Dick, 2015; Swerdlow, Gur, & Braff, 2015), and many of these share a conviction that endophenotypes are valuable for identifying genetic liability . However, despite their introduction to psychiatry by Gottesman and Shields (I. I. Gottesman & Shields, 1972) over four decades ago, the promise of endophenotypes for gene discovery has yet to be realized.
In this article, we evaluate this promise and recommend best practices for genetic endophenotype research that we believe can improve the quality of investigation. We are informed in this effort by our own experience conducting molecular-genetic investigations of more than a dozen psychophysiological measures (W. G. Iacono, Malone, Vaidyanathan, & Vrieze, 2014b; Malone et al., 2014; Vaidyanathan, Isen, et al., 2014; Vaidyanathan, Malone, Donnelly, et al., 2014b; Vaidyanathan, Malone, Miller, McGue, & Iacono, 2014; Vrieze et al., 2014a, 2014b), as well as by the lessons of the past decade or so of molecular genetic research, which have prompted researchers to think about the genetics of complex traits and diseases differently. We have learned that disorders like schizophrenia are caused by many genetic variants each of which has a small effect in the general population (Schizophrenia Working Group of the Psychiatric Genomics, 2014). In fact, neurodevelopmental disorders like schizophrenia, which only a few decades ago were thought to be caused primarily by the home rearing environment, are not all that different in their genetic architecture from complex medical diseases like coronary heart disease or type 2 diabetes (Visscher, Brown, McCarthy, & Yang, 2012). The small effect of any individual common genetic variant is undetectable without an adequately powered design. A longstanding hope is that individual genetic effects on endophenotypes will be larger, thereby increasing power to detect genetic variants related to disease and psychiatric disorders. However, relatively few reports put endophenotypes to the ultimate test, evaluating whether in fact an endophenotype can be used to identify molecular genetic variants associated with psychopathology. Those that exist have been largely restricted to candidate gene investigations or small sample reports that, if they have generated positive results, have not been verified. Our recent work, which constitutes the most comprehensive large sample molecular genetic investigation of electrophysiological endophenotypes undertaken to date (described in detail below, see W. G. Iacono, 2014a), provides little basis for optimism that endophenotypes will live up to the hope that they will lead to breakthroughs in the identification of psychopathology relevant genes. Hence, this is a good point to reconsider where the value of endophenotypes lies, and how endophenotypes can be profitably used to shed light on the etiology of psychiatric disorder.
Our focus is on electrophysiological endophenotypes, but the approach we recommend is broadly applicable to all endophenotypes. Advances in psychophysiology, such as improved signal processing and statistical methods, are obviously important, but we do not believe that the current level of sophistication used to quantify and process electrophysiological variables is a significant impediment to success in identifying genetic variants associated with endophenotypes. Electrophysiological variables have psychometric properties that are at least equivalent to and in many cases are better than those of phenotypes that have met with success in molecular genetic research. Our focus is on the best practices needed to promote a trait as a suitable endophenotype, and to demonstrate its utility for uncovering biological pathways to the development of psychiatric disorder. Although we believe our recommendations are supported by current practices governing molecular genetic investigations of complex traits, we recognize that there is apt to be some divergence of opinion regarding which practices are indeed optimal. We are not arguing that all endophenotype research must subscribe to our approach, but we do believe that deviation from our recommendations should be accompanied by sound justification.
We begin by reviewing the development of the endophenotype construct and update the criteria that endophenotypes should satisfy given the current state of knowledge regarding the genetics of complex traits. Next, we consider how to use molecular genetic methods and analytic techniques to gain insights into the biology of endophenotypes. These recommendations are derived in part from our experience evaluating the molecular genetic basis of 17 endophenotypes that we published as a special issue in Psychophysiology in December 2014. Included in the special issue is an article devoted to molecular genetic methodology (W. G. Iacono, Malone, Vaidyanathan, & Vrieze, 2014a). We recommend this methods article, which was written as a tutorial and included a glossary of technical terms, to our readers who desire better understanding of the molecular and statistical techniques we discuss here. We follow discussion of our special issue with a selective review and critique of electrophysiological endophenotypes in light of our recommended best practices. We conclude by considering future directions for endophenotype research.
2.0 Biomarkers and the Endophenotype Concept
Psychiatric research has had a longstanding interest in developing reliable biomarkers, clinically useful biological features that are associated with psychopathology. A valid biomarker could lead to improved diagnosis, prognosis, and treatment as well as provide clues to the nature of underlying pathophysiology (W. G. Iacono, 1985). Although biomarkers such as neurochemical metabolites or measures of inflammatory response have obvious face value, for the vast majority of disorders, there is no consensus regarding which molecules are likely to make the best marker targets. Absent a solid theoretical foundation from which a vast array of possible biochemical markers can be reduced to a few plausible candidates, identifying valid biomarkers remains a daunting empirical task. Part of the appeal of psychophysiological measures derives from their tapping central nervous system function broadly; if the integrity of any element of a brain system is compromised, an electrophysiological measure associated with that system may be affected. However, this advantage may be offset by the measure's spatial coarseness and inability to identify the exact locus of dysfunction or precise mechanism involved.
State-dependent biomarkers that are present only during exacerbation of symptoms constitute episode markers (W. G. Iacono, 1985). Episode markers can be useful for disorder identification, and for monitoring course and treatment effectiveness. Results from schizophrenia research have shown how psychophysiological measures can be used to “mark” the presence of psychotic symptoms. Using a vocalization paradigm adapted from primate research, Ford and colleagues (J.M. Ford, 2015; J. M. Ford et al., 2014) have identified an N1 event-related potential response in schizophrenia patients that, because it appears to assess the quality of neural processing associated with hallucinations, may index the state of psychosis in this disorder (Ford, 2016). Using magnetoencephalography (MEG) to examine functional connectivity across cortical regions, Hinkley et al. (Hinkley et al., 2011) also provide evidence of an electrocortical biomarker for schizophrenia. These investigators reported that diminished alpha band connectivity was associated with psychotic symptoms and impaired cognition, and posited that the observed neurophysiologic effect might be a useful treatment target.
Unlike state-dependent biomarkers, markers of environmentally induced susceptibility are temporally stable traits that identify those who have become vulnerable to disorder as a consequence of environmental exposure (W. G. Iacono, 1985). Acquired characteristics, such as those secondary to perinatal complications or substance abuse, or those arising subsequent to trauma, fall into this category. Investigations of posttraumatic stress disorder (PTSD) have shown that traumatic exposure may generate such susceptibility markers. Comparing monozygotic twins discordant for combat exposure, Orr et al. (2003) found that elevated heart rate response to startling sounds was evident only in the exposed twins, suggesting that the cardiac response represents an acquired marker of PTSD rather than a sign of pre-existing genetic vulnerability. In a MEG study, combat veterans with PTSD and resilient combat veterans exposed to trauma without developing PTSD showed distinctly different patterns of neural network activity that were interpreted as accounting for differences in how trauma was encoded in the brain (James et al., 2013). Such environmentally mediated heart rate and MEG responses have many of the qualities of an endophenotype, but because they are not a manifestation of genetic liability, they would have little value for identifying whatever genetic mechanisms are involved in the associated clinical disorder.
Psychophysiological measures have in common with other types of biomarkers the fact that the unit of measurement is biological. They, like many other types of biomarkers, depend on a psychological task or circumstance; understanding the paradigm used to elicit the physiological response is thus critical to the interpretation of the significance of the response. The eye blink startle electromyographic (EMG) response takes on quite different meaning if it is elicited by an intense unexpected event, the same event when part of a pre-pulse inhibition sequence, and the same event presented while viewing pleasant or aversive imagery. In only the first two of these paradigms is the response heritable, and only pre-pulse inhibition receives strong support as an endophenotype (Anokhin, Golosheykin, & Heath, 2007; Anokhin, Heath, Myers, Ralano, & Wood, 2003; Hasenkamp et al., 2010; Malone et al., 2014). Psychological context is thus important to the evaluation of a psychophysiological marker, a fact that is often not fully appreciated when an electrophysiological signal is interpreted as a neurophysiological biomarker as though the bioelectric response itself is all that matters.
Psychophysiological research that successfully identifies these different types of biomarkers may well have considerable public health significance. Endophenotypes are also biomarkers. They can be distinguished from episode markers and trait markers stemming from environmental exposure by their ability to index genetic liability for a psychiatrically relevant trait (W. G. Iacono, 1985).
3.0 The Research Domain Criteria (RDoC) Naturally Involve Endophenotypes
The US National Institute of Mental Health developed the Research Doman Criteria (RDoC) to encourage psychopathology research that is organized around a behavioral neuroscience framework rather than the clinical descriptions that characterize the DSM. RDoC emphasizes continuously distributed biobehavioral dimensions rather than categories based on descriptions of behavioral symptoms. Biomarker research in general and endophenotype research in particular fit the RDoC scheme which emphasizes taking advantage of knowledge in neuroscience, genomics, and behavioral science to gain insights into psychopathology-relevant dimensional constructs. As others have noted, the brain systems focus of RDoC “builds on a fundamentally psychophysiological outlook” (Cuthbert, 2014, p. 1205) that “brings the realm of endophenotypes to the foreground of the research enterprise” (Miller & Rockstroh, 2013, p. 201).
4.0 Endophenotype Construct Validation
We contend that best practice endophenotype research should be designed to address the topics listed in Table 1. These topics are grouped into three domains, representing three important aspects of construct validation (Cronbach & Meehl, 1955a) applied to endophenotypes. The first, threshold criteria, describe the minimum requirements any biomarker must satisfy to have standing as a putative endophenotype. The second, endophenotype verification, identifies the requirements necessary to remove the qualifier “putative” or “candidate” from an endophenotype's status. Compared to the first domain, there is far less endophenotype research targeting verification. The third domain deals with utility. While the topics listed here also enhance construct validity, they cover usefulness of an endophenotype, how it can be used as a research tool to advance knowledge regarding the biological etiology of psychiatric disorder. To prove its worth, an identified endophenotype should enable researchers to achieve the verification and utility topic goals. Thus far, few endophenotype investigations adequately tackle these aims.
Table 1.
Endophenotype Construct Validation
| I. Putative Endophenotype Threshold Criteria | 1. Associated with one or more relevant clinical phenotypes and |
| 2. Is heritable and/or | |
| 3. Is present in first degree relatives of those with the clinical phenotype and/or | |
| 4. Shares genetic variance with the clinical phenotype | |
| II. Molecular Genetic Endophenotype Verification | 5. Shows verified association with specific genetic variants |
| 6. These verified variants show robust association with the clinical phenotype | |
| III. Utility | 7. Predicts the development of the clinical phenotype |
| 8. Enhances theoretical understanding of the brain mechanisms accounting for endophenotype individual differences | |
| 9. Informs an animal model | |
| 10. Identifies genetic variants that have relatively large effect |
Note - Topics 1, 2, and 3 overlap with criteria in Gottesman & Gould (2003)
4.1 Threshold criteria
Despite ample evidence that psychiatric disorders involve dysfunctional brain systems, there are no biomarkers that can be used clinically to confirm a diagnosis or identify a given individual as at risk, and it is not clear that the candidate biomarkers that exist do a better job identifying cases than existing interview methods (although see (Clementz et al., 2015)). In addition, our understanding of the pathophysiology of psychiatric disorders remains fairly primitive. In the absence of a good theoretical framework regarding the mechanisms that give rise to abnormal behavior, it is difficult to develop hypotheses that generate new biomarker candidates, so much of the research conducted to date has involved an atheoretical empirical approach to biomarker discovery. Although this is true for endophenotypes as well as other biomarkers, the construct validity of a putative endophenotype is enhanced by the knowledge that psychopathology is heritable. Unlike other types of biomarkers which need only show an association to psychopathology to be elevated to candidate status, endophenotypes must show such association plus evidence that they are under genetic influence. As Table 1 indicates, these two criteria must be met for a biomarker to receive provisional consideration as an endophenotype.
Demonstrating an association between a candidate endophenotype and a clinical disorder or correlated clinical characteristic is necessary to establish the clinical relevance of the measure, but the measure need not show high sensitivity or specificity because DSM disorders are heterogeneous and overlapping. The value of the endophenotype is not to validate a DSM diagnosis, but to provide a biologically informed alternative avenue to uncover etiological factors relevant to the types of dysfunction those with a diagnosis experience. As Patrick and colleagues have emphasized (Patrick et al., 2013; Yancey, Venables, & Patrick, 2016), it is not reasonable to expect measures of constructs from different domains, such as a psychophysiological variable and a clinical interview assessment, to show more than a modest association, and correlations smaller than .20 are commonplace (W. G. Iacono, 2014b; W G Iacono, 2016). For this reason, relatively large sample investigations are required to quantify accurately the degree to which variance is shared between an endophenotype and an associated clinical phenotype (e.g., an N of 193 is required to have 80% power for detecting a Pearson correlation of .20 as significant at p<.05).
Investigations that recruit genetically informative samples, like twins, can be used to establish the heritability of an endophenotype. A general population sample can be used for this purpose for dimensional traits that are continuously distributed (which appears to be the case for the vast majority of candidate endophenotypes). Establishing heritability also establishes reliability, another critical endophenotype attribute. Monozygotic twins, who conceptually can be thought of as parallel forms of the same individual, are especially useful for demonstrating the psychometric soundness of an endophenotypic measure because they can be expected to be highly similar for virtually all human traits (Polderman et al., 2015). The relatives of affected individuals with the endophenotype should possess the trait. Affected relatives should be characterized by endophenotype values similar to those of probands. Unaffected relatives are of particular value. Because many, but not all, of the unaffected relatives are presumably at elevated risk for developing a heritable disorder, they should score midway between probands and healthy comparison subjects. These patterns should be evident in all first-degree affected and unaffected relatives - parents, siblings and offspring. Given that the genetic liability for a clinical phenotype is shared by a candidate endophenotype, shared genetic influence should be evident. The same genetically informative samples that are used to establish the heritability of an endophenotype can often also be used to establish the degree to which genetic covariance is shared across the endophenotype and the clinical phenotype.
Once these threshold criteria are satisfied, the nomological net that derives from the process of endophenotype construct validation (Cronbach & Meehl, 1955b) can be further developed by carrying out research that address topics in Sections II and III of Table 1.
4.2 Endophenotype verification requires molecular genetic data
Virtually all research conducted on endophenotypes address the threshold criteria in Section I of Table 1. These criteria have provided the default definition of an endophenotype for decades. However, virtually every human trait of significance is heritable (Polderman et al., 2015), so the current threshold for considering any biomarker as a putative endophenotype is low. The proliferation of candidate endophenotypes motivated primarily by meeting these threshold criteria has thus far contributed little to our understanding of the genetics of psychopathology. Given the current state of molecular genetic knowledge and methods, continued reliance on this default definition has outlived its usefulness; it is time to hold the definition of an endophenotype to a higher standard, and require that it demonstrate verifiable association with genetic variants. To achieve this criterion, the work supporting the molecular genetic basis of the endophenotype must be adequately powered, a topic we consider at length in subsequent sections. In addition, any molecular genetic finding must be supported through replication or meta-analysis. Once this is established, conducting investigations to determine if the same variants are associated with the relevant clinical phenotype should be relatively straightforward if power is also adequate. This accomplishment would speak directly to the hoped-to-be-realized promise of endophenotypes. However, and importantly, it is conceivable that this process would also work in reverse in that genetic variants associated with a clinical phenotype can be used to identify variants for an electrophysiological measure with which it is correlated.
Recent methodological advances make possible the use of molecular genetic data to estimate heritability and the degree to which genetic variants are shared between an endophenotype and its associated clinical phenotype, two of our criteria in Table 1, which were previously only possible in twin studies (although family studies can at least provide evidence of familial influence even if they cannot disentangle genes and common environment). Genomic-relatednesss-matrix restricted maximum likelihood (GREML) (Ge et al., 2015; Speed, Hemani, Johnson, & Balding, 2012; Yang et al., 2010), which is implemented in software tools such as GCTA (Yang, Lee, Goddard, & Visscher, 2011) enables researchers to estimate the heritability of a trait (Yang et al., 2010) in a sample of subjects who are unrelated by kinship. Heritability estimates (“SNP heritability”) derived from GREML, or other approaches with a similar aim (So, Li, & Sham, 2011), are based on the degree of genetic relatedness among individuals from different families of origin that is due to measured genetic variants, as opposed to the degree of genetic relatedness implied by different familial relationships (monozygotic twin siblings, dizygotic twins or other sibling pairs, parent-offspring pairs, and the like). GREML determines the degree to which genetic relatedness between pairs of individuals accounts for phenotypic variance, using a mixed model to estimate the genetic variance in the phenotype. Because subjects are not from the same family, phenotypic similarity between them cannot be due to the collection of factors that contribute to family resemblance, including shared environment. It must be due to genetic variants inherited independently by subjects who are unrelated by kinship that influence the trait under study or that are linked with causal variants. Endophenotype research can be extended by GREML in ways that are not possible with classic twin- or family-based methods, such as to partition heritability by different characteristics of the genome, such as chromosome, minor allele frequency, and functional annotation, a method of attaching relevant biological information to genomic elements. (See J. J. Lee, Vattikuti, & Chow, 2016, for a discussion.) GREML also can be used to derive estimates of the genetic correlation between traits (S. H. Lee, Yang, Goddard, Visscher, & Wray, 2012), as can cross-trait LD Score regression (Bulik-Sullivan et al., 2015), thus permitting endophenotype researchers to address criterion #4 in Table 1 using measured variants.
4.3 Utility
An underappreciated element of an endophenotype is its ability to identify youth without the manifest clinical phenotype who are nevertheless at high risk for developing it. This is a difficult criterion to establish because it requires longitudinal investigations in which a healthy sample of those with the endophenotype are followed for many years to determine their increased odds of developing psychopathology. Yoon et al. (Yoon, Malone, & Iacono, 2015) provided just such an example using reduced P300 event-related potential (ERP) amplitude, an endophenotype for externalizing disorders. This research team reported that healthy individuals assessed at age 17, who were subsequently diagnosed with a substance use disorder or antisocial personality disorder by age 29, had smaller P300 ERP amplitudes at age 17 than those who remained healthy. For each 1-microvolt decrease in age-17 P300 amplitude, the odds of an age-29 externalizing diagnosis were increased by 5%. Findings such as this point to the potential value of using endophenotypes to identify those at genetic high risk well before they become symptomatic. A longitudinal research strategy built from a sample enriched in this way makes it possible to study individuals in the prodromal period preceding disorder development. In turn, such an enriched sample provides an opportunity to identify factors that differentiate those at high risk who succumb from those at high risk who do not.
With respect to developmental continuity, it is not reasonable to expect mean level stability over the lifespan because brain growth and maturation vary by age, and thus age appropriate norms may be required. However, in longitudinal investigations, it is reasonable to expect rank order stability for an endophenotype over much of the life span. There may be age limits to stability; the P300 response of preschool-age children may not be related to psychological processes such as subjective probability in quite the same way as it does in subsequent developmental periods (Polich, Ladish, & Burns, 1990). The sensitivity of an endophenotype to changing developmental influences and the age at which it can be advantageously employed to assess genetic liability represent important aspects of construct validation. Developmental considerations aside, an endophenotype should show trait-like properties. For instance, it should show substantial test-retest reliability over short periods of time (W. G. Iacono & Lykken, 1981), and be present when clinical symptoms are inactive or during disorder remission (W. G. Iacono, Peloquin, Lumry, Valentine, & Tuason, 1982; e.g., W. G. Iacono, Tuason, & Johnson, 1981). An ideal endophenotype would be immune to the consequences of treatment; otherwise, it would not be possible to identify whether the genetic liability is present in treated individuals. However, if the hope that endophenotypes contribute to understanding etiologic mechanisms is realized and treatments that target the mechanism are developed, it is possible that treatment success could be indexed through the modification of the neurobehavioral trait.
Many if not most putative endophenotypes have been identified through research that is atheoretical and empirically driven. This is not surprising given that we are searching for a biomarker that taps into genetic risk at a time when we have scant knowledge of genetic or biological mechanisms associated with psychopathology. As a consequence, articles listing requirements for endophenotype identification emphasize empirical criteria, not how to generate hypotheses regarding how best to specify their operational definition given existing theory because our theories are not refined enough to offer such precise specification (Vaidyanathan, Vrieze, & Iacono, 2015). Nevertheless, endophenotypes offer strong clues as to the pathophysiology of disorder; the utility of an endophenotype ultimately will derive from the insight it provides into underlying mechanisms. Once a candidate endophenotype has been shown to have demonstrable construct validity, the nomological net should be expanded to include the results of theory building research designed to identify the biological processes involved.
Although this will necessarily include studies with humans, modeling similar phenomena in animals provides a potentially advantageous complement to investigations that attempt to model complex human behaviors like symptoms of disorder (e.g., anxiety, drug self-administration) and offers an avenue for studying processes related to clinical phenotypes that are without obvious animal analogs (e.g., psychosis). In addition to the fact that animal studies offer control over confounds that complicate human research, and thus may simplify the task of identifying relevant brain systems, they also facilitate study of gene function related to these systems (e.g., Rosen et al., 2015). Animal research that yields a gene finding for an analog endophenotype that involves a genetic homolog in humans has obvious merit: It can be used to generate a human candidate gene study that adopts a hypothesis backed by high biological plausibility. However, there are many candidate gene studies of clinical phenotypes in humans backed by this type of biological plausibility, and these studies have not yielded verified genetic variants. Thus, it remains to be seen whether endophenotypes would fare any better when following this approach.
If the promise of endophenotype research for gene finding is to be realized, the endophenotype must simplify the task in a way that a clinical phenotype cannot. Because endophenotypes are derived from laboratory measures that often require expensive equipment, time consuming procedures, and/or specialized technical expertise, obtaining the types of large samples now common to molecular genetic research of complex traits is difficult and expensive. Although neither the endophenotype nor its genetic basis needs to be simpler than the associated clinical phenotype, the task of identifying genetic variants has to be easier in some way for endophenotypes to be useful. For example, genetic variants associated with the endophenotype should have larger effect sizes than those associated with the corresponding clinical phenotype, making it possible to detect them using a relatively small sample. As desirable as this is, there is at present no compelling evidence that an endophenotype can achieve this objective. Nevertheless, we have retained this utility criterion because if an endophenotype is shown to have this property, it would be of utmost significance to the value of collecting expensive, labor intensive endophenotype data and to providing etiologically relevant insights into likely underlying mechanisms.
5.0 Lessons Learned from the Minnesota Twin Family Study Investigation of 17 Candidate Endophenotypes
5.1 Project overview
Launched in the 1990s and still ongoing, the Minnesota Twin Family Study (MTFS) is a longitudinal investigation of twin children and their parents (W. G. Iacono, 1998; W. G. Iacono & McGue, 2002; W. G. Iacono, McGue, & Krueger, 2006). Eligible families were identified using publicly available birth certificates indicating that they had twin children born in the state of Minnesota. All families with same-sex twins meeting basic inclusion criteria were considered eligible. Over 80% of eligible families participated, yielding a sample that was broadly representative of Minnesota families with children living at home based on the US Census of 2000. Parents and their offspring have been evaluated using psychophysiological paradigms selected for their potential to yield endophenotypes for substance use disorders, antisocial personality disorder, schizophrenia, and mood disorders (W. G. Iacono, 1998). Using data from 4905 of these participants, constituting the largest sample ever used for this purpose, we carried out in parallel a series of seven investigations designed to identify molecular genetic variants associated with 17 psychophysiological variables involving EEG frequency-based measures, P300 oddball visual event-related potential, antisaccade eye tracking, startle eye blink, and electrodermal activity (W. G. Iacono et al., 2014b; Malone et al., 2014; Vaidyanathan, Isen, et al., 2014; Vaidyanathan, Malone, Donnelly, et al., 2014b; Vaidyanathan, Malone, Miller, et al., 2014; Vrieze et al., 2014a, 2014b). The measures chosen represent a broad range of basic and complex psychological processes that tap into central and autonomic nervous system arousal, startle, orienting, habituation, emotion, cognition, and prepotent response inhibition.
In addition to the large sample size, our project had a number of strengths. We used an unscreened, epidemiological sample, meaning that the results were not conditional on inclusion/exclusion criteria, hence making the sample suitable for investigating all the endophenotypes. We adopted the same set of a priori analyses for all 17 endophenotypes and published the entire set of findings simultaneously. Our hope was to eliminate effects attributable to selective reporting of results, post hoc analysis leading to irreproducible findings, the need to report positive findings to justify publication, and piecemeal publication that would make it difficult to understand how the results varied from one endophenotype to another. Because our sample included twin families, we were able to determine the heritability of each endophenotype measure in the exact same sample used for molecular genetic analyses. We employed discovery based analyses to examine the association between each endophenotype and a) common variants (single nucleotide polymorphisms; SNPs) throughout the genome, b) autosomal genes, c) rare exonic variants, and d) rare and common variants throughout the genome. Our analyses took advantage of improved imputation with a powerful reference panel composed of >1000 moderate-depth whole-genome-sequenced individuals from our own sample. We also targeted specific SNPs, loci, and genes for which there were prior reports indicating they were associated with the endophenotypes, psychopathology related to the endophenotypes, or relevant brain and metabolic systems. Hence, in addition to conducting genome-wide discovery-based analyses, which required correcting for 1 million tests, we followed leads from the literature and tested “hypothesized” subsets of variants within candidate loci and corrected only for those, thus lessening substantially the p-value threshold required for a finding to be considered significant.
5.2 Key results
The main findings are highlighted in Table 2. In the end, across all analyses, we discovered only a handful of significant associations, none of which survived correction for multiple testing across the 17 endophenotypes, and each of which requires replication. Novel variant discovery in this sample was far from a resounding success and ready replication is not available for the hits we did have. Table 2 lists the largest GWAS effect sizes associated with any of the over 500,000 examined variants, only one of which was significant. To achieve 80% power to detect a SNP with the median effect size of .58 reported in the Table, a sample size of 6,808 would be required. If we accumulated a sample this big, it would still not be large enough to detect the largest SNP for half the endophenotypes listed in Table 2, and for the other half, it would detect very few variants, thus contributing little insight into the molecular genetic basis of individual differences in any of these measures.
Table 2.
Summary of biometric, GCTA, GWAS and rare variant results for 17 endophenotypes examined in the December 2014 issue of Psychophysiology,
| Heritability | Largest Effect | No. of Significant SNPs | |||||
|---|---|---|---|---|---|---|---|
| Endophenotype | Twin | Family | SNP | r2 | MAF | GWAS | Exome/Sequencing |
| P300 Measures (N=4,211) | |||||||
| P300 Amplitude | .497 | .602 | .290 | 0.51 | 0.49 | 0 | 0 |
| P3 Genetic Factor | 1.000 | 1.000 | .274 | 0.64 | 0.48 | 0 | 0 |
| Resting EEG (N=4,026) | |||||||
| Alpha peak frequency | .836 | .826 | .484 | 0.65 | 0.21 | 0 | 0 |
| Alpha power O1O2 | .772 | .781 | .450 | 0.63 | 0.20 | 0 | 0 |
| Alpha power Cz | .799 | .838 | .220 | 0.57 | 0.42 | 0 | 0 |
| Beta power Cz | .853 | .848 | .190 | 0.53 | 0.17 | 0 | 0 |
| Theta power Cz | .733 | .690 | .042 | 0.55 | 0.46 | 0 | 1 |
| Delta power Cz | .558 | .488 | .145 | 0.55 | 0.26 | 0 | 0 |
| Total power Cz | .782 | .757 | .069 | 0.50 | 0.11 | 0 | 0 |
| Skin Conductance (N=4,424) | |||||||
| SC Level | .656 | .627 | .232 | 0.58 | 0.29 | 0 | 0 |
| SCR Amplitude | .468 | .427 | .252 | 0.64 | 0.07 | 0 | 0 |
| SCR Frequency | .526 | .473 | .336 | 0.51 | 0.29 | 0 | 0 |
| SCR Factor | .578 | .520 | .349 | 0.47 | 0.18 | 0 | 0 |
| Startle and startle modulation (N=3,323) | |||||||
| Startle magnitude | .367 | .518 | .593 | 0.62 | 0.41 | 0 | 0 |
| Aversive–neutral | .000 | .109 | .000 | 0.86 | 0.06 | 0 | 0 |
| Pleasant–neutral | .014 | .052 | .000 | 0.75 | 0.21 | 0 | 0 |
| Antisaccade error (N=4,469) | |||||||
| Percent error | .510 | .489 | .468 | 0.65 | 0.24 | 1 | 0 |
| Median | .578 | .602 | .252 | 0.58 | 0.24 | 0 | 0 |
Note: SC is skin conductance, SCR skin conductance response. O1O2 represents an average over the O1 and O2 electrodes. Aversive–neutral and Pleasant–neutral are startle modulation (difference) scores. A SNP was considered significant if, for the identified endophenotype, the p-value was less than 5 × 10−8. Heritability estimates from different models are provided. Additive genetic variance was estimated from ACE models including only MZ and DZ twins as well as four-member families (mother, father, two twins). SNP heritability is the proportion of variance accounted for by all genotyped SNPs and thus in LD with them, as estimated by GCTA. Tabled numbers provide the median estimate across different methods and thresholds of genetic relatedness. The largest effects are given as percentages of variance accounted for (r2), and the MAF for each SNP is provided in the column, “MAF.” The SNP associated with the largest effect for antisaccade percent error was imputed, which means that an allele dosage was used in analyses in place of an allele count and the MAF is not available. We used the called SNP with the largest effect; the r2 for the imputed SNP was .67. GWAS gives the number of significant SNPs from genome-wide association scans, and Exome/sequencing provides the number of significant SNP associations from a whole-genome scan of nonsynonymous exonic variants or whole-genome sequencing analyses.
P300 amplitude provides an illuminating example. Since its discovery by Sutton and colleagues in the 1960s (Sutton, Braren, Zubin, & John, 1965), the P300 component has undoubtedly been one of the most widely studied ERP components. The initial report more than 30 years ago by Begleiter and colleagues that P300 amplitude was reduced in alcohol-naïve boys at risk for alcoholism (Begleiter, Porjesz, Bihari, & Kissin, 1984) has motivated a large literature exploring the notion that P300 amplitude reduction is associated with alcoholism and other forms of disinhibitory psychopathology (i.e., childhood disruptive disorders, antisociality, substance use disorders, and related traits like impulsivity, aggression, poor decision making, etc.). This body of work has produced many empirical reports and several meta-analyses, which in aggregate suggest that P300 amplitude reduction is a robust candidate endophenotype for disinhibitory psychopathology and behavior. Yet we failed to find a single variant associated with it. We also failed to confirm any associations reported in previous candidate gene studies (Vaidyanathan, Malone, Miller, et al., 2014). Thus, although P300 is arguably one of the best validated endophenotypes identified to date (Miller & Rockstroh, 2013), it did not lead to robust genetic discovery.
It is tempting to conclude that refining an endophenotype may help to identify specific variants. The P300 clearly represents activity from different neural sources that is partially overlapping in time, which is projected to the surface of the scalp. Its amplitude is determined to a significant degree by activity in specific frequency ranges, especially delta and theta (Karakas, Erzengin, & Basar, 2000; Kolev, Demiralp, Yordanova, Ademoglu, & Isoglu-Alkaç, 1997). One might think that time-frequency representations of P300-related activity yield candidate endophenotypes that are more fundamental in some way than the P300 and thus more sensitive to genetic effects (i.e., associated with larger effects). We therefore conducted a follow-up investigation in the same sample to examine power and inter-trial phase locking, a measure of consistency of the brain response across trials, of delta and theta activity in the P300 window (Malone, McGue, & Iacono, 2016). Although we obtained one genome-wide significant association, it has no obvious connection with brain activity, and unless replicated, it cannot be considered meaningful. Thus, decomposing P300 into simpler time-frequency components did not produce a fundamentally different result, and in general we suspect that refining endophenotypes or searching for supposedly simpler endophenotypes will not be a silver bullet for finding genes.
Our hypothesis-driven effort to follow-up candidate loci/variants reported previously in the literature was equally unsuccessful. We were unable to confirm any single-variant associations reported in the literature for any of the 17 endophenotypes, with gene-based tests faring only slightly better. Because we were unable to robustly corroborate any previous findings, additional fine-mapping efforts of these candidate loci and other investigations were deemed inappropriate at that time.
A couple of emblematic examples illustrate the challenges faced. Hodgkinson et al. (2010) performed a genome wide association study (GWAS) on alpha, beta, and theta power in a Native American cohort of 322 individuals and found that several SNPs in SGIP1 accounted for 8.8% of the variance in theta power. This association was replicated in a European American sample of 185, with one of the SNPs accounting for 3.5% of the variance in theta (although this finding did not survive correction for multiple testing). Effect sizes this large are exactly what we should hope endophenotypes to yield if they are to prove their value. Our study (Vrieze et al., 2014b), with almost ten times the sample size, was definitively powered to detect an effect this large. However, we obtained a nonsignificant p-value of .199 for the association between this gene and theta power using a gene-based test and we did not replicate findings at the level of individual SNPs within the gene. Greenwood et al. (2013) carried out a linkage study for antisaccade error in approximately 1000 individuals drawn from families with a schizophrenia proband. They reported linkage to a locus on chromosome 3p14. We examined a 10 Mb region around this locus, testing 39,000 markers for association with antisaccade error, and found no significant results or suggestive evidence that a genetic signal was present (Vaidyanathan, Isen, et al., 2014). One could generate a list of plausible reasons why we could not confirm these Hodgkinson and Greenwood findings in our MTFS sample. However, that would be missing the point. These failures to support previously reported findings represent only two of the many hundreds of leads we pursued in our project, none of which could be confirmed.
These results in aggregate cannot be easily attributed to measurement problems or a lack of heritable variance in this sample. Except for affectively modulated startle, which was neither similar in MZ twins nor appreciably heritable, MZ twin correlations were large for each of the 17 endophenotypes in our special issue, ranging from .53 to .86 (median .66). Table 2 provides the heritability estimates from twin- and family-based models for all endophenotypes. Excluding startle modulation, these ranged from .43 to .85 (median .59). Using GCTA for GREML analysis (Yang et al., 2011), we could show that the endophenotypes were associated with the combined effect of all genotyped SNPs from our GWAS chip, with “SNP heritabilities” (excluding affectively modulated startle) ranging from .04 to .59, median .25 (see Table 2). Hence it was not the case that we arrived at our results because we had no reliable genetic signal to detect.
As Table 2 shows, the degree to which our candidate endophenotypes were heritable had little discernible consequence. Their heritability ranged from virtually 0 to greater than .80, but the results did not vary appreciably. One endophenotype consisted of a P300 factor score that, because it captured the genetic covariance across multiple electrodes, had a heritability of 1.00. Despite this refinement to optimize capture of genetic variance, the genetic factor afforded no apparent advantage over other measures. Of note, even if genetic influences on a trait account for only a small fraction of trait variance, there is no reason why an adequately powered GWAS would not identify variants associated with what genetic effect exists. The crucial issue seems to be the genetic “architecture” of these electrophysiological endophenotypes. Our findings are consistent with a conclusion that they represent polygenic traits influenced by many genetic variants.
6.0 How Do Electrophysiological Endophenotypes Compare with Other Quantitative Traits?
A prevailing sentiment in the endophenotype literature is that psychiatric endophenotypes are somehow different from clinical phenotypes and other complex traits. Endophenotypes are promoted precisely because they are expected to be more proximal to gene action and thus more heavily influenced by particular genetic variants. That is, genetic effects are expected to be larger, much larger even, than the effects of more distal phenotypes. Although this may well prove to be true, the empirical literature available to date suggests otherwise.
6.1 Are endophenotype effect sizes larger than those of other phenotypes?
What constitutes a powerful study in genetic research is different than in many fields. For example, in the behavioral sciences a correlation of 0.1 is considered small, 0.3 medium, and 0.5 large (Cohen, 1988). On the r2 metric (variance accounted for) these correspond to r2 of 1%, 9%, and 25%. Consider by way of contrast SNPs located in the first intron of the FTO gene, which are well known to have effects on the common complex trait of body mass index (BMI) (Locke et al., 2015). This locus was the first associated with obesity through GWAS, and a PubMed search for “FTO and obesity” revealed 868 publications at the time of this writing. The effect size of the most strongly associated variants within this locus is 0.34% on the r2 metric in Europeans and even smaller in other ancestry groups (Loos & Yeo, 2014). The FTO variant effect is large by the standards of complex disease/trait genetic association standards but is tiny, ignorable even, by behavioral science standards. BMI is not unique in this respect. The average odds ratio of variants associated with Type II diabetes from a recent publication was 1.11 (Morris et al., 2012). The average effect of the nearly 700 GWAS-associated variants with height, one of the most accurately measured and highly heritable of all human quantitative traits, is around r2=0.03% (Wood et al., 2014). The SNP rs16969968 in CHRNA5 accounts for 0.5% of the variance in cigarettes per day (Furberg et al., 2010), one of the largest effects discovered between a common variant and a complex behavioral or psychiatric phenotype. Not only is r2=0.5% a very large effect, it is about the largest effect one now expects to find in a genetic association study of complex traits and common variants. Detecting effects of these sizes requires massively powered studies on a scale that until recently was not achieved in biomedical or behavioral science.
The effects described in the previous paragraph are for associations between genetic variants and genetically distal phenotypes like BMI, height, cigarettes per day, or type II diabetes. Our own work described in the previous section, using a discovery sample of 4,900 individuals, provides little reason to expect electrophysiological endophenotypes to be any different. We found only one significant GWAS hit, yet to be replicated, for any of the 17 endophenotypes investigated. The effect size of the most significant common variant we discovered (rs1868457) accounted for 0.67% of the variance in antisaccade eye tracking errors (p=3.3×10-9, Vaidyanathan, Malone, Donnelly, et al., 2014a), and that value is undoubtedly inflated due to “winner's curse” (Ioannidis, 2008). Replication attempts will only result in attenuated effect sizes for this variant. Brain structural measures, which arguably are more proximal to gene effects than electrophysiological measures recorded from the body surface, fare no better. The ENIGMA consortium (Thompson et al., 2014) combined GWAS data across multiple samples and conducted a meta-analysis of hippocampal and intracranial volume from structural MRI in a discovery sample of 7,795 individuals and multiple replication samples altogether totaling 21,151 (Stein et al., 2012). They found two genome-wide significant loci. The top hit explained 0.27% of the variance in hippocampal volume, slightly less than the 0.34% of the variance in BMI accounted for by the top hit in FTO described above. Perhaps it is unsurprising that genetic effects on the size of brain regions are similar in magnitude to genetic effects on the size of the entire body. Finally, perhaps the most widely studied electrophysiological endophenotype of relevance to psychopathology is resting heart rate. This endophenotype has been studied in 38,991 individuals, finding nine associated variants in six loci, accounting for between .083-.167% of variance in resting heart rate (Eijgelsheim et al., 2010), again less than the top hit in FTO for BMI.
To further illustrate the differences in effect sizes between endophenotype-like measures and regular phenotypes, we reviewed the complex trait GWAS meta-analysis studies described above for total cholesterol (Teslovich et al., 2010); BMI (Locke et al., 2015); height (Wood et al., 2014); brain volumes (Stein et al., 2012); resting heart rate (Eijgelsheim et al., 2010); and antisaccade error (Vaidyanathan, Isen, et al., 2014) plus GWAS results for two metabolites (Kottgen et al., 2013; Ware et al., 2016); bone mineral density (Estrada et al., 2012); diabetes (Scott et al., 2012); depressive symptoms , subjective well-being, and neuroticism (Okbay Baselmans, et al., 2016); and education level (Okbay Beauchamp, et al., 2016). We then plotted effect sizes of GWAS-significant variants from these studies in Figure 1 on the r2 metric (variance accounted for in the phenotype). For quantitative trait studies that did not directly report variance accounted for but did report standardized effects, we computed an approximation using the formula: r2 = 2β2(1 – MAF)MAF, where β is a standardized effect size estimate of the variant when the residual variance is ~1, and MAF is the minor allele frequency for the variant. Unstandardized effects were converted to r2 by converting the p-value to its implied t-distribution value, and then converting that with the formula r2 = t2/(t2 + df), where df was set equal to the sample size reported for each such genetic variant. While an exhaustive study of quantitative trait genetic architecture is infeasible here, we attempted to select broadly from the domain of quantitative phenotypes, ranging from heritable medical biomarkers (metabolites, cholesterol levels, bone mineral density), to endophenotypes (antisaccade eye movements, resting heart rate, intracranial volume, and hippocampal volume), to physical phenotypes (height, BMI), to psychological phenotypes (personality, education). Most of these studies have large enough samples (median sample size N~78,000; see Figure 1) to have detected the largest genetic effects, and are thus likely to provide only slightly overestimated effect sizes of these variants. The lone exception is the single antisaccade hit from our work, which we expect to be significantly overestimated. We did not include case-control results (e.g., schizophrenia, macular degeneration, etc.) because effects for binary phenotypes are estimated on different scales such as odds ratios and are not easily directly compared to quantitative effect metrics such as r-squared. As electrophysiological endophenotypes are frequently quantitative, we believed comparisons with other quantitative phenotypes was most appropriate.
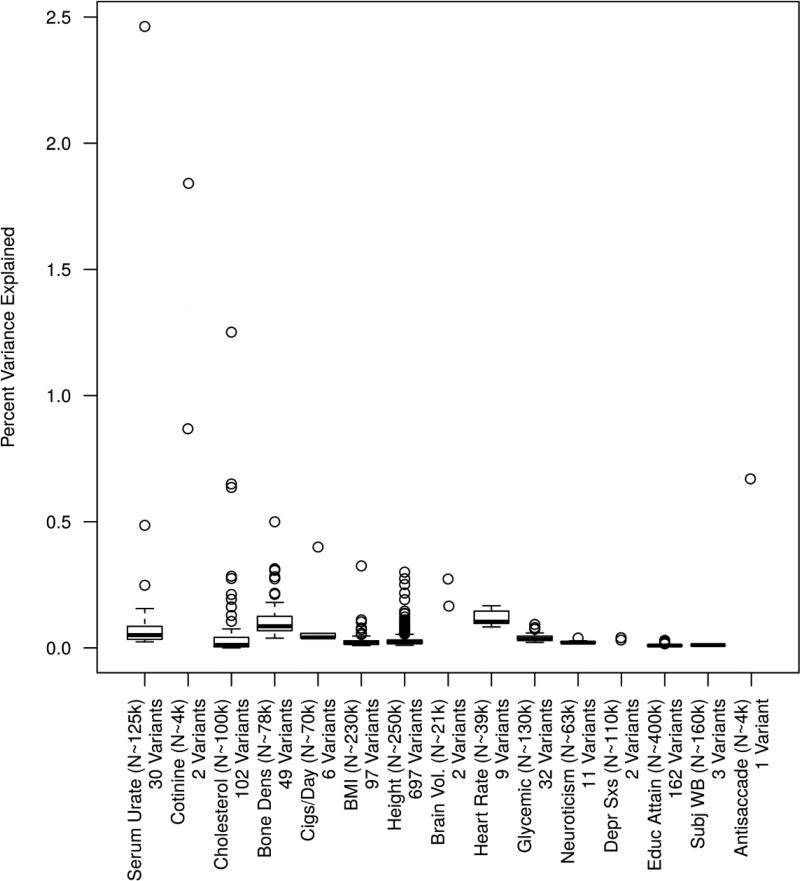
Figure 1. GWAS-significant Effect Sizes for Phenotypes, Endophenotypes, and Biomarkers.
Plotted here are GWAS-significant loci from large-scale GWAS meta-analyses of serum urate, cotinine levels (a nicotine metabolite) in smokers, total cholesterol, bone mineral density, cigarettes per day, BMI, height, brain anatomy volumes from structural MRI, resting heart rate, glycemic traits, neuroticism, depressive symptoms, subjective wellbeing, months of educational attainment, and antisaccade eye movements. Phenotypes are ordered by the maximum reported effect size except for Antisaccade, which was based on a single study and is undoubtedly an overestimate. The effect sizes for each trait illustrate the effect size distribution differences between the more “biological” measures such as cholesterol levels, brain volumes, and antisaccade eye movements, and genetically distal phenotypes such as BMI and height. Except for the three blood-derived phenotypes serum urate, cotinine and total cholesterol, all variants account for less than 1% of the variance in the corresponding trait.
From this snapshot of the GWAS literature, the distribution of effects for the more “biological” blood-derived phenotypes are not of materially greater magnitude as those observed for highly complex and distal phenotypes, including height and BMI. A few trends stand out. First, whereas some of the biomarkers tend to have much larger maximum effect sizes (~2% for serum urate and cotinine, 1.2% for cholesterol, 0.5% for bone marrow density), the largest effect sizes for the brain-based and heart rate endophenotypes are actually smaller than those observed for height and BMI. Second, the largest effect sizes observed for education levels and neuroticism were an order of magnitude smaller than the largest effect sizes for the brain-based and heart rate endophenotypes. Thus, endophenotypes may be associated with slightly larger effect sizes than distal psychological traits (like neuroticism), but these effects are still so small that massive sample sizes are still required to detect them (see discussion of power in section 7.3 below). Third, the most direct comparison available in Figure 1 is that between cigarettes per day and cotinine, the primary metabolite of nicotine and long a biomarker for nicotine consumption and addiction (Benowitz, 1996). The maximum effect sizes for both traits is from the same variant, rs16969968. The variant accounts for ~0.4% of the variation in cigarettes per day but 1.8% in cotinine, clearly a larger effect on the biomarker than the behavioral phenotype. Cotinine is not an endophenotype, however, and to our knowledge stands alone as a successful biomarker for a psychiatric condition, in this case nicotine addiction, largely thanks to our advanced understanding of nicotine pharmacokinetics. Fourth, the largest effect sizes for the blood-derived biomarkers listed here are 2-3x larger than those for physical traits, an order of magnitude larger than the brain volume measures, and many times larger than those for more psychological phenotypes. However, this applies only to the 1-2 largest effects. The vast majority of observed effects for all traits, whether blood-derived biomarker or years of education, is well below r2=0.1%.
6.2 Is the “genetic architecture” of endophenotypes different from that of other phenotypes?
The GREML approach alluded to earlier is very different from GWAS, in that it estimates the aggregate effect of all measured variants (and those in linkage disequilibrium with them) rather than estimating the effect of each individual variant in turn. Although GREML cannot therefore identify the specific variants influencing a trait, it can nevertheless establish the overall magnitude of genetic variance in the trait due to the tagging SNPs on GWAS arrays, which are predominantly common variants (those with MAFs of at least 1%). The initial study using this approach examined the SNP heritability of height, one of the most heritable human traits based on twin and family studies, with heritability estimates from those studies converging on ~80% (Yang et al., 2010). GREML analysis indicated that the SNP heritability of height was 45%. Although well short of the total heritability of 80% from twin and family studies, this was also much greater than the amount of variance in height accounted for by GWAS of tens of thousands of subjects, which at the time of this 2010 publication had identified approximately 50 loci accounting for about 5% of the variance in height in total. The difference between the variance accounted for by individual variants (5% in the case of height) and phenotypic heritability is called “missing heritability” (Manolio et al., 2009). The GREML analysis of height indicated that a substantial fraction of the total heritability in height was in fact captured by some unknown but presumably very large number of genotyped SNPs, indicating that much of the variance in height is therefore due to common SNPs.
We can use GREML-derived estimates of SNP heritability to ask whether common variants are likely to account for a greater proportion of variance in endophenotypes than clinical phenotypes or other complex phenotypes, which might in turn increase the likelihood of gene discovery. This might be the case if endophenotypes, by virtue of reflecting relatively fundamental psychological and neurobiological processes, are influenced primarily by common variants but psychiatric disorders are disproportionately influenced by mutations and rare variants (Singh et al., 2016). To address this, we draw again on results from our recent special issue of Psychophysiology that are provided in Table 2. Excluding startle modulation scores, which showed little evidence of being heritable (95% confidence intervals around our family-based heritabilities included 0), the ratio of SNP heritability to family-based heritability ranged widely, with a median of 0.54. This is almost exactly equal to the SNP heritability to biometric heritability ratio for height, which is 0.56 if we assume a heritability estimate of 0.8 for height. What these estimates rather convincingly demonstrate is that endophenotypes are far from immune to the missing heritability problem, and they do not differ from other phenotypes with respect to their overall genetic architecture.
6.3 Summary: Endophenotype genetic effect sizes and architecture resemble that seen for other complex traits
In summary, endophenotypes are like other complex traits; they are unlikely to be associated with variants that have large effect sizes, which is consistent with the findings from our special issue. Table 2 indicates that the maximum effect size we obtained for each endophenotype was small, but roughly in the range of what one might expect a large effect size to be for a biomedical or psychological trait. Yet we were underpowered to detect any as significant but one. Nor was it the case that the genetic signal was “hiding” in rare variants; our rare variant analyses yielded no more compelling result. In addition, GREML SNP heritability estimates indicate that the aggregate influence of all variants on most endophenotypes is substantial, and little different from what we see in other traits like height. This combination of findings highlights the fact that endophenotypes are massively polygenic, with many thousands of variants each contributing very small effects. Although our results could be unique to our set of endophenotypes and sample, the data available for other heritable quantitative traits, such as those listed in Figure 1, provide little reason to believe this to be the case. We are persuaded that these results in aggregate shift the burden of proof to those who would argue otherwise. Endophenotypes may ultimately confer some advantages in finding genes related to psychological outcomes, but those advantages are not likely to be great and are outweighed by the high relative cost of an electrophysiological lab session versus, for example, a questionnaire or interview.
7.0 Recommendations to Advance Endophenotype Genetics
If we resign ourselves to the idea that the effect sizes for popular endophenotypes are on average negligibly larger than for distal phenotypes, then an important question is how can we proceed to conduct the strongest studies possible, increasing our ability to understand the genetic bases of endophenotypes and associated clinical phenomena? Our most direct recommendations are listed in this section.
7.1 Candidate genes are poor candidates for genetic discovery
Selecting “candidate” genes has long been practiced in behavioral and psychiatric genetic research. Candidate gene often really means candidate variant, where a particular genotype within a gene is selected typically on the basis of functional studies in model organisms. One conclusion from GWAS has now been clear for some time – the expected effect size for such popular polymorphisms are no larger than effect sizes identified by GWAS. Even if a candidate polymorphism is truly associated with a phenotype, it will be at levels difficult to detect without a highly powered design. A recent intriguing test of the candidate gene design is reported in a paper by Farrell et al. (2015) in which the authors cross-referenced 108 schizophrenia-significant loci discovered by the Psychiatric Genomics Consortium (PGC) (Kamarajan & Porjesz, 2015; Schizophrenia Working Group of the Psychiatric Genomics, 2014) with a list of the 25 most popular schizophrenia candidate variants including COMT, DISC1, DTNBP1 and NRG1. A liberal interpretation of their findings is that four of the 25 candidate variants they investigated (16%) showed some evidence for association in the Psychiatric Genomics Consortium. In the end, only one of the 25 variants was genome-wide significant. When they expanded their attempt to validate any of the traditional candidate genes to include any variant within the candidate gene of interest, again they found some evidence for association in 4 of 25 genes (16%). The test reported in Farrell et al. makes clear that the traditional approach to candidate gene studies is highly fallible, and the traditional candidate variants within these candidate genes in psychiatry are poor places to search for genetic influences on psychiatric disorder and behavioral traits. Of further note is that Farrell et al. evaluated only the “best” available candidate genes. If one evaluates all of the SCGene database (www.sczgene.org), over 1700 studies have reported over 1000 genes to be associated with schizophrenia, but in this PGC investigation of 150,000 people, the largest and best powered study ever undertaken of schizophrenia genetics, the vast majority of these gene findings could not be confirmed. Even if a candidate polymorphism is truly associated with a phenotype, it will be at levels difficult to detect without a design powered adequately for finding what are likely to be very small effects.
The prevailing approach to candidate genes creates far more opportunities to produce untrustworthy findings than results that are likely to lead to genuine advances in scientific understanding. This is partly due to insufficient power, which creates the opportunity for false positives due to sampling variation. We return to the issue of power below, where we also provide a recommendation concerning defensible sample sizes. However, there is an additional difficulty with candidate gene studies. An appeal of candidate gene studies is that they are hypothesis-driven. We argue that this is something of an illusion given our current state of knowledge; in reality they are more exploratory than not. It is tempting to think that a particular candidate gene will somehow be different. Yet the history of molecular genetic research provides little evidence to suggest that this is true. Candidate genes are also often justified on the basis of initial findings that fail to hold up to the harsh light of replication. Imagine this hypothetical scenario: A researcher is interested in endophenotype X as an endophenotype for ADHD. Assume that two recent candidate gene studies have indicated that the COMT Val/Met polymorphism, which is thought to play an important role regulating dopamine availability in the prefrontal cortex (PFC), is related to ADHD. Our eager researcher conducts a candidate gene study of COMT in relation to endophenotype X, which turns out to yield a significant result: levels of endophenotype X are linearly related to the number of Val (Met) alleles. Excited about this result, s/he decides to conduct studies of other dopamine genes, some of which pan out whereas others do not. In the meantime, more studies examining the association between COMT and ADHD appear in the literature, many with negative results, and then a meta-analysis reports that there is no relationship between COMT and ADHD after all. Even worse, subsequent research indicates that the relationship between COMT and dopamine availability in PFC is weak. Now what does our researcher do? She has an interesting endophenotype finding based on a hypothesis that is no longer supported, and quite possibly several more that will suffer the same fate. Should this finding nevertheless be considered real? Is the field advanced by having published findings such as these?
We believe that the answer to both questions is “no”, or at least that it is incumbent upon our researcher and others pursuing this type of research to demonstrate otherwise. Although this particular example is hypothetical, this type of situation does in fact occur, which illustrates a difficulty that candidate gene studies can create even when they yield positive findings because the base rate expectation that those findings are actually false is extremely high. We argue in the next section that GWASs are clearly preferable to candidate gene studies, and we advocate a different approach to selecting candidate variants in section 10.3. We propose here that researchers should attempt to replicate a candidate gene-endophenotype association in an independent sample within the same report, reporting the meta-analytic effect size and p-value corrected for multiple testing as appropriate. That way, both positive and negative findings are more credible than they would be if based on a single sample.
Although we argue that the traditional approach to candidate gene studies is flawed, this does not necessarily mean that none of the traditional candidate genes will turn out to be robustly associated with psychophysiological endophenotypes. We are agnostic on this point, albeit somewhat skeptical. Amplitude of the event-related negativity (ERN) is associated with both internalizing and externalizing disorders (with different directions of effect). Several studies have demonstrated associations with candidate genes (summarized in Manoach & Agam, 2013). Nevertheless, as with P300 amplitude, another widely studied endophenotype, there have not been robust, validated findings. Models of ERN amplitude based in reinforcement learning hold that the ERN reflects phasic dopamine activity in response to an unexpected event or outcome (Nieuwenhuis, Holroyd, Mol, & Coles, 2004; Walsh & Anderson, 2012). This, of course, suggests that dopamine genes may influence ERN amplitude. To date, there has only been one replicated finding. Two studies have reported associations between ERN amplitude and a polymorphism of the DRD4 receptor gene (Agam et al., 2014; Kramer et al., 2007). However, whereas this SNP accounted for approximately 13% of the variance in ERN amplitude in the initial study of Kramer et al., it only accounted for about 3% of variance in the subsequent study of Agam and colleagues (the “winner's curse”; Ioannidis, 2008), and this latter effect was statistically significant only because a one-tailed test was used. In light of Figure 1, 3% is almost certainly a gross overestimate of the true effect. Moreover, subjects in Kramer et al. were selected from a larger group based on their genotypes (they were required to be homozygous for both the DRD4 receptor and COMT genes), creating a problem of interpretation, and the DRD4 gene was unrelated to BOLD activation of the dorsal anterior cingulate cortex (dACC) in Agam et al. In reinforcement learning accounts of the ERN, prediction error in the dACC is directly related to ERN amplitude (Holroyd & Coles, 2008). Failure to find an association between DRD4 and dACC activity throws the finding of association between DRD4 and ERN into doubt. Of course, it may turn out that the DRD4 receptor gene or another of the commonly studied candidate genes influences ERN amplitude. However, the history of candidate gene research leads us to be cautious. In addition, early attempts to identify specific molecular-genetic influences on substance abuse also focused on dopamine genes because so many substances of abuse influence dopaminergic activity. For the most part, these efforts did not bear fruit. We should be wary of falling into the trap of thinking that simply because a phenotype seems to be related to dopaminergic activity that the traditional candidate dopamine genes are certain to contain its molecular genetic secrets. We encourage researchers interested in the ERN and other endophenotypes to be more catholic in their approach to the problem of identifying molecular genetic variants, and to look beyond the usual candidates (see section 10.3).
7.2 GWAS to discover new variants associated with endophenotypes
In the effort to discover novel variants, we believe strongly that traditional candidate gene studies are to be discouraged in favor of GWAS. Genome-wide arrays are cheap, with suitable arrays selling for less than $50 per DNA sample including DNA preparation costs. Genome-wide arrays can be used effectively with DNA extracted from saliva or blood. They provide coverage of the entire genome, often with custom content including additional coverage of small gene sets. GWAS arrays leverage the fact that genetic variants are not independent. Recombination during meiosis creates segments of DNA that are inherited as a unit, creating a correlation among the base pairs within it, a phenomenon known as “linkage disequilibrium” or LD. Present day arrays use SNPs that are selected to carefully tag the LD structure of the genome, effectively allowing one to test the vast majority of common genetic variants in the human genome. Even better, genotype imputation works well with these SNPs to provide extremely dense and accurate measurements of nearly all common SNPs in the human genome. This increases statistical power for any particular variant and also facilitates GWAS meta-analysis by making it possible to easily combine data from multiple studies that happened to be genotyped on different arrays with slightly different tag SNPs.
It is important to note that genotype imputation is far more powerful than other common forms of imputation in the behavioral sciences. Typically, imputation techniques in the behavioral sciences, like multiple imputation or full information maximum likelihood, are meant to allow more accurate estimation of sampling error and statistical effects, but in the end one has no more information about a variable or dataset than was initially present. In genotype imputation, one obtains vast amounts of additional genetic information by leveraging a reference panel of additional individuals whose genomes are extensively measured through whole genome sequencing. The largest reference panel as of this writing contains 33,000 individuals and nearly 45 million variants with minor allele count ≥ 5 (Haplotype Reference Consortium, http://haplotype-reference-consortium.org); through genotype imputation this resource provides vast additional information about the genotypes in one's own dataset. In the next few years we anticipate reference panels of well over 100,000 individuals (200,000 haplotypes). Genotype imputation takes advantage of the fact that all individuals are related, if only slightly, and share short segments of their chromosomes. Imputation algorithms, typically hidden Markov models (Howie, Fuchsberger, Stephens, Marchini, & Abecasis, 2012), take advantage of this relatedness and probabilistically match chromosomes between the array-genotyped study sample and the sequence-based reference panel. When a match is found, the genotypes from the whole-genome-sequenced reference sample are imputed into the array-genotyped study sample. Imputed variants increase power to detect effects over tag SNPs alone, increase the precision with which an associated locus can be defined, and facilitate GWAS meta-analysis across studies by ensuring that all variants are measured or imputed in all studies (Li, Willer, Ding, Scheet, & Abecasis, 2010). Imputation (and the phasing required in order to carry out imputation) is now easier than ever, thanks to web services hosted at various academic institutions including the University of Michigan (https://imputationserver.sph.umich.edu). Users can upload their quality-controlled genotype files. Software on the server implements further quality checks, phases, imputes according to the reference panel of the user's choice, and makes imputed genotypes available for download.
Genome-wide array data also allow very precise estimation of each individual's ancestry, which must be controlled for in any genetic association study (A. L. Price et al., 2006). Ancestry is often confounded with phenotypic differences. Even among southern and northern Europeans, for example, there are differences in allele frequencies for many variants. If one were to conduct a GWAS of height in a study sample composed of 50% Italians and 50% Danes without addressing the ethnic composition of the sample, one would obtain artifactually decreased p-values for a large number of SNPs. This is only because northern Europeans are taller on average than southern Europeans and anything that distinguishes the two groups – including genetic variants – will also be associated with height. The standard controls for ancestry include principal components computed on genome-wide variants, or mixed model association tests using a genetic kinship matrix to account for genetic relatedness between all individuals (H. M. Kang et al., 2010; A. L. Price et al., 2006). Unlike GWAS, candidate gene studies of a handful of SNPs do not allow such corrections, making allelic stratification a serious confound in the candidate gene approach.
After quality control, phasing, imputation, and subsequent removal of poorly imputed or rare variants, a standard GWAS simply associates each of the variants with the quantitative phenotype. This essentially comprises millions of correlations in turn, one between the phenotype and each genetic variant. The next step involves determining which of the millions of variants are associated with the phenotype at a statistically significant level. The convention in GWASs of common variants (e.g., MAF > 1%) is to use the conventional alpha of .05 and a Bonferroni correction for 1 million tests, or .05/1,000,000 = 5×10−8. Testing all common variants, but then accounting for the non-independence among them due to linkage disequilibrium, is approximately equivalent to conducting 1 million independent tests; hence the convention.
But if common complex disorders/traits are highly polygenic (Chabris, Lee, Cesarini, Benjamin, & Laibson, 2015) with many variants of small effect, why would we restrict ourselves to consider as hits only the few most stringently selected variants? Why not relax the threshold and explore “suggestive” hits as well? Surely these are enriched for true signals. This is a seductive argument and we admit some level of sympathy. However, if the purpose of conducting a GWAS is to identify variants that are associated with a disease in order to conduct extremely expensive and time-consuming functional follow-up experiments to identify and understand biological mechanisms, then it is even more important to control the family-wise error rate, or the probability that at least one hit is a Type I error, and the convention since Fisher's time has been to control this rate at 1 in 20, or .05. This is what a Bonferroni correction of 5×10−8 does. A popular alternative to a Bonferroni correction is false discovery rate correction, which typically provides less stringent control of Type I errors in favor of controlling the expected proportion of Type I errors under a null and alternative distribution. We do not recommend using the false discovery rate in GWAS. Controlling the family-wise error rate instead allows for greater certainty in our findings before expensive functional follow-up experiments are conducted.
Unless one's sample is unusually large or unusually special, conducting a genetic association study of common variants and complex disorders/traits is expected to produce entirely null results or, on the rarest of occasions, it will produce significant results that then require extensive replication. There are multiple ways to conduct replication studies, and here we may differ with others in our recommendations. Our recommended approach is not to replicate, in the traditional sense where only the significant variant is replicated in an independent sample at p<.05, but to conduct GWAS meta-analyses. In this approach summary statistics (e.g., effects and p-values) are generated in each independent sample for every imputed variant, and then those summary statistics are combined into one meta-analytic result per variant. If each study imputes 45 million variants, then there will be 45 million results from a GWAS meta-analysis of those studies.
GWAS meta-analysis has multiple benefits over standard replication, just as standard replication has multiple benefits over a single study. Instead of replicating only a handful of significant variants from the original study, meta-analysis allows one to increase sample size for all variants. This increases the odds that novel variants not significant in the original study will be discovered and, once the summary statistics for all variants are released (e.g., to dbGaP, or hosted on a website), it becomes a valuable (quasi-) public resource for other investigators interested in the same trait/disorder. Meta-analysis is an elegant solution when multiple replication datasets exist. Instead of vote-counting or other crude methods (e.g., 1 replicated, 1 did not), meta-analysis provides an estimate of the overall effect size and p-value. Meta-analysis allows for estimating many secondary statistics of interest, such as effect size heterogeneity and effect size moderators. Finally, GWAS meta-analysis in genetic association studies has led to a sea change in how genetic association studies are done. Instead of lone investigators guarding their own valuable data, scientists have begun sharing their data to accumulate the large and powerful samples required to make progress in our understanding of the genetic basis of disorder and human behavior.
Indeed, in genetic association studies it often makes sense to begin not by conducting a GWAS in a single sample and then searching for other datasets to conduct GWAS meta-analysis using many samples. Rather, whenever possible, one should begin the entire process fully expecting that a meta-analysis of multiple samples will be necessary to obtain sufficient statistical power.
7.3 Adequate power to detect individual effects is crucial but almost never attained in existing endophenotype genetic association studies
7.3.1. Power and sampling schemes in GWAS
We suggest that, once an endophenotype has been chosen for GWAS, power to detect genetic variants can be increased in two primary ways, motivated by classic work in psychology (McClelland, 2000) and illustrated very simply by the well-known formula for calculating the standard error of the slope for arbitrary predictor X (e.g., a genetic variant) in multiple linear regression:
where MSE is the mean square error, n is the number of observations (individuals), VX is the variance of X, and () is the proportion of variation in X not shared with other variables in the model. Minimizing results in a smaller standard error and tighter confidence intervals; in short, more power. Clearly, doubling sample size, for example, will reduce and increase power, which illustrates the role of sample size in determining power, but doubling the other terms in the denominator (or halving MSE) will have exactly the same effect as doubling n. Thus, doubling the variance in X will have exactly the same influence on power as doubling the sample size. When X is the genetic variant increasing its variance is achieved by increasing its minor allele frequency – through sampling individuals from the phenotypic or genetic extremes (from diverse ancestries, for example). Doubling the variance in the predictor not shared with other predictors, by choosing independent covariate sets, for example, will have the same effect. Finally, we see the importance of phenotypic measurement, as more precise measures with less error will decrease MSE and thereby increase power. Although electrophysiological endophenotypes are often measured reliably compared to, for example, fMRI measures, error can be reduced through aggregating multiple measurements (Ford, 2014).
All these ways of increasing power should be considered to the extent possible in any study but, all else being fairly equal, the importance of sample size in genetic association studies cannot be overstated. Increasing sample size instead of increasing the variance of the predictor can be especially important in large-scale genetic association studies. In particular, extreme phenotypic sampling schemes to increase the variance of an associated genetic variant X (VX) are highly restrictive if one wants to evaluate a new phenotype. The original phenotypic sampling scheme to increase VX is not likely to increase the variance of other genetic variants associated with the new phenotype unless the correlation between the original phenotype, on which the sample selection was based, and the new phenotype is large. However, if the two phenotypes are highly correlated then sophisticated statistical procedures are required to help ensure that any genetic variant associated with the new phenotype is not spurious due to its confounding with the original phenotype (Liu & Leal, 2012).
Take a simple example: a phenotypic sample selection scheme to enroll individuals who have suffered from severe and recurrent depression versus controls who have never been depressed. All individuals are assessed for depression and also a depression-related endophenotype, such as EEG alpha frontal asymmetry. Because the selection scheme will increase the variation of disorder-related genetic variants, this study design is more powerful than an unselected sample for detecting variants associated with depressed mood. Because selected individuals will be at the extreme ends of the distribution of the endophenotype measure, this scheme is also a potentially powerful design for the depression endophenotype. However, any association between variant and endophenotype could easily be spurious, resulting from the correlation and confounding of depression, on which the sample was selected, with endophenotype level. Determining whether an endophenotype-associated variant is independently associated with the endophenotype is difficult, to say the least. Indeed, any other variable, including an endophenotype, that correlates with depression in this sample will be subject to the same difficulty, rendering this depression-based sampling scheme limited for understanding anything other than depression. A common approach is to conduct genetic associations studies for a new phenotype Y within each sampling cell (e.g., analyze separately those screened for high depression and the controls), and then meta-analyze the results, but then all the original advantages of selecting based on depression are lost.
A representative community-based sample, on the other hand, allows one to investigate any number of (endo) phenotypes without this problem. If the purpose of the field then is to discover genes associated with a wide variety of endophenotypes and their associated clinical disorders, spending scarce resources on a few large community-based samples with many phenotypes can be a more efficient strategy than spending those resources on many smaller extreme sampling designs, each for a handful of phenotypes. This is especially true for endophenotypes that can be efficiently assayed (e.g., resting heart rate) in large numbers of individuals at low cost.
Power is complex and depends on a variety of factors, including those in equation 1, but also on the true effect size, which is often not known until research is conducted. We evaluated how power would change under a variety of favorable circumstances (indeed, unrealistically favorable). In particular, we calculated power for the largest effect sizes reported for selected phenotypes in Figure 1 and then recalculated power assuming the investigator implemented a highly effective extreme sampling design and highly precise measures (see Table 3). First, we calculated the sample size required to have 80% power to detect the original reported effect. As can be seen from Table 3, sample sizes that are unheard of in electrophysiological research would be required. Under realistic circumstances, such as that reported by ENIGMA for hippocampal volume (Stein et al., 2012), 15,258 total samples would be required for 80% power to detect a single genome-wide effect. Next, we estimated the sample size required if one were to maximize the variance of X (VX) or to reduce measurement error from 50% to 25%. We assume that the predictor X is a biallelic SNP, its variance is determined by the variance of the binomial distribution, or 2*MAF(1-MAF), which is maximized when MAF =.5. To reduce measurement error we simply assumed that the variance in the phenotype was produced in accord with classical test theory: var(P) = var(T) + var(E), where P is the phenotypic score, T is the true score, and E is the measurement error. We conservatively assumed that measurement error accounted for half the variance in the phenotype, and that one could then reduce measurement error by half. As Table 3 indicates, even under the most unrealistically optimistic of circumstances, when one assumes the effect is as large as the largest effect reported for the nicotine metabolite cotinine, error in measuring the phenotype is reduced by half, and MAF for the variant is maximized at 50%, 1,465 participants are still required to have 80% power to detect a single genome-wide effect. The worst-case scenario is observed for educational attainment, where N=198,302 is required to return a single genome-wide significant effect for 80% power. N=148,475 is still required after error and MAF are optimized. Discovering more than a single effect clearly will require much, much larger samples.
Table 3.
Minimum Sample Size Recommendations for Independent Samples under an Additive Model
| Sample sizes for 80% power in original studies for largest r2 in GWAS or GWAS Meta-Analyses | Relative sample sizes for 80% power to detect same Betas under alternative (and unrealistic) power-maximizing scenarios. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GWAS discovery with α=5e−8 | Association follow-up with α=.05 | |||||||||||
| GWAS Hit | Phenotype | r2 | MAF | Est. Beta | N for 80% power (α=5e−8) | N for 80% power (α=.05) | MAF=.5 (Max VX) | Error reduced by halfb | MAF & Error optimized | MAF=.5 (Max VX) | Error reduced by halfb | MAF & Error optimized |
| rs10851907 | Cotinine | 1.84% | .41 | .20 | 2,026 | 402 | 1,960 | 1,515 | 1,465 | 389 | 300 | 290 |
| rs964184 | Cholesterol | 1.25% | .13 | .24 | 3,020 | 598 | 1,355 | 2,260 | 1,011 | 269 | 448 | 200 |
| rs9533090 | Bone Mineral Density | 0.50% | .49 | .10 | 7,904 | 1,566 | 7,900 | 5,920 | 5,920 | 1,566 | 1,174 | 1,173 |
| rs10851907 | Cigarettes per Day | 0.40% | .41 | .09 | 10,086 | 1,999 | 9,758 | 7,559 | 7,313 | 1,934 | 1,498 | 1,449 |
| rs1558902 | BMI | 0.33% | .42 | .08 | 12,681 | 2,513 | 12,355 | 9,505 | 9,261 | 2,449 | 1,884 | 1,836 |
| rs724016 | Height | 0.30% | .45 | .08 | 12,481 | 2,474 | 12,355 | 9,355 | 9,261 | 2,449 | 1,854 | 1,836 |
| rs7294919 | Hippocampal Volume | 0.26% | .10 | .12 | 15,258 | 3,024 | 5,480 | 11,438 | 4,105 | 1,086 | 2,267 | 814 |
| rs9398652 | Heart Rate | 0.17% | .10 | .10 | 23,275 | 4,613 | 8,366 | 17,450 | 6,269 | 1,658 | 3,459 | 1,243 |
| rs9401593 | Educational Attainment | 0.03% | .48 | .02 | 198,302 | 39,303 | 197,985 | 148,713 | 148,475 | 39,240 | 29,475 | 26,117 |
| rs1868457 | Antisaccade | 0.67%a | .31 | .13 | 5,891 | 1,168 | 4,667 | 4,088 | 3,495 | 925 | 810 | 693 |
This table indicates the minimum sample sizes necessary to detect only the largest effects reported in the GWAS literature for each of the listed phenotypes, ordered as a function of the phenotypic effect on the r2 metric. Antisaccade is listed last, as it is based on a single sample GWAS and is undoubtedly an overestimated effect size. The sample sizes are optimistic even under the most fortunate circumstances, such as a sampling design that has greatly inflated the MAF or a measurement protocol that drastically reduces measurement error. r^2 is the coefficient of determination (i.e., variance accounted for in the phenotype), Est. Beta is the effect size estimated from the r2 and MAF and used to generate all power calculations in the table, is the standardized regression coefficient, MAF is the minor allele frequency, Error is the measurement error in the phenotype.
This value is based on a single GWAS study and is undoubtedly an overestimate of the true effect.
Assuming error is 50% of the phenotypic variance.
The reason we include power calculations under “special” circumstances in the right-hand side of Table 3 is not because we believe that it is easy or realistic to cut measurement error in half or increase variant MAF to 50%. In fact, we have argued throughout this article that the failure to find genetic variants associated with electrophysiological candidate endophenotypes is unlikely to be due to their operational definitions or the precision with which they are measured, and it is clear that phenotypic selection schemes will not result in great MAF enrichment given how small the associations between genotype and phenotype are. The purpose of Table 3, instead, is to respond directly to the argument that some particular study sample is special and therefore extremely powerful. To create a caricature of the argument: a small study has huge statistical power because it used the best and most expensive measures collected in-person over the course of days in multiple waves on a very special sample of phenotypically extreme individuals from a population that is very hard to access and has not been studied. This argument can be extended then to rebut later failed replications because the replication study was not—in some cases, could not be—designed in the same way. This argument can be reformulated as an argument about statistical power: extreme sampling of a special population increases variance of the predictor, or MAF of the variant in question; measurement accuracy, time-consuming in-person assessments, and multiple waves of assessment serve to decrease measurement error; and the “best” measures are expected to have large effect sizes associated with them. However, as we have shown, even in the most unrealistically optimistic of such scenarios, it is very difficult or even impossible to overcome the inherent limitations of small samples to discover variants with the effect sizes we now expect for complex traits including endophenotypes. GWAS of complex traits, even in a unique sample assessed in the best way imaginable, will not be powerful without thousands of individuals and, more likely, tens to hundreds of thousands of individuals. Consequently, even studies of very special samples using refined phenotypes require extremely large samples to be credible.
One can imagine exceptions to our general conclusion, but these exceptions are not observed in the current literature. For example, some rare variant may have a large effect on the phenotype, such that carrying it greatly increases one's risk for disease or score on a disease-related endophenotype. In a community representative sample, or a sample selected on the basis of phenotype, the rare variant will only affect risk in the few carriers who harbor it, and statistical power to detect the effect will remain low (because VX is necessarily low due to low minor allele frequency [MAF]). However, if one selects participants based on genotype or ancestry, such that carriers of the rare variant are oversampled, the MAF of the variant in the sample and thus statistical power can be greatly increased. This kind of study design has traditionally involved large pedigrees with high incidence of disorder. Classic examples include the first report of linkage in Huntington's disease (Gusella et al., 1983) and even the Sherrington et al paper (Sherrington et al., 1988) described at the outset of this article. Novel designs to sample rare variant carriers are now possible with the advent of large-scale biobanks (e.g., The Nord-Trøndelag Health Study HUNT Biobank, the UK Biobank, Million Veterans Program, and many others) from which individuals could be selected on the basis of their (rare) genotype and perhaps augmented with family members not already in the biobank but who are also likely to carry the rare allele of interest. This novel selection strategy, and many others that will be possible with population-level sequencing, may be leveraged to greatly increase variance in the predictor (VX) by increasing the MAF of the genetic variant, thereby increasing power for testing association. Keep in mind, however, that this entire strategy depends on knowing which rare variants are important, out of the many hundreds of millions of rare variants that exist. We have argued in this section that GWAS is one recommended way to determine which genomic regions, or loci, contain genetic variants that influence risk for disorder. Determining which variants within a locus are causal is an important step in understanding the genetic basis of the (endo) phenotype. Knowing the causal variants is essential for selection strategies based on genotype but, more importantly, for understanding the biological mechanisms by which the variant influences the (endo) phenotype.
7.3.2 Power in GREML
Like genetic association studies, GREML also requires large samples. Figure 2 provides sample size estimates for univariate SNP heritability estimates (panel A) and genetic correlations between pairs of traits (panel B), based on R code made publicly available by Jian Yang, the developer of the GCTA software, which introduced the GREML approach (Yang et al., 2010). (An online power calculator is also available at http://cnsgenomics.com/shiny/gctaPower/.) Panel A plots the power for finding that SNP heritability is significant for a range of sample sizes and a range of levels of trait SNP heritability (h2 = .20, .40, and .60). To have adequate power to find a significant SNP heritability estimate requires 1,500 subjects if heritability is high (.60), and 4,450 if heritability is relatively low (.20). In panel B of Figure 2 we show power required to estimate bivariate genetic correlations. For the sake of simplicity, power estimates for genetic correlations assume that the heritability of both traits is the same, using the same range of true heritability values as in panel A. Power is plotted for phenotypic correlations between the traits ranging from .10 to .40, and we assume that the genetic correlation between traits accounts for 80% of the phenotypic correlation. A sample of 10,000 is not nearly large enough to detect a genetic correlation of .08 (phenotypic correlation of .10), even if both traits are highly heritable (h2 = .60). If the heritability of the traits is relatively small in magnitude (h2 = .20), then nearly 10,000 subjects are required to detect the largest genetic correlation. If trait heritability is high and the phenotypic correlations are large, obviously an optimistic scenario, more than 3,000 subjects are still required. Factors such as the proportion of the phenotypic correlation accounted for by the genetic correlation and the degree to which the heritability of the two traits is similar affect power both positively and negatively, but the need for large samples is clear. When sufficiently powered, GREML analyses have the potential to provide insight into the genetic architecture of endophenotypes and the nature of their association with clinical phenotypes, but because neither individual SNPS nor genes are identified, they do not shed light on the specific nature of the genetic etiology.
Figure 2.
Power calculations for GREML analyses of SNP heritability and genetic correlations. In Panel A, power is plotted against sample size for three di_erent levels of SNP heritability (the total phenotypic variance accounted for by measured SNPs and SNPs in LD with them): h2 of 0.20 (plotted in red), 0.40 (plotted in blue), and 0.60 (plotted in green). The dashed horizontal line represents power of 80%. Dropping an imaginary vertical line to the x-axis from the point where each curve crosses this line provides an estimate of the sample size needed to have adequate power (80% power) to detect a SNP heritability of the corresponding magnitude. Panel B plots power against sample size for detecting genetic correlations, the proportion of variance shared by two phenotypes due to measured SNPs. The SNP heritability is assumed to be the same for both phenotypes, and the same three levels are used as in Panel A. Power is estimated for four di_erent phenotypic correlations, r = .10 to r = .40. The true genetic correlation is assumed to account for 80% of the phenotypic correlation. All power estimates were conducted using R code provided by Jian Yang on the GCTA software discussion board (http://gcta.freeforums.net/board/1/gctadiscussion-board).
7.4 Summary of recommendations
The above recommendations for variant discovery are simple but important; our key points are annotated in Table 4. Traditional candidate gene studies, where genes are selected a priori based on animal models or non-genetic neurobiological findings in humans, are mired in replication difficulties and have not provided clear and robust associations. Genome-wide arrays are now more affordable than individually genotyping many candidate genes and should be preferred in any new genotyping effort. Genome-wide arrays allow for standard ancestry corrections and can be imputed, which makes genome-wide meta-analysis easy. Power to detect novel associations is small because effects are small, so researchers should consider this at the outset and plan to build the largest sample possible and, in perhaps many cases, realize they will be unable to conduct an adequately powered study. Using GREML to study the genetic architecture of endophenotypes and their genetic relationship to clinical phenotypes also requires large samples. If the goal is to discover and understand the biological pathways by which a gene affects a phenotype, then a Bonferroni correction that controls family-wise error is essential to control the proliferation of low-confidence findings.
Table 4.
Recommendations for Novel Variant Discovery Efforts
| Recommendation | Rationale |
|---|---|
| Use genome-wide array | Tests entire genome. Can be imputed. Capable of easy meta-analysis. Straightforward ancestry correction. Helps avoid costly false-positives. |
| Imputation | Increases association power. Easy replication by other groups. Improves ability to finely map an association locus. Allows tests of non-SNP genetic variants. |
| Power | Sample sizes for GWAS of endophenotypes will require >10,000 samples to make robust discoveries because the effects of common variants on endophenotypes will be small, almost certainly less than r2=0.005 and probably less than r2=0.0005. This recommendation holds even for so-called “enriched” studies of phenotypic extremes or highly precise measurements. Using GREML to investigate genetic architecture and the covariance between the endophenotype and clinical phenotype requires smaller but still quite large samples. |
| Bonferroni threshold | If purpose is to identify variants for follow-up functional testing, a Bonferroni threshold of 5×10−8 will control family-wise error rate and reduce costly false positives. |
| Meta-analysis | The simplest way to increase statistical power is to join forces and data with like-minded people with similar data. |
| Replication | If outright meta-analysis of all variants is not possible, then attempt to meta-analyze top hits from your study. |
8.0 Moving from GWAS-implicated Loci to Causal Variants
Up to now we have focused primarily on recommendations for improving the discovery of new genes associated with endophenotypes, and therefore genes that are presumably important for clinical disorders. Let us assume now, contrary to available evidence, that we have been successful in discovering a variant. What next?
GWAS takes advantage of the LD structure of the genome, which reflects the fact that through recombination we inherit chromosomes from our parents that are mosaics of our grandparent's chromosomes. Recombination creates a correlation among variants within these segments. LD is both a strength and weakness of GWAS. It allows one to efficiently test all common variation within the genome with a few hundred thousand carefully selected markers, but any particular variant significantly associated with a phenotype is far from guaranteed to be a causal variant. Rather, the associated variant implicates a genomic locus, which might span millions of nucleotide bases, thousands of variants, and dozens of genes. A major hurdle lies in figuring out which of these variants are causal, and which are only spuriously associated with the phenotype through LD with causal variants.
A common technique to understand a locus is to fine map it, or genotype/sequence all variants within the implicated locus. According to the latest findings from the 1000 Genomes Project Consortium, there have been nearly 90 million genetic variants discovered in humans (1000 Genomes Project Consortium, 2015). However, any particular individual differs from the extensively curated human reference genome at only ~5 million sites. The difference between 5 and 90 million lies in the fact that most variants identified to date are rare – the allele not observed on the reference genome only exists in a tiny fraction of individuals and any particular individual only has a small number of these rare alleles distributed throughout their genome. Traditional GWAS studies use a genome-wide array that genotypes a few hundred thousand up to a few million common genetic variants. Imputation methods can now fairly accurately impute variants down to ~0.1% minor allele frequency, the frequency with which the rare allele (often but not always the non-reference allele) is observed. Observing variants with even lower frequency currently requires direct genotyping or genome sequencing techniques.
Once most or even all variants within a locus have been genotyped (e.g., through sequencing), one can systematically test these variants for association. A common approach is to conduct stepwise forward selection conditional tests to exhaustively test all variants within a region conditional on all other associated variants. This can be done precisely when raw genotypes are available or approximated through tests that model the covariance among variant effects using LD as a proxy (Yang et al., 2012).
A potentially more powerful approach to disentangle LD from truly independently associated effects is to leverage different ancestral groups in a process known as trans-ethnic fine mapping (Kichaev & Pasaniuc, 2015). Trans-ethnic fine mapping exploits the fact that different ancestral groups (e.g., East Asian versus Native American ancestry) have different LD patterns across the genome. It is well known, for example, that regions of LD in individuals of European ancestry are large relative to individuals of sub-Saharan African ancestry. If a locus identified through GWAS shows different LD patterns in individuals of different ancestries, then conducting an association analysis in both ancestries can narrow the locus, thus reducing the set of credible candidate causal variants. For this reason, electrophysiological studies of different ethnic groups, such as those carried out with Native and Mexican American communities in the US (Ehlers & Gizer, 2013; Ehlers & Phillips, 2007; Ehlers, Wills, Phillips, & Havstad, 2015; Norden-Krichmar et al., 2015), are especially valuable even if they are not expected to greatly increase power to detect novel associations, nor do they obviate the need for large samples (e.g., see Table 3).
Another approach is to test putatively functional variants within a locus for association. One expects, for example, that coding variants inducing a nonsense or missense mutation are more likely to have functional effects on gene transcription and, by the same token, larger effects on downstream phenotypes. An excellent example of the power of functional annotation was recently reported for the intronic variants in FTO associated with BMI, as discussed above. While these variants were first discovered over a decade ago, their mechanism of action was unknown until 2015. In the end, FTO is not the relevant gene affecting BMI levels. Instead, the intronic locus harboring the BMI-associated variants is an enhancer—a functional unit in the genome that influences expression of a gene—for nearby genes, IRX3 and IRX5, over 1 million bases away from FTO. The finding was originally suggested by epigenomic annotations (e.g., from ENCODE or NIH Roadmap) indicating that the intronic locus in FTO contained an enhancer for IRX3/IRX5, the phenotypic effects of which were subsequently confirmed through mouse and patient tissue samples (Claussnitzer et al., 2015). Understanding the function of associated loci can be a powerful tool to understand which variants to prioritize for later experiments, and also to elucidate biological mechanisms of action.
9.0 Selective Review of Electrophysiological Biomarkers as Candidate Endophenotypes
In this section we review the literature on electrophysiological biomarkers, with an emphasis on those that are candidate endophenotypes. We begin by addressing how well putative endophenotypes satisfy the threshold criteria in Table 1. Some have been the focus of years of research, whereas investigations into others have only recently been undertaken. However, with the exception of resting heart rate, none of the candidate endophenotypes meet the criteria in Section II of Table 1 for molecular genetic verification. Of note, they do not satisfy criterion #5, which requires demonstration of a verified association with a specific genetic variant. Given this failure, they of course cannot satisfy criterion #6, which requires the verified variant to also be associated with the clinical phenotype. As with our investigation of 17 endophenotypes described in our special issue of Psychophysiology, we do not feel that it is likely that the failure of endophenotypes in general to lead to verified molecular genetic discovery is due to poor measurement. For one thing, it is unlikely that the candidate endophenotypes listed in Table 5 are uniformly poorly measured. In addition, most are heritable, and we argue in section 5.2 that large MZ twin correlations can be viewed as a form of reliability. Endophenotype researchers should always be concerned with the psychometric properties of their measures (e.g., see W. G. Iacono & Malone, 2011). However, the results of our review suggest that a change in perspective is what is needed more than (just) improved measurement reliability and increased methodological sophistication. Despite the fact that electrophysiological endophenotypes have to date largely failed to meet the rigorous criteria we enumerated in Table 1, there are nevertheless molecular genetic endophenotype studies that stand out from the rest because they represent a step in the right direction.
Table 5.
Degree to which electrophysiological biomarkers satisfy threshold criteria for a putative endophenotype
| Measure | Evidence | Clinical Phenotype | Phenotypic Association | Observed in Unaffected Relatives |
Shared Genetic Liability |
Heritability |
|---|---|---|---|---|---|---|
| Spontaneous EEG activity (power) | ||||||
| Very low frequency | S | ADHD | Cooper 2014; Helps 2008; Tye 2012 | Tye 2012 | Tye 2012 | |
| Delta | S | Schizophrenia/Bipolar ↑ | Moran 2011; Narayanan 2014 | Narayanan 2014 | Malone 2014a; Smit 2005; Zietsch 2007 | |
| S | ADHD ↑ | Rudo-Hutt 2015 | ||||
| S | Binge drinking ↑ | Courtney 2010 | ||||
| S | Autism ↑ | Wang 2013 | ||||
| Theta | M | Schizophrenia/Bipolar ↑ | Hong 2012; Moran 2011; Narayanan 2014 | Hong 2012 | Tye 2014 | |
| S | Externalizing ↑ | Rudo-Hutt 2015 | ||||
| S | ADHD ↑ | Barry 2003a; Loo 2010; Rudo-Hutt 2015 | ||||
| S | Alcoholism ↑ | Rangaswamy 2003 | ||||
| S | Depression ↑ | Fingelkurts 2015 (frontal sites) | ||||
| S | Autism ↑ | Rommelse 2011; Wang 2013 | ||||
| Alpha | S | Schizophrenia/Bipolar ↑ | Narayanan 2014 | Narayanan 2014 | ||
| S+ | ASB/Aggression ↓ | Barry 2003a; Rudo-Hutt 2015 | Niv 2015 | |||
| S | Alcoholism ↓ | Ehlers 2007; Enoch 1999 | ||||
| S | Autism ↓ | Rommelse 2011; Wang 2013 | ||||
| S | Depression ↑ | Fingelkurts 2015 | ||||
| Beta | S | Schizophrenia/Bipolar ↑ | Kam 2013; Narayanan 2014 | Narayanan 2014 | ||
| S+ | Externalizing ↓ | Rudo-Hutt 2015 | Gilmore 2010 | |||
| S | ADHD ↓ | Barry 2003a; Loo 2010; Rudo-Hutt 2015 | ||||
| S | Alcoholism ↑ | Ehlers 2010; Rangaswamy 2002 | Rangaswamy 2004 | |||
| S | Binge drinking ↑ | Courtney 2010 | ||||
| S | Autism ↑ | Rommelse 2011; Wang 2013 | ||||
| S | Depression ↑ | Fingelkurts 2015 | ||||
| Gamma | S | Autism ↑ | Wang 2013 | Ehlers 2010 | ||
| S | Alcoholism ↑ | Ehlers 2010 | ||||
| Low-voltage alpha | S | Alcoholism-related, anxiety | Ehlers 2015; Enoch 1995 | Anokhin 1992; Vogel 1970 | ||
| Theta/beta ratio | B | ADHD | Arns 2013; Snyder 2006 | |||
| Frontal alpha asymmetry | S+ | Depression, anxiety | Allen 2015; Goldstein 2014; Thibodeau 2006 | Jacobs 2015 | Anokhin 2006; Smit 2007b | |
| S+ | Autism | Wang 2013 | Gabard-Durnam 2015 | |||
| S | Aggression | Harmon-Jones 2007; Keune 2012 | ||||
| Event-related EEG amplitude or power | ||||||
| Theta or delta power | M | Alcoholism | Chen 2009; Jones 2006; Jones 2004 | Kamarajan 2015; Kamarajan 2006; Rangaswamy 2007 | Gilmore 2010; Jones 2004; Zlojutro 2011 | |
| S+ | Externalizing | Bernat 2011; Gilmore 2010; Yoon 2013 | Gilmore 2010 | |||
| S | Schizophrenia | Ethridge 2012 | Ethridge 2012 | |||
| Beta or gamma power | M | Schizophrenia | Hall 2011b; Uhlhaas 2010 | Clementz 1998; Hall 2011b; Leicht 2011 | Hall 2011b | |
| S | Bipolar disorder | Kam 2013 | ||||
| S+ | Alcoholism | Padmanabhapillai 2006a | Padmanabhapillai 2006b | |||
| S+ | Autism | Uhlhaas 2006 | ||||
| S | Depression ↑ | Webb 2015 | ||||
| ITPC | S | Externalizing | Burwell 2014 | Hall 2011b; Malone 2016 | ||
| S+ | ADHD | McLoughlin 2014 | McLoughlin 2014 | |||
| S+ | Schizophrenia | Hall 2011b; Leicht 2015; Uhlhaas 2010 | Hall 2011b; Leicht 2011 | Hall 2011b | ||
| S | Autism | Rojas 2008 | Rojas 2008 | |||
| Connectivity | B | PTSD | Georgopoulos 2010; James 2015 | |||
| S | Depression | Fingelkurts 2007; Leuchter 2012; Pizzagalli 2003 | Smit 2010 | |||
| S | Schizophrenia | Ford 2002; Kam 2013; Micheloyannis 2006; Winterer 2003 | ||||
| S | Autism | Vissers 2012; Wass 2011 | ||||
| PFC broadband noise | S | Schizophrenia | Winterer 2004 | Winterer 2004 | ||
| P50 sensory gating | M | Schizophrenia | Adler 1982; Bramon 2004; de Wilde 2007; Freedman 1983; Patterson 2008; Siegel 1984 | de Wilde 2007; Thaker 2008 | M. Hall 2007 | Anokhin 2007b; Aukes 2008; Hall 2006; Young 1996 |
| S+ | Bipolar disorder | Schulze 2007; Thaker 2008 | Schulze 2007 | |||
| S | PTSD | Karl 2006; Neylan 1999 | ||||
| N1/P2 amplitude | S | Schizophrenia | Ethridge 2012; Salisbury 2010 | Sponheim 2006 | ||
| Oddball N2 amplitude | S | Alcoholism | Cristini 2003; Realmuto 1993 | Smit 2007a | ||
| Flanker/Nogo N2 amplitude | S+ | ADHD | Albrecht 2008; McLoughlin 2009 | Albrecht 2008; McLoughlin 2009 | Anokhin 2004 | |
| S | Alcoholism | Cristini 2003; Pandey 2012 | ||||
| P300 amplitude | S+ | Antisocial behavior | Gao 2009 | Viana-Wackermann 2007 | ||
| P | SUDs/Externalizing | Euser 2012; Iacono 2002; Iacono 2011 | Begleiter 1984; Iacono 2011; Polich 1994 | Gilmore 2010 | Malone 2014b; van Beijsterveldt 2002 | |
| P | Schizophrenia | Bramon 2004; Chen 2014; Jeon 2003; Qiu 2014 | Bramon 2005; Ethridge 2012 | M. Hall 2007 | ||
| S | ADHD | Barry 2003b; Szuromi 2011 | ||||
| M | Bipolar disorder | Hall 2009; Schulze 2008; Thaker 2008 | Hall 2009; Schulze 2008; Thaker 2008 | |||
| S | PTSD | Johnson 2013; Karl 2006; Kimble 2000; McFarlane 1993 | ||||
| P300 latency | P | Schizophrenia | Bramon 2004; Qiu 2014 | Bramon 2005 | M. Hall 2007 | Hall 2009; van Beijsterveldt 2002 |
| M | Bipolar disorder | Hall 2009; Lenox 2002; Muir 1991; Schulze 2008; Thaker 2008 | Hall 2009; Schulze 2008 | Hall 2009 | ||
| ERN amplitude | S+ | OCD, anxiety ↑ | Hajcak 2002; Olvet 2008; Moser 2016 (sex specific) | Olvet 2008 | Anokhin 2008 | |
| M | ADHD ↓ | Albrecht 2008; Geburek 2013 | Albrecht 2010; Albrecht 2008; McLoughlin 2009 | |||
| S | Externalizing ↓ | J. Hall 2007 | ||||
| S+ | SUDs ↓ | Franken 2007 | Euser 2013 | |||
| FRN amplitude | M | Depression | Moran 2016; Foti 2009 | Foti 2011; Kujawa 2015; Weinberg 2015 | ||
| Reward positivity | S | Depression | Proudfit 2015 | Proudfit 2015 | ||
| MMN amplitude | S | Schizophrenia | Erickson 2016; Haigh 2016; Shelley 1991; Takahashi 2013; Umbricht 2005 | Haigh 2016 (NS) | M. Hall 2007 | Hall 2009 |
| S | Psychosis | Ranlund 2015 | Ranlund 2015 | |||
| S | Alcoholism | Inconsistent findings | ||||
| M | Dyslexia (speech stimuli) | Kraus 1996; Schulte-Korne 2001 | Hommet 2009; Maurer 2009; Maurer 2003 | |||
| N400 amplitude | S | Alcoholism | Roopesh 2010 | Roopesh 2009 | ||
| N170/N250 amplitude | B | Schizophrenia | Feuerriegel 2015; McCleery 2015 | |||
| N170 latency | B | Autism | Dawson 2005a | |||
| Atypical lateralization | S | Autism | Dawson 2005a; Seery 2013 | Dawson 2005b | ||
| B | Depression | Trinkl 2015 | ||||
| ANS and electromyographic measures | ||||||
| Acoustic startle | S | PTSD/anxiety | Pole 2007 | Anokhin 2003; Hasenkamp 2010; Vaidyanathan 2014b | ||
| S | Schizophrenia | Braff 1992; Light 2012; Quednow 2008 | ||||
| Startle modulation | S | Psychopathy | Benning 2005; Patrick 1993 | Weak h2 (Anokhin 2007a; Vaidyanathan 2014c) | ||
| S+ | Anxiety/fearfulness | Cuthbert 2003; Grillon 1993; Grillon 2003; Vaidyanathan 2009 | Grillon 1997 | |||
| S | Borderline personality | Hazlett 2007 | ||||
| S | Bipolar disorder | Giakoumaki 2010 | Giakoumaki 2010 | |||
| Prepulse inhibition | M | Schizophrenia | Braff 1978; Braff 2010; Swerdlow 2014; Turetsky 2007 | Cadenhead 2000; Kumari 2005 | Anokhin 2003; Greenwood 2015 | |
| M | Bipolar disorder | Thaker 2008 | Thaker 2008 | |||
| Resting HR | M | ASB/psychopathy ↓ | Lorber 2004; Ortiz 2004 | Mednick 1972, cited in Venables 1987 | Baker 2009 | Wang 2015 |
| S | PTSD ↑ | Pole 2007 | ||||
| ED activity/reactivity | S | Externalizing ↓ | Isen 2013; Isen 2012 | Herpertz 2007 | Hettema 2003; Tuvblad 2012; Vaidyanathan 2014a | |
| S+ | PTSD/anxiety ↑ | Pole 2007 | Balle 2013 | |||
| S | Schizophrenia ↑ | Iacono 1999 | Iacono 1999 | |||
| RSA patterns | S | Depression | Yaroslavsky 2014 | Yaroslavsky 2014 | ||
| B | ADHD | Beauchaine 2015 | ||||
| B | Antisocial behavior | Beauchaine 2015 | ||||
| EDA modulation | B | SUDs | Taylor 2009 | |||
| Eye tracking | ||||||
| Antisaccade error rate | M | Schizophrenia | Clementz 1998; Fukushima 1988; Turetsky 2007 | Calkins 2008; Levy 2004 | Greenwood 2007; Vaidyanathan 2014b | |
| S | Psychosis | Reilly 2014 | Reilly 2014 | |||
| S | OCD | Lennertz 2012 | Lennertz 2012 | |||
| Smooth pursuit tracking | M | Schizophrenia | Calkins 2000; Hong 2008; Levy 1993; O'Driscoll 2008; Thaker 2008 | Calkins 2008; Hong 2008; Ross 2002; Thaker 2008 | Blekher 1997; Katsanis 2000 | |
Note: References are provided in the Supplementary Material. Columns represent the threshold criteria for an endophenotype (see Section I, Table 1). Citations include seminal papers establishing associations between biomarker and clinical phenotype, as well as meta-analytic and narrative reviews (in bold face and italics, respectively) of the literature concerning a biomarker and evidence that it is a candidate endophenotype. First author and publication year are listed for each citation. The strength of the empirical evidence supporting a measure as a putative endophenotype was coded as follows: P = persuasive, M = moderate, S = suggestive (S+ = strongly suggestive), B = biomarker only, evidence does not support measure as a putative endophenotype. NS indicates a nonsignificant effect (e.g., in a meta-analysis).
Abbreviations: ADHD, attention deficit-hyperactivity disorder; ASB, antisocial behavior; PTSD, posttraumatic stress disorder; OCD, obsessive-compulsive disorder; SUDs, substance use disorders; ITPC, inter-trial phase consistency; FRN, feedback-related negativity; MMN, mismatch negativity; HR, heart rate; ED, electrodermal; EDA, electrodermal activity; RSA, respiratory sinus arhythmia
9.1 A review of how well candidate endophenotypes satisfy threshold criteria
Table 5 lists a variety of electrophysiological measures that are either considered biomarkers or endophenotypes. We searched PubMed using variations of “endophenotype,” “intermediate phenotype,” and “biomarker” together with “electrophysiology,” “psychophysiology,” and terms specifying more specific physiological measures or classes of measures (e.g., “event-related potential or ERP,” “autonomic,” “EEG”, and so on). The table provides citations for studies that support each measure as a biomarker or endophenotype, particularly meta-analyses, which are indicated by bold face, and reviews, indicated by italics, as well as key papers. Table 5 addresses the Threshold Criteria in Table 1 that we identified as necessary for treating a phenotype as a candidate endophenotype: association with a clinical phenotype, heritability, presence in unaffected relatives, and evidence for a shared genetic liability. We have attempted to be systematic and thorough in our search of PubMed, although it is not possible to be exhaustive. We did not include measures reflecting neurobiological systems that appear to be disrupted by environmental adversity and stress, such as EEG asymmetry in infants and children associated with psychosocial risk. We also did not include putative endophenotypes relevant for different levels of analysis, such as developmental trajectories (Tierney, Gabard-Durnam, Vogel-Farley, Tager-Flusberg, & Nelson, 2012), or weighted composites (Clementz et al., 2015) and multivariate endophenotypes (Gilmore, Malone, & Iacono, 2010; W. G. Iacono, Carlson, & Malone, 2000; G. W. Price et al., 2006). There is invariably a certain degree of subjective judgment involved in how to efficiently and fairly summarize so much research. The table is nevertheless useful for drawing broad inferences about what we are learning to date from psychophysiological endophenotype research.
The sheer number of entries in Table 5 attests to the level of interest in endophenotypes and biomarkers. What is perhaps less apparent is that interest is increasing, judging from the number of papers published in recent years. The majority of work has focused on addressing the threshold criteria, which is clearly a desirable first step to establishing a given measure as a putative endophenotype. The viability of some measures as endophenotypes is stronger than for others in that it is supported by meta-analysis or at least the results of a narrative review, whereas one or two empirical reports is all that we could find for others. This may be because interest in a particular measure as an endophenotype is recent, as is the case with feedback-related negativity, reward positivity, measures of connectivity or phase synchrony, broadband noise, and other measures that are more recently available to endophenotype researchers. In other cases, such as the oddball N2 amplitude, interest seems to have faded. There have been a number of twin studies, which establish broadly that heritable individual differences are evident for the majority of putative endophenotypes. Many studies have also been conducted of healthy first-degree relatives of probands with a clinical disorder, who presumably share the genetic liability for the disorder despite not having manifested it. Somewhat fewer studies have been conducted with children or youth at high risk for a disorder by virtue of a positive family history.
The second column of Table 5 contains our evaluation of the strength of evidence for considering the biomarker in each row an endophenotype, based on how well it meets the Section I Threshold Criteria in Table 1. We classified measures into one of four categories: biomarker (represented by B), suggestive evidence (S), moderate evidence (M), and persuasive evidence (P). Putative endophenotypes meeting only criterion #1 in Table 1 – evidence of a phenotypic association with a clinical phenotype, without evidence of genetic influence -- are classified as biomarkers. For instance, the theta/beta ratio of resting EEG power, thought to be a marker of ADHD, and disrupted neural synchrony in PTSD, do not appear to be genetically influenced. Evidence of genetic influence is most commonly provided by heritability studies. However, evidence for genetic influence on a few measures comes from findings in healthy first-degree relatives, such as PFC broadband noise and the feedback positivity. We considered the evidence as “suggestive” if there was evidence of heritability but neither criterion #3 or #4 was met. For us to consider the evidence as “moderate,” a putative endophenotype must meet criteria #3 and #4 or there must be more than one report showing evidence that it meets one or the other. If it meets one of the two criteria, we assigned an “S+.” However, if evidence that a putative endophenotype meets either of these criteria came from the same report that also established its criterion validity, we considered this an “S.” For instance, Iacono (1999) found that electrodermal activity (EDA) was increased in patients with schizophrenia and first-degree relatives. In cases such as this we considered the evidence as merely suggestive (“S”) rather than “S+.” For evidence to be considered persuasive, the putative endophenotype must meet both criteria (#3 and #4) with more than a single empirical report and preferably meta-analysis or narrative review providing evidence relevant to each criterion.
While necessarily imprecise and somewhat subjective, we feel that this scheme is nevertheless useful. It indicates that the majority of endophenotypes supported by moderate or persuasive evidence are endophenotypes for schizophrenia and bipolar disorder (11 out of 16). This undoubtedly reflects in part the fact that there is a long history of interest in endophenotypes for schizophrenia in particular. This scheme also indicates that, whereas the ratio of theta to beta resting EEG power may constitute a biomarker for ADHD, it is amplitude of the error-related negativity (ERN; rated “M”) that is the best putative endophenotype for ADHD. Although a focus of research only for a few years, the feedback-related negativity shows moderate evidence as an endophenotype for depression. P300 amplitude is a persuasive endophenotype for SUDs and externalizing disorders and for schizophrenia, and the evidence is moderate that it is an endophenotype for bipolar disorder as well. P300 latency is a strong endophenotype for schizophrenia and moderate for bipolar disorder. Indeed, P300 amplitude and latency are the only candidate endophenotypes for which the evidence is persuasive.
9.2 Selective review of molecular genetic studies of endophenotypes
The vast majority of molecular genetic studies of putative endophenotypes have been candidate gene studies with small samples. The performance of candidate gene studies for complex traits in the past decade gives us little reason to believe that their results are verifiable. Nevertheless, we review in this section all studies, whether candidate gene studies or GWAS, with a sample size of at least 400 or that included a replication sample in the original report. This criterion includes studies with a discovery and replication sample as well as the few studies that combined GWAS results across multiple independent samples through meta-analysis. We chose 400 subjects as a minimum sample size because it provides 80% power to detect effects accounting for 2% of the variance in an endophenotype. As we outlined earlier, effect sizes of this magnitude are substantially larger than we expect of true effect sizes, and this targeted minimum sample size should not be taken as a recommendation. It simply allows us to highlight studies that stand out from the majority of molecular genetic studies because they are more highly, although not sufficiently, powered.
Well over a hundred studies have been published that assessed whether measured genetic variants are associated with candidate endophenotypes. Of these, only a dozen, summarized in Table 6, meet our admittedly liberal criteria. Despite representing research that is commendable in certain respects, the studies in Table 6 illustrate some of the reasons that candidate gene studies are so fallible, as we discussed in section 7.1. Although 12 studies are listed in the table, nine of them were contributed by three research groups. The studies reported within each group involve overlapping or identical samples, and thus should not be considered independent reports despite the fact that they examine different candidate genes or candidate endophenotypes. These overlapping reports also do not conduct project wide correction for multiple testing, so they do not adequately adjust for Type I error, a problem that is now well recognized in molecular genetic research of complex traits. Although the table indicates effects that were nominally significant by the criteria used (p < .05 or p < .01), none reach significance if all variants within all genes reported across all these studies were used in the multiple testing correction. This may seem like an unfair standard because the reports were conducted under the candidate gene paradigm, where a priori evidence is used to select candidate genes based on “theory”. However, we maintain the position stated in section 7.1 that candidate gene selection based on theory is highly fallible and that candidate gene studies conducted in the traditional sense where candidate genes are selected based on psychobiological theory and not statistical evidence, are in fact more exploratory than not. Thus, when multiple candidate gene findings are described in multiple publications from the same dataset, they should be held to higher statistical standards than within-study corrections, the now widely adopted standard for genome wide significance. None of the associations in Table 6 constitutes a verified finding.
Table 6.
Candidate gene endophenotype studies with large samples or a replication sample
| Measure | Lead Author | Candidate Gene (MAF) | N (replication N) | Largest Effect |
|---|---|---|---|---|
| PPI | Petrovsky (2010)1 | CHRNA3 (0.35) | 107 (73) | 11.4% (7.5%) |
| PPI | Quednow (2011)1 | TCF4 (0.06) | 107 (73) | 4.4% (12.1%) |
| Startle reactivity | Roussos (2011)2 |
CACNA1C (0.27) ANK3 (0.04) |
445 | 9.55% 4.42% |
| PPI | Roussos (2011)2 | NRG1 (0.37) | 445 | 2.65% |
| PPI | Roussos (2011)2 | DAO (0.35) | 445 | 1.38% |
| P50 | Quednow (2012)1 | TCF4 (0.06) | 1,821 | 0.91% |
| P300 latency | Xu (2010) | miR-30e (0.03) | 2,190 | 0.63% |
| SPEM (gain, saccade frequency) | Smyrnis (2011)3 | NRG1 (0.41) | 1,502 | 0.34% |
| Antisaccade error SPEM | Kattoulos (2012)3 | RGS4 (0.48) | 1,530 | 0.33% |
| Antisaccade error | Stefanis (2008)3 | RGS4 (0.48) | 1,532 | 0.20% |
| P50 | Shaikh (2011) |
COMT (0.34) BDNF (0.17) NRG1 (0.36 – 0.39) |
451 |
—
— — |
| P50 | Cabranes (2013) | CHRNA7 (0.27 – 0.49) | 375 | — |
Germany-Great Britain collaboration
Learning on Genetics of Schizophrenia (LOGOS)
Athens Study for Psychosis Proneness and Incidence of Schizophrenia (ASPIS)
Note: The MAF is given for each candidate gene studied where the published data permitted calculating it (i.e., if homozygotes for the rare allele were not combined with heterozygotes), or a range of MAFs if more than one SNP from a gene was examined. The column labeled “N (Replication N)” lists the number of subjects in each study, or in the Stage 1 sample if the study included a replication study. The number of subjects in the replication sample is provided in parentheses if applicable. The effect size of the largest association is provided in the last column, as a percentage of endophenotype variance accounted for. Associations that were not reported as statistically significant are indicated by a dash in place of the effect size. For studies including a replication sample (and analyzing discovery and replication samples separately), the variance accounted for in the replication by the variant is provided in parentheses.
In light of the fact that Type I error was not adequately controlled, whether within a study or across studies within the same research group, it cannot be surprising that several of the findings listed in Table 6 have not replicated. Roussos and colleagues divided the LOGOS sample into a discovery and replication sample and conducted a GWAS of PPI and startle reactivity (Roussos et al., 2015). Although they did obtain unreplicated novel genome-wide significant hits for two, they did not obtain significant associations between any candidate genes previously found to be associated with PPI, including those listed in Table 6 which they themselves had previously obtained.
The GWAS by Roussos and colleagues is listed in Table 7, along with GWAS or studies similarly using a genome-wide scan, representing seven electrophysiological endophenotypes in all. To avoid redundancy, we did not add to this table the GWAS results from the 17 endophenotypes examined in our special issue of Psychophysiology that are included in Table 2, none of which are covered in Table 7, or the results we obtained for event-related theta power described in section 5.2, a measure that is included in Table 7. Examples of the challenges that we faced and discussed in section 5.0 are also apparent in several of these studies. Zlojutro and colleagues with the Consortium on the Genetics of Alcoholism (COGA) conducted a GWAS of event-related EEG theta activity in a discovery sample of 1,064 subjects, from which 42 SNPs were prioritized for replication in a second, family-based sample of 1,095 subjects from 242 families with many alcohol-dependent individuals (Zlojutro et al., 2011). None of the variants were genome-wide significant. Table 7 lists a second GWAS of event-related theta activity conducted by COGA, which seems to have expanded on the family sample of Zlojutro et al., studying 1,560 subjects in 117 densely alcoholic families (S. J. Kang et al., 2012). A genome-wide significant signal was observed in chromosome 21 from multiple SNPs in KCNJ6. This study did not replicate the Zlojutro et al. finding. A subsequent study from our group with a sample of approximately 4,100 (see Section 5.2 and Table 2), failed to confirm either one. These studies from our group, Roussos et al., and COGA represent among the best currently available in electrophysiological endophenotype research; they show that electrophysiological endophenotypes have not protected researchers from the problem of false positives that have been widely observed to plague underpowered studies of complex traits.
Table 7.
Genome-wide studies with large samples, replication samples, or both
| Measure | Lead Author (Year) | N (replication N) | Largest Effect |
|---|---|---|---|
| GWAS | |||
| MMN | Roeske (2011) | 200 (184) | 10.53% |
| Resting EEG theta power | Hodgkinson (2010) | 322 (185) | 8.8% (3.5%) |
| P50 | Hall (2015) | 392 | 9.05% |
| PPI | Roussos (2015) | 792 (405) | ? |
| Event-related theta power | Zlojutro (2011) | 1,064 (1,095) | 2.26% (0.42%) |
| Event-related theta power | Kang (2012) | 1,560 | 0.91% |
| Resting heart rate | Deo (2013) | 13,372 | 0.42% |
| Resting heart rate | Holm (2010) | 23,112 | 0.28% |
| Resting heart rate | Cho (2009) | 17,899 | 0.22% |
| Resting heart rate | Eijgelsheim (2010) | 38,991 | 0.17% |
| Exome chip | |||
| N4S response to pre-pulse | Norden-Krichmar (2015) | 420 | 1.97% |
Note: The column labeled “N (Replication N)” lists the number of subjects in each study, or in the Stage 1 sample if the study included a replication study. The number of subjects in the replication sample is provided in parentheses if applicable. The effect size of the largest association is provided in the last column, as a percentage of endophenotype variance accounted for. For studies including a replication sample (and analyzing discovery and replication samples separately), the variance accounted for in the replication by the variant is provided in parentheses. To avoid redundancy, we did not include GWAS results from the 17 endophenotypes examined in our special issue of Psychophysiology, which are summarized in Table 2, despite representing a different set of endophenotypes. In addition, we did not include results we obtained for event-related theta power described in section 5.2, although this measure that appears in this table.
Fueled by substantially larger sample sizes, resting heart rate provides a picture in stark contrast. It is the only candidate endophenotype to fully meet our threshold for verified discovery. Although a putative endophenotype for antisocial behavior and perhaps PTSD (see Table 5), resting heart rate has not received as much attention as an endophenotype as other candidates in Table 5. Yet, as we discussed in section 6.1, GWAS meta-analyses of heart rate and related measures, such as Q-T interval, have identified several loci with genome-wide significant associations with heart rate in samples comprising subjects from different racial or ethnic backgrounds (European, Icelandic, Asian, African-American) (Cho et al., 2009; Deo et al., 2013; Eijgelsheim et al., 2010; Holm et al., 2010). There have been replicated findings, and several of the variants discovered are nonsynonymous and biologically plausible. The effect sizes are small, accounting for no more than 1 beat per minute. For instance, one variant accounted for 0.4% of the variance in heart rate (shortening the R-R interval by 12.6 ms) (Deo et al., 2013). Other studies report somewhat smaller effects (see Figure 1 and Table 4).
Examining Tables 6 and 7 reveals that the largest effect sizes are reported by studies with the smallest samples. In fact, sample size is inversely correlated with the magnitude of effect obtained. Unsurprisingly, only those studies with the largest samples obtained small effects; large samples are necessary in order to provide adequate power to detect such effects, which is precisely why they are needed (see Table 3). In addition, however, what is most striking about the studies with large samples is what they do not report: large effects. This is obviously not due to lack of power. That studies with the largest samples have not obtained large effects argues that the large effects from studies with small samples are almost certainly due to sampling variation or allelic stratification and are not true associations. To put this another way, a large effect size should be disquieting rather than reassuring; it almost surely signals that a finding is a false positive rather than confirming that it is real.
10.0 Where Do We Go From Here?
Our special issue in Psychophysiology was a step forward in the systematic evaluation of genetic associations for electrophysiological endophenotypes, but it is far from the last word. Electrophysiological endophenotypes are not dead, despite their apparent inability to dramatically increase power for genetic association studies. Endophenotypes, by their definition, are biological lab-based measures of genetically-influenced components of clinical disorders. As such, they provide insight into the nature of mental disorders and brain function. Of interest, then, is how to optimize their potential contribution.
10.1 Data sharing
The wave of consortia dedicated to the identification of phenotype-genotype associations has been driven in our opinion by one basic fact: no single investigator has access to a large enough sample size to profitably conduct genetic association studies of complex traits. To make progress, she must share her data and combine it with other data. In the world of neurobehavioral endophenotypes, the way has been led thus far by the ENIGMA consortium, which now is branching out into electrophysiology with the hope of building meta-samples in the tens of thousands of individuals.
Sharing data, broadly, without embargo, and with open consents, is our surest path to success. The NIH recognizes this fact and is encouraging it through new genomic data sharing policies (https://gds.nih.gov/) requiring researchers to obtain more open consents to allow data sharing with repositories, such as the National Institute of Health's database for genotypes and phenotypes (dbGaP).
However, we encourage researchers to take things one step further and make their data more broadly available than through repositories like dbGaP. Whereas dbGaP is good for genotypes, available phenotype data are often highly limited, and are only a tiny fraction of all the phenotypes a particular study collects. Those phenotypes, and the potential genetic associations arising from them, are never shared. In the saddest cases, they are never even analyzed by the original investigative team. Direct collaboration between like-minded investigators with similar data is a powerful way to aggregate data and increase statistical power and generalizability. In those cases where raw data cannot be shared, or if genetic data is at some point in the future considered identifiable data, it is still possible to share association summary statistics, which has been widely successful in GWAS meta-analysis.
10.2 Consortia
One clear path forward is to build consortia to aggregate data across many studies for genetic association analyses. Genetic association consortia have worked for medical disease, psychiatric traits, and normal range behaviors. They guarantee increased power through increased sample size and, when successful, they provide many new opportunities for additional analyses and hypotheses that cannot be answered in a single dataset. Consortia are also one way in the current funding climate to obtain genotyping funds. There are many studies with electrophysiological data but not genome-wide genotypes. By banding together and building total sample size into the tens of thousands it may become feasible to request funding to genotype all available non-genotyped samples because, once done, there is reason to think a genetic association study of the large sample would be successful. This particular approach has been successful in funding genotyping in the Psychiatric Genomics Consortium (PGC; Sullivan, 2010). PGC comprises consortia to study the genetics of many disorders in addition to schizophrenia, such as autism, ADHD, substance abuse, major depressive disorder, bipolar disorder, PTSD, OCD, and anorexia nervosa (https://www.med.unc.edu/pgc). There are other relevant consortia as well, such as GSCAN for addiction (http://gscan.sph.umich.edu), the Social Science Genetic Association Consortium (SSGAC; http://www.ssgac.org/Home.php), which is currently investigating educational attainment, subjective well being, and fertility, and ENIGMA for imaging and psychophysiology (http://enigma.ini.usc.edu/; Thompson et al., 2014).
10.3 A different approach to selecting candidate genes
In light of the many difficulties in candidate gene studies as they have typically been conducted, we suggest conducting such studies only when there is compelling evidence supporting a particular gene's candidacy. We suggest a two-dimensional spectrum of acceptability, diagrammed in Figure 3. First, we propose a spectrum of statistical association evidence. On one end are variants that have been indubitably associated with the clinical outcome(s) of interest or the endophenotype of interest. On the other end are variants that have mixed evidence from low-powered non-GWAS studies. The second spectrum concerns whether the variant is predicted to affect a genomic or molecular mechanism. Variants with relatively clear mechanisms (e.g., changes to coding sequence or gene expression) will be easier to interpret and functionally characterize, whereas variants with unclear mechanisms, such as intergenic variants with no functional signatures, may be more difficult to characterize. Variants can be prioritized for follow-up in proportion to the degree of evidence for association. Variants that show strong evidence of association but also implicate a known mechanism should receive highest priority.
Figure 3. Prioritizing Candidate Genes/Variants for Follow-up Study.
The usual set of candidate variants studied in psychiatric genetics and psychiatric endophenotype candidate gene research is represented in the upper left-hand corner. They are variants with plausible mechanisms based on behavioral neuroscience but inconsistent evidence for association. All candidates with high evidence for association are worthy of followup, especially those with highly plausible mechanisms of effect.
We believe the candidate gene design is changing. Instead of basing candidate gene selection on the usual (fallible) suspects (5-HTTLPR, DRD2, DRD4, COMT, BDNF, MAO-A, to name a few), which have largely led to dead ends or confusion, or on results from infrahuman model systems with unknown generalizability, we now know from GWAS what variants are truly associated with human disorders and traits under a controlled family-wise error rate. For schizophrenia, for example, we now have evidence for the association of 128 variants within 108 loci. These “candidate variants” have strong support for their role in schizophrenia and are prima facie good candidates for schizophrenia endophenotypes. In addition, large consortia have formed to identify variants associated with phenotypes other than schizophrenia, such as personality, depression, and educational attainment. Researchers studying endophenotypes related to these distal and clinical outcomes will be able to take advantage of discoveries being made in these areas. The endophenotype then can inform us about the function of those specific markers. For instance, two recently published studies examined a number of these markers now known to be related to schizophrenia as well as markers related to other disorders, and each reported an association between a marker and P300 amplitude that survived correction for multiple testing (Del Re et al., 2014; Hall et al., 2014). If replicated, these findings may ultimately shed light on mechanisms governing poor attention allocation and working memory in schizophrenia.
An alternative to examining the association between individual markers and endophenotypes is to construct a polygenic risk score, which captures the aggregate effect of many variants. Such scores are weighted composites of allele counts, with the weights consisting of the regression coefficients associated with the endophenotype from GWAS or meta-analysis. The development, or training, sample and test sample should ideally be independent, of course. If not, then cross-validation techniques (e.g., k-fold or leave-one-out cross-validation) should be used. Weights can be based on the results from the consortia that are forming. The regression coefficients may thus be for predicting a related clinical phenotype, rather than an endophenotype. The simplest approach is to use all markers to generate a risk score, but researchers more commonly construct a set of risk scores by using increasingly stringent p-value thresholds are commonly used to identify “significant” markers, such as p-values ranging from .50 to very small values. At a minimum, significant associations between such risk scores based on clinical outcomes and endophenotypes attest to the construct validity of the endophenotype for the clinical outcome in question. Studies have begun to appear that relate risk scores derived from PGC, for instance, to psychophysiological measures, such as PPI (Hall et al., 2015; Roussos et al., 2015).
The extent to which these types of studies yield findings that can be replicated and ultimately advance our understanding of the endophenotype and its associated disorder remains to be seen. However, they can certainly be used to confirm the construct validity of endophenotypes. The gain in power that derives from testing a small number of well-validated markers, or a single risk score based on many markers in aggregate, increases power to detect whether and how two phenotypes are related, although at the expense of being able to identify specific biological mechanisms. Moreover, molecular-genetic researchers are developing ways to optimize polygenic risk scores (e.g., Vilhjalmsson et al., 2015), which should make them ever more powerful. Psychophysiological researchers can contribute to this type of endeavor in a unique way by developing endophenotype risk scores. For instance, a P300 amplitude reduction (P3AR) “risk score” might be developed from GWAS or meta-analytic results and tested for its association with externalizing disorders, which would provide even more direct evidence of the validity of the notion that P3AR is an endophenotype for externalizing psychopathology (W. G. Iacono, Carlson, Malone, & McGue, 2002; W. G. Iacono, Malone, & McGue, 2008). Because P3AR is also associated with other disorders, like schizophrenia, the risk score can also be expected to show association with any disorder where this effect is observed.
Perhaps more importantly, identifying variants convincingly associated with a given clinical phenotype will suggest biological mechanisms and new endophenotypes. That is, it may be fruitful to develop endophenotypes based on the biological function of validated polymorphisms. This is now beginning, with follow-up studies of the 108 loci implicated by the PGC consortium. A recent article fine-mapped structural variation within the major histocompatibility complex, discovering CNVs that severely affect gene expression of C4A and C4B in the brain (Sekar et al., 2016). When taken to a mouse model C4 activity mediated synaptic pruning during postnatal development. This GWAS-based finding has been translated to a neurobiological mechanism which, if true, would implicate C4 expression and possibly synaptic pruning in the etiology of schizophrenia. As the C4-related mechanisms are better understood, it should be possible to derive candidate endophenotypes for them. Indeed, we may come to think of our current arsenal of endophenotypes as obsolete and unlinked to genetic variants of measureable effects, replacing it with a new set of endophenotypes developed on the basis of known genetic effects.
It is important to note that even the highest priority candidate variants may be difficult to study. If the follow-up study involves association analysis of the candidate variant with another complex (endo) phenotype samples of over 1000 unrelated individuals will be required to obtain sufficient power (see Table 3). Molecular work, for example studies of gene expression or cellular processes affected by a candidate variant, may result in much larger expected effect sizes and require a fraction of the observations as for complex phenotypes. In either case recent success in GWAS will only spur additional focused analyses of candidate genes and genomic regions, except now we will know that these regions harbor genetic variation that actually affect disorder risk in humans, a critical advance in genetic research.
10.4 More research on the utility of an endophenotype
10.4.1 Predictive utility and development
Studies concerning the utility of an endophenotype, criteria 7 through 10 in Table 1, were hard to find, and are certainly fewer in number than those focused on establishing whether a putative endophenotype meets threshold criteria utility. The majority concern criterion #7 that an endophenotype should predict development of a disorder. This includes studies examining a candidate endophenotype's longitudinal stability during development. For instance, P300 amplitude (Carlson & Iacono, 2006; Yoon et al., 2015), amplitude of the error-related negativity (ERN) (Meyer, Weinberg, Klein, & Hajcak, 2012) or feedback negativity (FN) (Bress, Meyer, & Proudfit, 2015), and resting heart rate (Baker et al., 2009) are stable during transitions from childhood to adolescence or from adolescence into young adulthood. A number of studies have evaluated the predictive utility of candidate endophenotypes. For instance, EEG alpha power in childhood has been found to predict antisocial behavior in adolescent male twins, which was due to genetic liability shared between endophenotype and outcome (Niv et al., 2015); alpha asymmetry has been found to predict depression (Mitchell & Possel, 2012); reduced delta and theta event-related power (Yoon et al., 2015) and P300 amplitude (Carlson, Iacono, & McGue, 2004; W. G. Iacono et al., 2002; Yoon et al., 2015) are reported to predict externalizing disorders; reduced high-frequency power has been found to predict autism; increased ERN is reported to predict the onset of anxiety disorders (Meyer, Hajcak, Torpey-Newman, Kujawa, & Klein, 2015); reduced feedback positivity is reported to predict subsequent depression (Proudfit, 2015); reduced MMN amplitude has been found to predict the development of psychosis in at-risk individuals (Shaikh et al., 2015); and reduced resting heart rate in childhood has been found to predict antisocial behavior in adulthood (Raine, Venables, & Williams, 1990). This latter finding was recently corroborated and extended in a longitudinal study of more than 700,000 men in Sweden, which found that lower resting heart rate in late adolescence among military conscripts was associated with significantly elevated hazard of being convicted of violent and nonviolent crimes as well as assault and unintentional injuries in adulthood (Latvala, Kuja-Halkola, Almqvist, Larsson, & Lichtenstein, 2015). Although it is encouraging to see the number of such studies growing, more are needed.
Moreover, even if heritability by itself does not translate into successful gene finding, the fact that a valid endophenotype reflects genetic risk can be used in novel ways. For instance, longitudinal designs can characterize trajectories of change in endophenotypes and factors that influence such trajectories, including environmental factors, which may improve our ability to predict who is going to develop a disorder as well as enhance our understanding of the genetic risk captured by the endophenotype. Understanding the potentially dynamic relationship between endophenotype and clinical phenotype and the parameters affecting it is important. Anxiety appears to relate to increased ERN differently in young children relative to older ones (Meyer et al., 2012). The reduction in P300 amplitude characteristic of children and youth at risk for substance abuse appears to become attenuated in early adulthood (Carlson & Iacono, 2008; Hill et al., 1999), suggesting that the utility of this endophenotype may be limited in this developmental period. Because gene expression is itself dynamic, it is also important to consider age and development when searching for genetic variants influencing a candidate endophenotype; the relationship between gene and endophenotype may not be constant across development. The relationship between person and environment is also dynamic. On Waddington's model of the epigenetic landscape (Waddington, 1956), earlier periods of development are characterized by a flatter epigenetic landscape, permitting deviations in developmental trajectory more readily than later periods, when trajectories become increasingly canalized and constrained. Longitudinal designs may be particularly useful because they allow one to describe and characterize developmental trajectories. Empirical Bayes estimates of growth curve parameters characterizing different aspects of individual subject's developmental trajectories may well be more sensitive targets of gene discovery than are single measures, just as reducing measurement error through aggregating multiple measurements increases power. If understanding gene effects on developmental trajectories themselves is illuminating, they may also be more appropriate targets.
10.4.2 Enhancing our understanding of brain mechanisms
We are arguing here that, in light of what we have learned from a decade of GWAS, endophenotype research needs to move beyond merely establishing that a candidate endophenotype marks genetic risk, with the underlying assumption that this will lead to genetic discoveries. Although genetic discovery is possible, the available evidence strongly suggests that it is unlikely without larger sample sizes or a technological revolution in our ability to conduct electrophysiological research much more efficiently. Nevertheless, endophenotype research can enhance our understanding of brain mechanisms accounting for individual differences in endophenotype level or configuration (criterion #8). Parametric manipulations of experimental parameters (e.g., Curtis, Calkins, Grove, Feil, & Iacono, 2001; Salisbury et al., 1994) may be able to provide experimental evidence about parameters affecting an endophenotype. For instance, a modified version of the Eriksen flanker task has been used in which target-distractor incompatibility was parametrically varied in order to probe the sensitivity of the N2 ERP component to the degree of response conflict subjects experience when presented with a particular variant of the stimulus array (Forster, Carter, Cohen, & Cho, 2011). The nogo-N2 is considered a candidate endophenotype for alcoholism (Kamarajan & Porjesz, 2015), and similar types of experimental manipulations are likely to inform us about the nature of individual differences in this putative endophenotype. Lenartowicz and colleagues (Lenartowicz et al., 2014) used a visual-spatial version of the Sternberg working memory test in a sample of children with ADHD and typically developing controls to parametrically vary task load and thereby try to isolate the source of the difficulties experienced by children with ADHD. Studies such as these can help to isolate the psychological and neural processes reflected in putative endophenotypes for ADHD such as the P2 ERP component and thus the precise nature of genetic risk reflected in the endophenotype. Testing for associations with genetic variants is not a necessary part of this type of research. However, studies that provide a better characterization of the mechanisms underlying a putative endophenotype and the latent genetic risk it reflects will also ultimately be more informative about the neurobehavioral processes associated with a variant that can be verified for the endophenotype-associated disorder or proxy phenotype.
Of course, we ultimately want to trace the path from endophenotype to specific genes. Research that provides a more detailed understanding of the endophenotype nevertheless is also potentially informative about the nature of unobserved, latent genetic risk reflected in the endophenotype. For instance, administering ketamine, an antagonist of the NMDA receptor for glutamate, to healthy subjects results in mean reduced MMN amplitude and performance on the CPT-AX task that resembles that of patients with schizophrenia (Umbricht et al., 2000). This suggests that compromised NMDAR function may be a characteristic of the disorder. Reduction of the amplitude of the P300 ERP potential is a robust endophenotype for externalizing disorders (W. G. Iacono & Malone, 2011; W. G. Iacono et al., 2008). However, “the” P300 is not a unitary phenomenon, as we have discussed in an earlier section. Although we argued that decomposing it into supposedly simpler elements is not likely to lead to significant genetic discovery, this type of effort can nevertheless lead to an increasingly elaborated understanding of the genetic risk for externalizing. For instance, Ford, Mathalon, and colleagues found that the consistency of inter-trial phase locking (ITPC) of gamma activity during the peak of the P300 response was correlated with P300 amplitude in control subjects but not patients with schizophrenia, suggesting a breakdown of the normal (partial) dependence of P300 amplitude on gamma synchrony in schizophrenia (J. M. Ford, Roach, Hoffman, & Mathalon, 2008). We have similarly found that ITPC of theta activity is reduced in externalizing disorders, and in fact partially mediates P300 amplitude reduction (Burwell, Malone, Bernat, & Iacono, 2014). To be clear, we are not advocating ITPC as yet another candidate endophenotype; we are suggesting that endophenotypes can lead to further understanding of the neurobehavioral mechanisms contributing to a clinical outcome as well as new targets for genetic association analysis.
10.4.3 Informing animal models
Criterion #9 in Table 1 notes that an endophenotype can inform an animal model, which can help us to understand the neurobehavioral mechanisms captured by an endophenotype as well as associated gene function. For instance, a number of animal models of PPI deficits have been developed, with the hope of understanding the etiology and pathophysiology of schizophrenia. Reduced PPI in rats is associated with increased perseverative responding during task switching (Freudenberg, Dieckmann, Winter, Koch, & Schwabe, 2007), which is often characteristic of schizophrenic patients during tasks such as the Wisconsin Card Sorting Test, which also requires behavioral flexibility and switching. Reduced levels of methylation of NRG1 have been reported in rats bred for reduced levels of PPI, and methylation levels are reduced in brain regions associated with both PPI and schizophrenia, such as medial PFC, hippocampus, nucleus accumbens (Rhein et al., 2013). This type of finding may lead to understanding epigenetic changes associated with schizophrenia as well as potential therapeutic targets. Oscillatory activity in the EEG of mice selectively bred to respond positively to alcohol administration resembles event-related time-frequency findings in humans in some respects (although not others), lending credence to mouse models of alcoholism (Criado & Ehlers, 2009) and suggesting routes to understanding time-frequency endophenotypes in humans. In addition, delayed latency of the N1 EEG or MEG response is a putative endophenotype for autism spectrum disorders (ASD). A recent investigation found that the N1 response in the right hemisphere was delayed in children with ASD relative to typically developing children, as expected. Investigators also observed a latency delay of virtually the same magnitude in mice treated prenatally with valproic acid (VPA), an insult-based mouse model of autism. This latency delay was inversely correlated with inter-trial phase locking of gamma activity in both species. Moreover, expression levels of messenger RNA for the autism risk gene neuroligin-3 (NLGN3), which are reduced in mice exposed to VPA, were associated with gamma phase locking in mice as well. Thus, this study suggests the possibility that an autism endophenotype may be related to an autism risk gene through a specific mechanism (gamma phase locking), which would help to understand the gene function reflected in the mediating mechanism as well as the specific nature of the genetic risk conveyed by the candidate gene and candidate endophenotype.
11.0 Summary and conclusions
To conclude, we offer the following list as our take home message to encourage best practices and avoiding past pitfalls that have plagued molecular genetic investigations of complex traits. We believe that future endophenotype research will be most profitable if investigators consider these points in the design and conduct of their research.
Most endophenotypes only meet threshold criteria. Most endophenotype research is confined to establishing whether an electrophysiological variable meets the threshold criteria in Section I of Table 1. Hence, other than increasing the list of variables with endophenotype potential (see Table 5), endophenotype research has underperformed in its promise to deliver insights into the etiology of psychiatric disorders.
The very definition of an endophenotype should evolve. The Gottesman & Gould (Irving I Gottesman & Gould, 2003) endophenotype criteria have proved monumentally valuable for stimulating research on endophenotypes, but we believe the time is ripe to adopt the criteria in Table 1 as an aid to further advance the field. It is time to define an endophenotype as a biobehavioral trait that not only identifies genetic liability for a disorder, but one that has demonstrated robust, verifiable association with specific genetic variants that in turn are associated with a clinically relevant behavioral phenotype (as noted in Section II of Table 1). At present, only one electrophysiological variable, resting heart rate, has reached either of these thresholds (see Tables 2, 3, & 7).
Endophenotypes are the same as other complex traits in their genetic architecture. There is no compelling evidence that endophenotypes are associated with genetic variants of large effect or that they are substantially different than other complex phenotypes (see Figure 1). They appear to be polygenic traits influenced by many thousands of genetic variants, all contributing small effects. They do not appear to be any better than clinical phenotypes in their potential to assist gene finding
Sample size, sample size, sample size. How the electrophysiological variable is operationalized and measured is certainly important, and improving measurement improves reliability and thus statistical power, but the failure of endophenotypes to uncover genes is not due to poor measurement or particular sampling approaches, and improving measurement or sampling from extremes of the population will not overcome the need for very large samples (see Table 3).
Candidate gene studies involving endophenotypes have not uncovered verified genetic variants (see Table 6). They should be discouraged in favor of using large sample GWAS studies and GWAS meta-analysis for genetic discovery.
Data sharing. Using endophenotypes to discover novel genetic variants will require sample sizes that number in the thousands or more likely in the tens of thousands, sample sizes that will not be attainable unless investigators share data and establish consortia to pool genotyped samples and harmonize measures.
Use endophenotypes to understand mechanisms, not to discover genes. It is now clear that large sample studies needed to identify verified genetic variants associated with psychopathology can be obtained by pooling those with a harmonized measure of psychopathology. In this context, a key value of endophenotypes will not be to identify novel genetic associations, but rather to identify and characterize neural mechanisms associated with verified genetic variants associated with psychopathology and relevant psychological traits (see Figure 3).
Use genetic signals to develop new (and better) endophenotypes. Once verified genetic variants associated with psychopathology are identified, research is encouraged to capitalize on what is known about the likely functional significance of the variants to develop new endophenotypes. An implication of this is that, if in 10 years P300 amplitude reduction is still the most widely studied electrophysiological endophenotype for disinhibitory psychopathology and schizophrenia (see Table 5), we will have to conclude that the endophenotype molecular genetic research program has failed.
Endophenotypes are of value even if they do not help find genes. They are underappreciated for their likely predictive utility, ability to enhance understanding of brain mechanisms, and potential to inform animal models (see Section III of Table 1).
Supplementary Material
Highlights.
The potential of endophenotypes for identifying psychopathology-related genes has been oversold
Endophenotypes have largely failed to produce verified molecular genetic associations
Endophenotypes are complex and influenced by many genes each with very small effect
Extremely large samples are necessary to discover variants associated with endophenotypes
Endophenotype research should be informed by molecular genetic findings
Endophenotypes are valuable even without producing molecular genetic hits
Acknowledgments
This work was supported through National Institute of health Grants DA05147, DA036216, AA091367, DA024417, AA023974, DA037904, and DA040177
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
William G. Iacono, University of Minnesota
Stephen M. Malone, University of Minnesota
Scott I. Vrieze, University of Colorado Boulder
References
- 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–+. doi: 10.1038/nature15393. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agam Y, Vangel M, Roffman JL, Gallagher PJ, Chaponis J, Haddad S, Manoach DS. Dissociable genetic contributions to error processing: a multimodal neuroimaging study. PLoS One. 2014;9:e101784. doi: 10.1371/journal.pone.0101784. doi: 10.1371/journal.pone.0101784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokhin AP. Genetic psychophysiology: advances, problems, and future directions. Int J Psychophysiol. 2014;93:173–197. doi: 10.1016/j.ijpsycho.2014.04.003. doi: 10.1016/j.ijpsycho.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokhin AP, Golosheykin S, Heath AC. Genetic and environmental influences on emotion-modulated startle reflex: a twin study. Psychophysiology. 2007;44:106–112. doi: 10.1111/j.1469-8986.2006.00486.x. doi: 10.1111/j.1469-8986.2006.00486.x. [DOI] [PubMed] [Google Scholar]
- Anokhin AP, Heath AC, Myers E, Ralano A, Wood S. Genetic influences on prepulse inhibition of startle reflex in humans. Neurosci Lett. 2003;353:45–48. doi: 10.1016/j.neulet.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Baker LA, Tuvblad C, Reynolds C, Zheng M, Lozano DI, Raine A. Resting heart rate and the development of antisocial behavior from age 9 to 14: genetic and environmental influences. Dev Psychopathol. 2009;21:939–960. doi: 10.1017/S0954579409000509. doi: 10.1017/S0954579409000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchaine TP. The Role of Biomarkers and Endophenotypes in Prevention and Treatment of Psychopathological Disorders. Biomark Med. 2009;3:1–3. doi: 10.2217/17520363.3.1.1. doi: 10.2217/17520363.3.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Bihari B, Kissin B. Event-related brain potentials in boys at risk for alcoholism. Science. 1984;225:1493–1496. doi: 10.1126/science.6474187. [DOI] [PubMed] [Google Scholar]
- Benowitz NL. Cotinine as a biomarker of environmental tobacco smoke exposure. Epidemiologic Reviews. 1996;18:188–204. doi: 10.1093/oxfordjournals.epirev.a017925. [DOI] [PubMed] [Google Scholar]
- Bress JN, Meyer A, Proudfit GH. The stability of the feedback negativity and its relationship with depression during childhood and adolescence. Dev Psychopathol. 2015;27:1285–1294. doi: 10.1017/S0954579414001400. doi: 10.1017/S0954579414001400. [DOI] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Loh PR, Neale BM. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton BK, Hjorthoj C, Jepsen JR, Thorup A, Nordentoft M, Plessen KJ. Research Review: Do motor deficits during development represent an endophenotype for schizophrenia? A meta-analysis. J Child Psychol Psychiatry. 2015 doi: 10.1111/jcpp.12479. doi: 10.1111/jcpp.12479. [DOI] [PubMed] [Google Scholar]
- Burwell SJ, Malone SM, Bernat EM, Iacono WG. Does electroencephalogram phase variability account for reduced P3 brain potential in externalizing disorders? Clin Neurophysiol. 2014;125:2007–2015. doi: 10.1016/j.clinph.2014.02.020. doi: 10.1016/j.clinph.2014.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabranes JA, Ancin I, Santos JL, Sanchez-Morla E, Garcia-Jimenez MA, Lopez-Ibor JJ, Barabash A. No effect of polymorphisms in the non-duplicated region of the CHRNA7 gene on sensory gating P50 ratios in patients with schizophrenia and bipolar disorder. Psychiatry Res. 2013;205:276–278. doi: 10.1016/j.psychres.2012.08.015. doi: 10.1016/j.psychres.2012.08.015. [DOI] [PubMed] [Google Scholar]
- Campanella S, Pogarell O, Boutros N. Event-related potentials in substance use disorders: a narrative review based on articles from 1984 to 2012. Clin EEG Neurosci. 2014;45:67–76. doi: 10.1177/1550059413495533. doi: 10.1177/1550059413495533. [DOI] [PubMed] [Google Scholar]
- Carlson SR, Iacono WG. Heritability of P300 amplitude development from adolescence to adulthood. Psychophysiology. 2006;43:470–480. doi: 10.1111/j.1469-8986.2006.00450.x. doi: 10.1111/j.1469-8986.2006.00450.x. [DOI] [PubMed] [Google Scholar]
- Carlson SR, Iacono WG. Deviant P300 amplitude development in males is associated with paternal externalizing psychopathology. J Abnorm Psychol. 2008;117:910–923. doi: 10.1037/a0013443. doi: 2008-16252-016 [pii] 10.1037/a0013443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson SR, Iacono WG, McGue M. P300 amplitude in nonalcoholic adolescent twin pairs who become discordant for alcoholism as adults. Psychophysiology. 2004;41:841–844. doi: 10.1111/j.1469-8986.2004.00238.x. doi: 10.1111/j.0048-5772.2004.00238.x. [DOI] [PubMed] [Google Scholar]
- Chabris CF, Lee JJ, Cesarini D, Benjamin DJ, Laibson DI. The Fourth Law of Behavior Genetics. Curr Dir Psychol Sci. 2015;24:304–312. doi: 10.1177/0963721415580430. doi: 10.1177/0963721415580430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Kim HL. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41:527–534. doi: 10.1038/ng.357. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim KH, Quon G, Meuleman W, Haugen C, Kellis M. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz BA, Sweeney JA, Hamm JP, Ivleva EI, Ethridge LE, Pearlson GD, Tamminga CA. Identification of Distinct Psychosis Biotypes Using Brain-Based Biomarkers. Am J Psychiatry. 2015:appiajp201514091200. doi: 10.1176/appi.ajp.2015.14091200. doi: 10.1176/appi.ajp.2015.14091200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. Statistical power analysis for the behavioral sciences. 2nd ed. Lawrence Erlbaum; Hillsdale, New Jersey: 1988. [Google Scholar]
- Criado JR, Ehlers CL. Event-related oscillations as risk markers in genetic mouse models of high alcohol preference. Neuroscience. 2009;163:506–523. doi: 10.1016/j.neuroscience.2009.06.039. doi: 10.1016/j.neuroscience.2009.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronbach LJ, Meehl PE. Construct validity in psychological tests. Psychological Bulletin. 1955a;52:281–302. doi: 10.1037/h0040957. [DOI] [PubMed] [Google Scholar]
- Cronbach LJ, Meehl PE. Construct validity in psychological tests. Psychological Bulletin. 1955b;52:281–302. doi: 10.1037/h0040957. [DOI] [PubMed] [Google Scholar]
- Curtis CE, Calkins ME, Grove WM, Feil KJ, Iacono WG. Saccadic disinhibition in patients with acute and remitted schizophrenia and their first-degree biological relatives. American Journal of Psychiatry. 2001;158:100–106. doi: 10.1176/appi.ajp.158.1.100. [DOI] [PubMed] [Google Scholar]
- Cuthbert BN. Translating Intermediate Phenotypes to Psychopathology: The NIMH Research Domain Criteria. Psychophysiology. 2014 doi: 10.1111/psyp.12342. [DOI] [PubMed] [Google Scholar]
- Del Re EC, Bergen SE, Mesholam-Gately RI, Niznikiewicz MA, Goldstein JM, Woo TU, Petryshen TL. Analysis of schizophrenia-related genes and electrophysiological measures reveals ZNF804A association with amplitude of P300b elicited by novel sounds. Transl Psychiatry. 2014;4:e346. doi: 10.1038/tp.2013.117. doi: 10.1038/tp.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo R, Nalls MA, Avery CL, Smith JG, Evans DS, Keller MF, Whitsel EA. Common genetic variation near the connexin-43 gene is associated with resting heart rate in African Americans: a genome-wide association study of 13,372 participants. Heart Rhythm. 2013;10:401–408. doi: 10.1016/j.hrthm.2012.11.014. doi: 10.1016/j.hrthm.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Gizer IR. Evidence for a genetic component for substance dependence in Native Americans. Am J Psychiatry. 2013;170:154–164. doi: 10.1176/appi.ajp.2012.12010113. doi: 10.1176/appi.ajp.2012.12010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Phillips E. Association of EEG alpha variants and alpha power with alcohol dependence in Mexican American young adults. Alcohol. 2007;41:13–20. doi: 10.1016/j.alcohol.2007.02.001. doi: 10.1016/j.alcohol.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Wills DN, Phillips E, Havstad J. Low voltage alpha EEG phenotype is associated with reduced amplitudes of alpha event-related oscillations, increased cortical phase synchrony, and a low level of response to alcohol. Int J Psychophysiol. 2015;98:65–75. doi: 10.1016/j.ijpsycho.2015.07.002. doi: 10.1016/j.ijpsycho.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eijgelsheim M, Newton-Cheh C, Sotoodehnia N, de Bakker PI, Muller M, Morrison AC, O'Donnell CJ. Genome-wide association analysis identifies multiple loci related to resting heart rate. Hum Mol Genet. 2010;19:3885–3894. doi: 10.1093/hmg/ddq303. doi: 10.1093/hmg/ddq303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Rivadeneira F. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501. doi: 10.1038/ng.2249. doi: 10.1038/ng.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euser AS, Arends LR, Evans BE, Greaves-Lord K, Huizink AC, Franken IH. The P300 event-related brain potential as a neurobiological endophenotype for substance use disorders: a meta-analytic investigation. Neurosci Biobehav Rev. 2012;36:572–603. doi: 10.1016/j.neubiorev.2011.09.002. doi: 10.1016/j.neubiorev.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Faraone SV, Bonvicini C, Scassellati C. Biomarkers in the diagnosis of ADHD--promising directions. Curr Psychiatry Rep. 2014;16:497. doi: 10.1007/s11920-014-0497-1. doi: 10.1007/s11920-014-0497-1. [DOI] [PubMed] [Google Scholar]
- Farrell MS, Werge T, Sklar P, Owen MJ, Ophoff RA, O'Donovan MC, Sullivan PF. Evaluating historical candidate genes for schizophrenia. Molecular Psychiatry. 2015;20:555–562. doi: 10.1038/mp.2015.16. doi: 10.1038/mp.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarelli F. Endophenotypes and biological markers of schizophrenia: from biological signs of illness to novel treatment targets. Curr Pharm Des. 2013;19:6462–6479. doi: 10.2174/13816128113199990554. [DOI] [PubMed] [Google Scholar]
- Ford JM. Studying auditory verbal hallucinations using the RDoC framework. Psychophysiology. 2015 doi: 10.1111/psyp.12457. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JM, Morris SE, Hoffman RE, Sommer I, Waters F, McCarthy-Jones S, Cuthbert BN. Studying hallucinations within the NIMH RDoC framework. Schizophr Bull. 2014;40(Suppl 4):S295–304. doi: 10.1093/schbul/sbu011. doi: 10.1093/schbul/sbu011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JM, Roach BJ, Hoffman RS, Mathalon DH. The dependence of P300 amplitude on gamma synchrony breaks down in schizophrenia. Brain Res. 2008;1235:133–142. doi: 10.1016/j.brainres.2008.06.048. doi: 10.1016/j.brainres.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster SE, Carter CS, Cohen JD, Cho RY. Parametric manipulation of the conflict signal and control-state adaptation. J Cogn Neurosci. 2011;23:923–935. doi: 10.1162/jocn.2010.21458. doi: 10.1162/jocn.2010.21458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudenberg F, Dieckmann M, Winter S, Koch M, Schwabe K. Selective breeding for deficient sensorimotor gating is accompanied by increased perseveration in rats. Neuroscience. 2007;148:612–622. doi: 10.1016/j.neuroscience.2007.06.034. doi: 10.1016/j.neuroscience.2007.06.034. [DOI] [PubMed] [Google Scholar]
- Furberg H, Kim Y, Dackor J, Boerwinkle E, Franceschini N, Ardissino D, Sullivan PF. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature Genetics. 2010;42:441–U134. doi: 10.1038/ng.571. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge T, Nichols TE, Lee PH, Holmes AJ, Roffman JL, Buckner RL, Smoller JW. Massively expedited genome-wide heritability analysis (MEGHA). Proc Natl Acad Sci U S A. 2015;112:2479–2484. doi: 10.1073/pnas.1415603112. doi: 10.1073/pnas.1415603112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore CS, Malone SM, Iacono WG. Brain electrophysiological endophenotypes for externalizing psychopathology: a multivariate approach. Behav Genet. 2010;40:186–200. doi: 10.1007/s10519-010-9343-3. doi: 10.1007/s10519-010-9343-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Knowles EE, McKay DR, Sprooten E, Raventos H, Blangero J, Almasy L. Arguments for the sake of endophenotypes: examining common misconceptions about the use of endophenotypes in psychiatric genetics. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:122–130. doi: 10.1002/ajmg.b.32221. doi: 10.1002/ajmg.b.32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein BL, Klein DN. A review of selected candidate endophenotypes for depression. Clin Psychol Rev. 2014;34:417–427. doi: 10.1016/j.cpr.2014.06.003. doi: 10.1016/j.cpr.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. American Journal of Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Shields J. Schizophrenia and genetics. Academic Press; New York, NY: 1972. [Google Scholar]
- Greenwood TA, Swerdlow NR, Gur RE, Cadenhead KS, Calkins ME, Dobie DJ, Lazzeroni LC. Genome-wide linkage analyses of 12 endophenotypes for schizophrenia from the consortium on the genetics of schizophrenia. American Journal of Psychiatry. 2013;170:521–532. doi: 10.1176/appi.ajp.2012.12020186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella JF, Wexler NS, Conneally PM, Naylor SL, Anderson MA, Tanzi RE, et al. A polymorphic DNA marker genetically linked to Huntington's disease. Nature. 1983;306:234–238. doi: 10.1038/306234a0. [DOI] [PubMed] [Google Scholar]
- Hall MH, Chen CY, Cohen BM, Spencer KM, Levy DL, Ongur D, Smoller JW. Genomewide association analyses of electrophysiological endophenotypes for schizophrenia and psychotic bipolar disorders: a preliminary report. Am J Med Genet B Neuropsychiatr Genet. 2015;168B:151–161. doi: 10.1002/ajmg.b.32298. doi: 10.1002/ajmg.b.32298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MH, Levy DL, Salisbury DF, Haddad S, Gallagher P, Lohan M, Smoller JW. Neurophysiologic effect of GWAS derived schizophrenia and bipolar risk variants. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:9–18. doi: 10.1002/ajmg.b.32212. doi: 10.1002/ajmg.b.32212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasenkamp W, Epstein MP, Green A, Wilcox L, Boshoven W, Lewison B, Duncan E. Heritability of acoustic startle magnitude, prepulse inhibition, and startle latency in schizophrenia and control families. Psychiatry Res. 2010;178:236–243. doi: 10.1016/j.psychres.2009.11.012. doi: 10.1016/j.psychres.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Shen S, Locke J, Steinhauer SR, Konicky C, Lowers L, Connolly J. Developmental delay in P300 production in children at high risk for developing alcohol-related disorders. Biological Psychiatry. 1999;46:970–981. doi: 10.1016/s0006-3223(99)00032-3. [DOI] [PubMed] [Google Scholar]
- Hinkley LB, Vinogradov S, Guggisberg AG, Fisher M, Findlay AM, Nagarajan SS. Clinical symptoms and alpha band resting-state functional connectivity imaging in patients with schizophrenia: implications for novel approaches to treatment. Biol Psychiatry. 2011;70:1134–1142. doi: 10.1016/j.biopsych.2011.06.029. doi: 10.1016/j.biopsych.2011.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CA, Enoch MA, Srivastava V, Cummins-Oman JS, Ferrier C, Iarikova P, Goldman D. Genome-wide association identifies candidate genes that influence the human electroencephalogram. Proc Natl Acad Sci U S A. 2010;107:8695–8700. doi: 10.1073/pnas.0908134107. doi: 10.1073/pnas.0908134107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Stefansson K. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- Holroyd CB, Coles MG. Dorsal anterior cingulate cortex integrates reinforcement history to guide voluntary behavior. Cortex. 2008;44:548–559. doi: 10.1016/j.cortex.2007.08.013. doi: 10.1016/j.cortex.2007.08.013. [DOI] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nature Genetics. 2012;44:955–+. doi: 10.1038/ng.2354. doi: Doi 10.1038/Ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG. Psychophysiologic markers of psychopathology: A review. Canadian Psychology/Psychologie canadienne. 1985;26:96–112. [Google Scholar]
- Iacono WG. Identifying psychophysiological risk for psychopathology: examples from substance abuse and schizophrenia research. Psychophysiology. 1998;35:621–637. [PubMed] [Google Scholar]
- Iacono WG. Genome-wide scans of genetic variants for psychophysiological endophenotypes: introduction to this special issue of Psychophysiology. Psychophysiology. 2014a;51:1201–1202. doi: 10.1111/psyp.12340. doi: 10.1111/psyp.12340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG. Neurobehavioral aspects of multidimensional psychopathology. Am J Psychiatry. 2014b;171:1236–1239. doi: 10.1176/appi.ajp.2014.14091132. doi: 10.1176/appi.ajp.2014.14091132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG. Achieving Success with the Research Domain Criteria (RDoC): Going beyond the Matrix. Psychophysiology. 2016 doi: 10.1111/psyp.12584. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Carlson SR, Malone SM. Identifying a multivariate endophenotype for substance use disorders using psychophysiological measures. Int J Psychophysiol. 2000;38:81–96. doi: 10.1016/s0167-8760(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Carlson SR, Malone SM, McGue M. P3 event-related potential amplitude and the risk for disinhibitory disorders in adolescent boys. Arch Gen Psychiatry. 2002;59:750–757. doi: 10.1001/archpsyc.59.8.750. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Lykken DT. Two-year retest stability of eye tracking performance and a comparison of electro-oculographic and infrared recording techniques: evidence of EEG in the electro-oculogram. Psychophysiology. 1981;18:49–55. doi: 10.1111/j.1469-8986.1981.tb01542.x. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Malone SM. Developmental Endophenotypes: Indexing Genetic Risk for Substance Abuse with the P300 Brain Event-Related Potential. Child Development Perspectives. 2011;5:239–247. doi: 10.1111/j.1750-8606.2011.00205.x. doi: 10.1111/j.1750-8606.2011.00205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, McGue M. Behavioral disinhibition and the development of early-onset addiction: common and specific influences. Annu Rev Clin Psychol. 2008;4:325–348. doi: 10.1146/annurev.clinpsy.4.022007.141157. doi: 10.1146/annurev.clinpsy.4.022007.141157. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, Vaidyanathan U, Vrieze SI. Genome wide scans of genetic variants for psychophysiological endophenotypes: A methodological overview. Psychophysiology. 2014a doi: 10.1111/psyp.12343. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Malone SM, Vaidyanathan U, Vrieze SI. Genome-wide scans of genetic variants for psychophysiological endophenotypes: a methodological overview. Psychophysiology. 2014b;51:1207–1224. doi: 10.1111/psyp.12343. doi: 10.1111/psyp.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, McGue M. Minnesota Twin Family Study. Twin Research. 2002;5:482–487. doi: 10.1375/136905202320906327. [DOI] [PubMed] [Google Scholar]
- Iacono WG, McGue M, Krueger RF. Minnesota Center for Twin and Family Research. Twin Research and Human Genetics. 2006;9:978–984. doi: 10.1375/183242706779462642. doi: 10.1375/183242706779462642. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Peloquin LJ, Lumry AE, Valentine RH, Tuason VB. Eye tracking in patients with unipolar and bipolar affective disorders in remission. J Abnorm Psychol. 1982;91:35–44. doi: 10.1037//0021-843x.91.1.35. [DOI] [PubMed] [Google Scholar]
- Iacono WG, Tuason VB, Johnson RA. Dissociation of smooth-pursuit and saccadic eye tracking in remitted schizophrenics. An ocular reaction time task that schizophrenic perform well. Arch Gen Psychiatry. 1981;38:991–996. doi: 10.1001/archpsyc.1981.01780340043005. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–648. doi: 10.1097/EDE.0b013e31818131e7. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- James LM, Engdahl BE, Leuthold AC, Lewis SM, Van Kampen E, Georgopoulos AP. Neural network modulation by trauma as a marker of resilience: differences between veterans with posttraumatic stress disorder and resilient controls. JAMA Psychiatry. 2013;70:410–418. doi: 10.1001/jamapsychiatry.2013.878. doi: 10.1001/jamapsychiatry.2013.878. [DOI] [PubMed] [Google Scholar]
- Kamarajan C, Porjesz B. Advances in electrophysiological research. Alcohol Res. 2015;37:53–87. [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SJ, Rangaswamy M, Manz N, Wang JC, Wetherill L, Hinrichs T, Dick D. Family-based genome-wide association study of frontal theta oscillations identifies potassium channel gene KCNJ6. Genes, Brain and Behavior. 2012;11:712–719. doi: 10.1111/j.1601-183X.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas S, Erzengin OU, Basar E. The genesis of human event-related responses explained through the theory of oscillatory neural assemblies. Neurosci Lett. 2000;285:45–48. doi: 10.1016/s0304-3940(00)01022-3. doi: S0304-3940(00)01022-3 [pii] [DOI] [PubMed] [Google Scholar]
- Kattoulas E, Stefanis NC, Avramopoulos D, Stefanis CN, Evdokimidis I, Smyrnis N. Schizophrenia-related RGS4 gene variations specifically disrupt prefrontal control of saccadic eye movements. Psychological Medicine. 2012;42:757–767. doi: 10.1017/S003329171100167X. doi: S003329171100167X [pii] 10.1017/S003329171100167X. [DOI] [PubMed] [Google Scholar]
- Kichaev G, Pasaniuc B. Leveraging Functional-Annotation Data in Trans-ethnic Fine-Mapping Studies. Am J Hum Genet. 2015;97:260–271. doi: 10.1016/j.ajhg.2015.06.007. doi: 10.1016/j.ajhg.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolev V, Demiralp T, Yordanova J, Ademoglu A, Isoglu-Alkaç Ü. Time-frequency analysis reveals multiple functional components during oddball P300. Neuroreport. 1997;8:2061–2065. doi: 10.1097/00001756-199705260-00050. [DOI] [PubMed] [Google Scholar]
- Kottgen A, Albrecht E, Teumer A, Vitart V, Krumsiek J, Hundertmark C, Gieger C. Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet. 2013;45:145–154. doi: 10.1038/ng.2500. doi: 10.1038/ng.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer UM, Cunillera T, Camara E, Marco-Pallares J, Cucurell D, Nager W, Munte TF. The impact of catechol-O-methyltransferase and dopamine D4 receptor genotypes on neurophysiological markers of performance monitoring. J Neurosci. 2007;27:14190–14198. doi: 10.1523/JNEUROSCI.4229-07.2007. doi: 10.1523/JNEUROSCI.4229-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latvala A, Kuja-Halkola R, Almqvist C, Larsson H, Lichtenstein P. A Longitudinal Study of Resting Heart Rate and Violent Criminality in More Than 700000 Men. JAMA Psychiatry. 2015;72:971–978. doi: 10.1001/jamapsychiatry.2015.1165. doi: 10.1001/jamapsychiatry.2015.1165. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Vattikuti S, Chow CC. Uncovering the Genetic Architectures of Quantitative Traits. Computational and Structural Biotechnology Journal. 2016;14:28–34. doi: 10.1016/j.csbj.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28:2540–2542. doi: 10.1093/bioinformatics/bts474. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenartowicz A, Delorme A, Walshaw PD, Cho AL, Bilder RM, McGough JJ, Loo SK. Electroencephalography correlates of spatial working memory deficits in attention-deficit/hyperactivity disorder: vigilance, encoding, and maintenance. J Neurosci. 2014;34:1171–1182. doi: 10.1523/JNEUROSCI.1765-13.2014. doi: 10.1523/JNEUROSCI.1765-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzenweger MF. Thinking clearly about the endophenotype-intermediate phenotypebiomarker distinctions in developmental psychopathology research. Dev Psychopathol. 2013;25:1347–1357. doi: 10.1017/S0954579413000655. doi: 10.1017/S0954579413000655. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: Using Sequence and Genotype Data to Estimate Haplotypes and Unobserved Genotypes. Genetic Epidemiology. 2010;34:816–834. doi: 10.1002/gepi.20533. doi: Doi 10.1002/Gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu DJ, Leal SM. A unified method for detecting secondary trait associations with rare variants: application to sequence data. PLoS Genet. 2012;8:e1003075. doi: 10.1371/journal.pgen.1003075. doi: 10.1371/journal.pgen.1003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Speliotes EK. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo SK, Lenartowicz A, Makeig S. Research Review: use of EEG biomarkers in child psychiatry research - current state and future directions. J Child Psychol Psychiatry. 2015 doi: 10.1111/jcpp.12435. doi: 10.1111/jcpp.12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos RJF, Yeo GSH. The bigger picture of FTO-the first GWAS-identified obesity gene. Nature Reviews Endocrinology. 2014;10:51–61. doi: 10.1038/nrendo.2013.227. doi: 10.1038/nrendo.2013.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone SM, McGue M, Iacono WG. What can time-frequency and phase coherence measures tell us about the genetic basis of P3 amplitude? International Journal of Psychophysiology. 2016 doi: 10.1016/j.ijpsycho.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone SM, Vaidyanathan U, Basu S, Miller MB, McGue M, Iacono WG. Heritability and molecular-genetic basis of the P3 event-related brain potential: a genome-wide association study. Psychophysiology. 2014;51:1246–1258. doi: 10.1111/psyp.12345. doi: 10.1111/psyp.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoach DS, Agam Y. Neural markers of errors as endophenotypes in neuropsychiatric disorders. Front Hum Neurosci. 2013;7:350. doi: 10.3389/fnhum.2013.00350. doi: 10.3389/fnhum.2013.00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland GH. Increasing statistical power without increasing sample size. American Psychologist. 2000;55:963–964. doi: Doi 10.1037/0003-066x.55.8.963. [Google Scholar]
- Meyer A, Hajcak G, Torpey-Newman DC, Kujawa A, Klein DN. Enhanced error-related brain activity in children predicts the onset of anxiety disorders between the ages of 6 and 9. J Abnorm Psychol. 2015;124:266–274. doi: 10.1037/abn0000044. doi: 10.1037/abn0000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Weinberg A, Klein DN, Hajcak G. The development of the error-related negativity (ERN) and its relationship with anxiety: evidence from 8 to 13 year-olds. Dev Cogn Neurosci. 2012;2:152–161. doi: 10.1016/j.dcn.2011.09.005. doi: 10.1016/j.dcn.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GA, Rockstroh B. Endophenotypes in psychopathology research: where do we stand? Annual Review of Clinical Psychology. 2013;9:177–213. doi: 10.1146/annurev-clinpsy-050212-185540. doi: 10.1146/annurev-clinpsy-050212-185540. [DOI] [PubMed] [Google Scholar]
- Mitchell AM, Possel P. Frontal brain activity pattern predicts depression in adolescent boys. Biol Psychol. 2012;89:525–527. doi: 10.1016/j.biopsycho.2011.12.008. doi: 10.1016/j.biopsycho.2011.12.008. [DOI] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Replication DG. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nature Genetics. 2012;44:981–+. doi: 10.1038/ng.2383. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses-Kolko EL, Horner MS, Phillips ML, Hipwell AE, Swain JE. In search of neural endophenotypes of postpartum psychopathology and disrupted maternal caregiving. J Neuroendocrinol. 2014;26:665–684. doi: 10.1111/jne.12183. doi: 10.1111/jne.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis S, Holroyd CB, Mol N, Coles MG. Reinforcement-related brain potentials from medial frontal cortex: origins and functional significance. Neurosci Biobehav Rev. 2004;28:441–448. doi: 10.1016/j.neubiorev.2004.05.003. doi: 10.1016/j.neubiorev.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Niv S, Ashrafulla S, Tuvblad C, Joshi A, Raine A, Leahy R, Baker LA. Childhood EEG frontal alpha power as a predictor of adolescent antisocial behavior: a twin heritability study. Biol Psychol. 2015;105:72–76. doi: 10.1016/j.biopsycho.2014.11.010. doi: 10.1016/j.biopsycho.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden-Krichmar TM, Gizer IR, Phillips E, Wilhelmsen KC, Schork NJ, Ehlers CL. Variants Near CCK Receptors are Associated With Electrophysiological Responses to Pre-pulse Startle Stimuli in a Mexican American Cohort. Twin Res Hum Genet. 2015;18:727–737. doi: 10.1017/thg.2015.77. doi: 10.1017/thg.2015.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA, Cesarini D. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet. 2016;48:624–633. doi: 10.1038/ng.3552. doi: 10.1038/ng.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, Benjamin DJ. Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 2016;533:539–542. doi: 10.1038/nature17671. doi: 10.1038/nature17671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr SP, Metzger LJ, Lasko NB, Macklin ML, Hu FB, Shalev AY, Harvard/Veterans Affairs Post-traumatic Stress Disorder Twin Study, I. Physiologic responses to sudden, loud tones in monozygotic twins discordant for combat exposure: association with posttraumatic stress disorder. Arch Gen Psychiatry. 2003;60:283–288. doi: 10.1001/archpsyc.60.3.283. [DOI] [PubMed] [Google Scholar]
- Owens EM, Bachman P, Glahn DC, Bearden CE. Electrophysiological Endophenotypes for Schizophrenia. Harv Rev Psychiatry. 2016;24:129–147. doi: 10.1097/HRP.0000000000000110. doi: 10.1097/HRP.0000000000000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick CJ, Venables NC, Yancey JR, Hicks BM, Nelson LD, Kramer MD. A construct-network approach to bridging diagnostic and physiological domains: application to assessment of externalizing psychopathology. J Abnorm Psychol. 2013;122:902–916. doi: 10.1037/a0032807. doi: 10.1037/a0032807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlson GD. Etiologic, phenomenologic, and endophenotypic overlap of schizophrenia and bipolar disorder. Annu Rev Clin Psychol. 2015;11:251–281. doi: 10.1146/annurev-clinpsy-032814-112915. doi: 10.1146/annurev-clinpsy-032814-112915. [DOI] [PubMed] [Google Scholar]
- Petrovsky N, Quednow BB, Ettinger U, Schmechtig A, Mossner R, Collier DA, Kumari V. Sensorimotor gating is associated with CHRNA3 polymorphisms in schizophrenia and healthy volunteers. Neuropsychopharmacology. 2010;35:1429–1439. doi: 10.1038/npp.2010.12. doi: 10.1038/npp.2010.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman TJ, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, Posthuma D. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet. 2015;47:702–709. doi: 10.1038/ng.3285. doi: 10.1038/ng.3285. [DOI] [PubMed] [Google Scholar]
- Polich J, Ladish C, Burns T. Normal variation of P300 in children: age, memory span, and head size. Int J Psychophysiol. 1990;9:237–248. doi: 10.1016/0167-8760(90)90056-j. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Price GW, Michie PT, Johnston J, Innes-Brown H, Kent A, Clissa P, Jablensky AV. A multivariate electrophysiological endophenotype, from a unitary cohort, shows greater research utility than any single feature in the Western Australian family study of schizophrenia. Biol Psychiatry. 2006;60:1–10. doi: 10.1016/j.biopsych.2005.09.010. doi: 10.1016/j.biopsych.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Proudfit GH. The reward positivity: from basic research on reward to a biomarker for depression. Psychophysiology. 2015;52:449–459. doi: 10.1111/psyp.12370. doi: 10.1111/psyp.12370. [DOI] [PubMed] [Google Scholar]
- Quednow BB, Brinkmeyer J, Mobascher A, Nothnagel M, Musso F, Grunder G, Winterer G. Schizophrenia risk polymorphisms in the TCF4 gene interact with smoking in the modulation of auditory sensory gating. Proc Natl Acad Sci U S A. 2012;109:6271–6276. doi: 10.1073/pnas.1118051109. doi: 10.1073/pnas.1118051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quednow BB, Ettinger U, Mossner R, Rujescu D, Giegling I, Collier DA, Kumari V. The schizophrenia risk allele C of the TCF4 rs9960767 polymorphism disrupts sensorimotor gating in schizophrenia spectrum and healthy volunteers. J Neurosci. 2011;31:6684–6691. doi: 10.1523/JNEUROSCI.0526-11.2011. doi: 10.1523/jneurosci.0526-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine A, Venables PH, Williams M. Relationships between central and autonomic measures of arousal at age 15 years and criminality at age 24 years. Arch Gen Psychiatry. 1990;47:1003–1007. doi: 10.1001/archpsyc.1990.01810230019003. [DOI] [PubMed] [Google Scholar]
- Rhein M, Muschler MR, Krauss JK, Bleich S, Frieling H, Schwabe K. Hypomethylation of neuregulin in rats selectively bred for reduced sensorimotor gating. Schizophr Res. 2013;150:262–265. doi: 10.1016/j.schres.2013.07.012. doi: 10.1016/j.schres.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Roeske D, Ludwig KU, Neuhoff N, Becker J, Bartling J, Bruder J, Schulte-Korne G. First genome-wide association scan on neurophysiological endophenotypes points to trans-regulation effects on SLC2A3 in dyslexic children. Mol Psychiatry. 2011;16:97–107. doi: 10.1038/mp.2009.102. doi: 10.1038/mp.2009.102. [DOI] [PubMed] [Google Scholar]
- Rosen AM, Spellman T, Gordon JA. Electrophysiological endophenotypes in rodent models of schizophrenia and psychosis. Biol Psychiatry. 2015;77:1041–1049. doi: 10.1016/j.biopsych.2015.03.021. doi: 10.1016/j.biopsych.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Giakoumaki SG, Adamaki E, Bitsios P. The influence of schizophrenia-related neuregulin-1 polymorphisms on sensorimotor gating in healthy males. Biol Psychiatry. 2011;69:479–486. doi: 10.1016/j.biopsych.2010.09.009. doi: 10.1016/j.biopsych.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Roussos P, Giakoumaki SG, Adamaki E, Georgakopoulos A, Robakis NK, Bitsios P. The association of schizophrenia risk D-amino acid oxidase polymorphisms with sensorimotor gating, working memory and personality in healthy males. Neuropsychopharmacology. 2011;36:1677–1688. doi: 10.1038/npp.2011.49. doi: 10.1038/npp.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Giakoumaki SG, Zouraraki C, Fullard JF, Karagiorga VE, Tsapakis EM, Bitsios P. The Relationship of Common Risk Variants and Polygenic Risk for Schizophrenia to Sensorimotor Gating. Biol Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.06.019. doi: 10.1016/j.biopsych.2015.06.019. [DOI] [PubMed] [Google Scholar]
- Rubenstein E, Wiggins LD, Lee LC. A Review of the Differences in Developmental, Psychiatric, and Medical Endophenotypes Between Males and Females with Autism Spectrum Disorder. J Dev Phys Disabil. 2015;27:119–139. doi: 10.1007/s10882-014-9397-x. doi: 10.1007/s10882-014-9397-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury DF, O'Donnell BF, McCarley RW, Nestor PG, Faux SF, Smith RS. Parametric manipulations of auditory stimuli differentially affect P3 amplitude in schizophrenics and controls. Psychophysiology. 1994;31:29–36. doi: 10.1111/j.1469-8986.1994.tb01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore JE, Gottesman II, Dick DM. Endophenotypes for Alcohol Use Disorder: An Update on the Field. Curr Addict Rep. 2015;2:76–90. doi: 10.1007/s40429-015-0046-y. doi: 10.1007/s40429-015-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics, C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, Barroso I. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, McCarroll SA. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh M, Dutt A, Broome MR, Vozmediano AG, Ranlund S, Diez A, Bramon E. Sensory gating deficits in the attenuated psychosis syndrome. Schizophr Res. 2015;161:277–282. doi: 10.1016/j.schres.2014.12.021. doi: 10.1016/j.schres.2014.12.021. [DOI] [PubMed] [Google Scholar]
- Shaikh M, Hall MH, Schulze K, Dutt A, Walshe M, Williams I, Bramon E. Do COMT, BDNF and NRG1 polymorphisms influence P50 sensory gating in psychosis? Psychol Med. 2011;41:263–276. doi: 10.1017/S003329170999239X. doi: 10.1017/s003329170999239x. [DOI] [PubMed] [Google Scholar]
- Sherrington R, Brynjolfsson J, Petursson H, Potter M, Dudleston K, Barraclough B, Gurling H. Localization of a susceptibility locus for schizophrenia on chromosome 5. Nature. 1988;336:164–167. doi: 10.1038/336164a0. doi: 10.1038/336164a0. [DOI] [PubMed] [Google Scholar]
- Singh T, Kurki MI, Curtis D, Purcell SM, Crooks L, McRae J, Barrett JC. Rare loss-of-function variants in SETD1A are associated with schizophrenia and developmental disorders. Nat Neurosci. 2016 doi: 10.1038/nn.4267. doi: 10.1038/nn.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyrnis N, Kattoulas E, Stefanis NC, Avramopoulos D, Stefanis CN, Evdokimidis I. Schizophrenia-related neuregulin-1 single-nucleotide polymorphisms lead to deficient smooth eye pursuit in a large sample of young men. Schizophr Bull. 2011;37:822–831. doi: 10.1093/schbul/sbp150. doi: 10.1093/schbul/sbp150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So HC, Li M, Sham PC. Uncovering the total heritability explained by all true susceptibility variants in a genome-wide association study. Genet Epidemiol. 2011;35:447–456. doi: 10.1002/gepi.20593. doi: 10.1002/gepi.20593. [DOI] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. Am J Hum Genet. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanis NC, Trikalinos TA, Avramopoulos D, Smyrnis N, Evdokimidis I, Ntzani EE, Stefanis CN. Association of RGS4 variants with schizotypy and cognitive endophenotypes at the population level. Behav Brain Funct. 2008;4:46. doi: 10.1186/1744-9081-4-46. doi: 10.1186/1744-9081-4-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM, Enhancing Neuro Imaging Genetics through Meta-Analysis, C. Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet. 2012;44:552–561. doi: 10.1038/ng.2250. doi: 10.1038/ng.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron. 2010;68:182–186. doi: 10.1016/j.neuron.2010.10.003. doi: 10.1016/j.neuron.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton S, Braren M, Zubin J, John ER. Evoked-potential correlates of stimulus uncertainty. Science. 1965;150:1187–1188. doi: 10.1126/science.150.3700.1187. [DOI] [PubMed] [Google Scholar]
- Swerdlow NR, Gur RE, Braff DL. Consortium on the Genetics of Schizophrenia (COGS) assessment of endophenotypes for schizophrenia: an introduction to this Special Issue of Schizophrenia Research. Schizophr Res. 2015;163:9–16. doi: 10.1016/j.schres.2014.09.047. doi: 10.1016/j.schres.2014.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. doi: Doi 10.1038/Nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, Alzheimer's Disease Neuroimaging Initiative, E. C. I. C. S. Y. S. G. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014;8:153–182. doi: 10.1007/s11682-013-9269-5. doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney AL, Gabard-Durnam L, Vogel-Farley V, Tager-Flusberg H, Nelson CA. Developmental trajectories of resting EEG power: an endophenotype of autism spectrum disorder. PLoS One. 2012;7:e39127. doi: 10.1371/journal.pone.0039127. doi: 10.1371/journal.pone.0039127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbricht D, Schmid L, Koller R, Vollenweider FX, Hell D, Javitt DC. Ketamine-induced deficits in auditory and visual context-dependent processing in healthy volunteers: implications for models of cognitive deficits in schizophrenia. Arch Gen Psychiatry. 2000;57:1139–1147. doi: 10.1001/archpsyc.57.12.1139. [DOI] [PubMed] [Google Scholar]
- Vaidyanathan U, Isen JD, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of electrodermal activity: a genome-wide association study. Psychophysiology. 2014;51:1259–1271. doi: 10.1111/psyp.12346. doi: 10.1111/psyp.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Donnelly JM, Hammer MA, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of antisaccade eye tracking error rate: A genome wide association study. Psychophysiology. 2014a doi: 10.1111/psyp.12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Donnelly JM, Hammer MA, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of antisaccade eye tracking error rate: a genome-wide association study. Psychophysiology. 2014b;51:1272–1284. doi: 10.1111/psyp.12347. doi: 10.1111/psyp.12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Malone SM, Miller MB, McGue M, Iacono WG. Heritability and molecular genetic basis of acoustic startle eye blink and affectively modulated startle response: a genome-wide association study. Psychophysiology. 2014;51:1285–1299. doi: 10.1111/psyp.12348. doi: 10.1111/psyp.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan U, Vrieze SI, Iacono WG. The power of theory, research design, and transdisciplinary integration in moving psychopathology forward. Psychological Inquiry. 2015;26:209–230. doi: 10.1080/1047840X.2015.1015367. doi: 10.1080/1047840X.2015.1015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilhjalmsson BJ, Yang J, Finucane HK, Gusev A, Lindstrom S, Ripke S, Price AL. Modeling Linkage Disequilibrium Increases Accuracy of Polygenic Risk Scores. Am J Hum Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Pankratz N, Vaidyanathan U, Miller MB, Kang HM, Iacono WG. Genetic associations of nonsynonymous exonic variants with psychophysiological endophenotypes. Psychophysiology. 2014a doi: 10.1111/psyp.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze SI, Malone SM, Pankratz N, Vaidyanathan U, Miller MB, Kang HM, Iacono WG. Genetic associations of nonsynonymous exonic variants with psychophysiological endophenotypes. Psychophysiology. 2014b;51:1300–1308. doi: 10.1111/psyp.12349. doi: 10.1111/psyp.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Principles of Embryology. George Allen & Unwin; London: 1956. [Google Scholar]
- Walsh MM, Anderson JR. Learning from experience: event-related potential correlates of reward processing, neural adaptation, and behavioral choice. Neurosci Biobehav Rev. 2012;36:1870–1884. doi: 10.1016/j.neubiorev.2012.05.008. doi: 10.1016/j.neubiorev.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware JJ, Chen X, Vink J, Loukola A, Minica C, Pool R, Munafo MR. Genome-Wide Meta-Analysis of Cotinine Levels in Cigarette Smokers Identifies Locus at 4q13.2. Sci Rep. 2016;6:20092. doi: 10.1038/srep20092. doi: 10.1038/srep20092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Gustafsson S, Frayling TM. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Liu H, Li F, Sun N, Ren Y, Liu Z, Zhang K. A polymorphism in the microRNA-30e precursor associated with major depressive disorder risk and P300 waveform. J Affect Disord. 2010;127:332–336. doi: 10.1016/j.jad.2010.05.019. doi: 10.1016/j.jad.2010.05.019. [DOI] [PubMed] [Google Scholar]
- Yancey JR, Venables NC, Patrick CJ. Psychoneurometric operationalization of threat sensitivity: relations with clinical symptom and physiological response criteria. Psychophysiology. 2016 doi: 10.1111/psyp.12512. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Ferreira T, Morris AP, Medland SE, Genetic Investigation of ATC, Replication DIG, Visscher PM. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. S361–363. doi: 10.1038/ng.2213. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HH, Malone SM, Iacono WG. Longitudinal stability and predictive utility of the visual P3 response in adults with externalizing psychopathology. Psychophysiology. 2015 doi: 10.1111/psyp.12548. doi: 10.1111/psyp.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlojutro M, Manz N, Rangaswamy M, Xuei X, Flury-Wetherill L, Koller D, Almasy L. Genome-wide association study of theta band event-related oscillations identifies serotonin receptor gene HTR7 influencing risk of alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:44–58. doi: 10.1002/ajmg.b.31136. doi: 10.1002/ajmg.b.31136. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.