Abstract
Retroelements constitute a large portion of our genomes. One class of these elements, the human endogenous retroviruses (HERVs), is comprised of remnants of ancient exogenous retroviruses that have gained access to the germ line. After integration, most proviruses have been the subject of numerous amplifications and have suffered extensive deletions and mutations. Nevertheless, HERV-derived transcripts and proteins have been detected in healthy and diseased human tissues, and HERV-K, the youngest, most conserved family, is able to form virus-like particles. Although it is generally accepted that the integration of retroelements can cause significant harm by disrupting or disregulating essential genes, the role of HERV expression in the etiology of malignancies and autoimmune and neurologic diseases remains controversial. In recent years, striking evidence has accumulated indicating that some proviral sequences and HERV proteins might even serve the needs of the host and are therefore under positive selection. The remarkable progress in the analysis of host genomes has brought to light the significant impact of HERVs and other retroelements on genetic variation, genome evolution, and gene regulation.
Almost half of the mammalian genome is derived from ancient transposable elements. The two general types, (DNA)-transposons and retroelements, often regarded as “selfish DNA parasites or junk DNA,” encompass ≈2.8% and 42.2% of the human genome, respectively (1, 2). This striking finding is one of the many insights from recent large-scale sequencing projects that have provided the most valuable information in this field since the initial discovery of mobile elements in 1956 by Barbara McClintock (3, 4). Whereas DNA-transposons amplify without an RNA intermediate, retroelements rely on an RNA transcript that is retrotranscribed by a reverse transcriptase before integration in the genome. Here, we briefly review the characteristics of retroelements, their present classification, and the available evidence for their biological significance and function in normal and pathological processes. The focus is on human endogenous retroviruses (HERVs), the remnants of ancient germ-cell infections. Although most of the HERV proviruses have undergone extensive deletions and mutations, some have retained ORFs coding for functional proteins. A few families, including the HERV-K (HML-2) group, have been shown to form viral particles (5, 6), and an apparently intact provirus has recently been discovered in a small fraction of the human population, indicating a very recent acquisition (5–7).
Classification of Retroelements
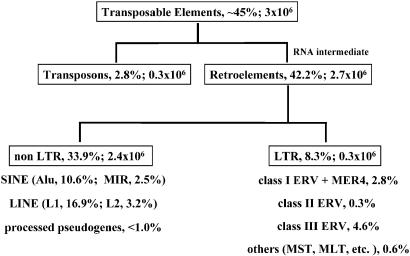
Retroelements constitute 90% of the ≈3 million transposable elements present in the human genome (1). They are split into two large groups, the non-LTR and LTR elements (Fig. 1). Two of the non-LTR members are present in extremely high copy numbers in the mammalian germ line: the short interspersed elements (SINE) with the prominent Alu and MIR repeats and the long-terminal interspersed elements (LINE) containing the autonomous L1 and L2 sequences (8). SINEs have no protein coding capacity and depend on LINE elements for their amplification. The LTR class elements make up 8% of human chromosomes and include retrotransposons, endogenous retroviruses (ERVs), and repeat elements with HERV origin, such as SINE-R (Fig. 2). The SINE-R retroposon family contains a partial sequence of a LTR of HERV-K. The env gene of ERVs confers the potential to spread between cells and individuals. Solitary LTRs of ERVs and retrotransposons, generated by LTR-based homologous recombination processes, are usually one or two orders of magnitude more abundant than preserved or partially complete proviruses (9). In total, >200 families of LTR-containing retroelements are defined in Repbase (10) although, according to Medstrand and coworkers (8), six superfamilies can be defined (Table 1). Whereas class I and II ERVs presumably entered the germ line of primitive primates as infectious retroviruses and subsequently have been subject to multiple amplification and transposition bursts at several time points during primate evolution, the other superfamilies most likely represent ancient retrotransposons that amplified at several stages in earlier mammalian evolution. Most of the retroelements appear to be deeply fixed in the primate genomes and virus free alleles are not known. The rate of new human germ line insertions is presently at an extremely low level compared to earlier periods of evolutionary history or to the rate in some other mammals. At this time, only a small fraction of the youngest subtypes of Alu and L1 non-LTR-elements are still actively retrotransposing in humans (8). It has been estimated that ≈1 in every 100 human births has a de novo insertion of such a retroelement. There is, however, a great deal of uncertainty in these kind of estimations (1). No current transposition activity of HERVs or endogenization of human exogenous retroviruses has been documented so far. Although unlikely, the continuation of such events in our species cannot be completely excluded per se, because examples of probable ongoing endogenization processes are known from other mammals. For example, mouse mammary tumor virus (MMTV) and murine leukemia viruses in mice, Jaagsiekte sheep retrovirus (JSRV) in sheep, porcine endogenous retroviruses (PERV) in pigs, avian leukemia viruses (ALV) in chicken, and feline leukemia virus (FeLV) in cats have presently both endogenous and exogenous strains (11). Moreover, several of the known class II HERV-K proviruses are human specific and a few loci are polymorphic, indicating very recent activity in evolutionary terms (7, 12).
Fig. 1.
Classification of transposable elements. The percentage of each element in the genome and the estimated number of the elements of the main groups are indicated.
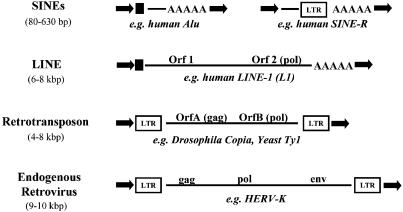
Fig. 2.
Structural features of important retroelements. Arrows indicate repeat sequences generated during the integration process. Filled squares represent promoter regions, and A-runs indicate original polyadenylation. The usual length of the respective element is shown in parentheses.
Table 1. Superfamilies of LTR-containing retroelements.
| Element | Characteristics |
|---|---|
| Class I ERV | Similar to type C or γ-retroviruses |
| Class II ERV | Similar to type B or β-retroviruses |
| Class III ERV | Distantly related to spuma retroviruses |
| MER4 | Nonautonomous class I-related ERVs |
| MST | Named for a common MstII restriction site |
| MLT | Mammalian LTR transposons |
The six superfamilies are grouped according to ref. 8.
The taxonomy of HERVs is still a source of confusion. The preferred systematic nomenclature uses the amino acid specificity of the tRNA that hybridizes to the primer-binding site. The name is defined by adding its one-letter code as a suffix to the acronym HERV (13). HERV-K, for example, uses a lysine-specific tRNA as primer for the initiation of the reverse transcription reaction. The limitations of this approach include the fact that very distantly related viruses use the same tRNAs, and that incomplete information about this short region due to deletions or mutations make classification of these retroviral sequences almost impossible. In addition, many HERV proviruses have been given arbitrary laboratory names or extensions.
HERV Discovery
The driving force behind most efforts to identify new HERVs was the discovery of replication competent endogenous retroviruses that were associated with some forms of cancer in mice, sheep, and other mammals. Most of these efforts relied on low stringency screenings of human genomic libraries with probes derived from conserved pol regions of various animal retroelements (14, 15). By using such a pol fragment of Syrian hamster intracisternal type A particles as a probe in Southern blot analyses, Ono et al. (15) detected the first HERV-K sequence in the human genome. Subsequently, pol-gene homologies together with the primer-binding region were used as a major criterion to classify related HERV elements. Others have used degenerated primers based on known sequences in diverse PCR approaches to search for new endogenized retroviruses (16). A large number of retroelements were found by chance during analysis of specific gene loci and chromosomal regions (17). Recently, most new HERVs or their remnants were identified by searches of the fast growing genomic databases. Very few of the identified HERV loci contain proviruses with preserved ORFs for the three major structural proteins Gag, Pol, and Env (7, 18, 19). All of these most complete proviruses belong to the youngest family of HERV-K elements, which are exclusively present in the genomes of catarrhines (Old World monkeys, apes, and humans), dating the beginning of their endogenization process to ≈45 million years ago (7). Viruses of this family were active both before and after the evolutionary separation of humans and chimpanzees ≈5 million years ago. A few HERV-K loci are exclusive to humans, indicating an integration after the divergence of these two lineages (12, 20). Recently, the youngest known HERV-K provirus, HERV-K113, was identified, and its age was estimated to be <200,000 years (7). Such estimations can be made by comparing the sequence of the two LTRs and knowing that HERVs accumulate mutations at a rate of ≈2.3 × 10-9 to 5 × 10-9 substitutions per site per year (21). These values translate to one difference per LTR every 200,000–450,000 years. Keeping in mind that, at the time of a standard retroviral integration process, both LTRs are identical, one can estimate the time of integration at the given locus. The HERV-K113 provirus, located on chromosome 19p13.11, is not completely fixed in the human population. Genotyping of a small number of genetically diverse individuals has shown an overall allelic frequency of 19%, and the allelic prevalence seems to depend on ethnic origin. Whereas the provirus is rare in Caucasians, it is more abundant in African, Asian, and Polynesian populations. The provirus contains ORFs for all retroviral genes, and all known functional motifs are preserved (7). HERV-K113 is the best candidate for a potentially active provirus in humans today. Allelic polymorphism has also been documented for another human-specific HERV-K, the HERV-K (HML-2.HOM) located on chromosome 7. However, all allelic variants identified so far contain deletions or premature stop codons in the structural genes or, alternatively, a defective reverse transcriptase carrying a mutation in the functionally essential YXDD motif (18). Nevertheless, it remains conceivable that a completely intact, potentially active allele of this or other HERVs are present in the human population at a low allelic frequency or at a higher prevalence in genetically separated ethnic groups.
Proviral Organization and Transcripts of HERV-K
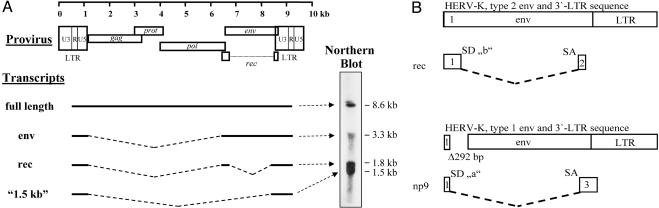
In the human genome, two types of the 30–50 HERV-K proviruses exist. Their genomic organization is shown in Fig. 3. Compared to type 2 sequences, type 1 proviruses are missing a 292-nt stretch of the env gene and their pol and env genes are fused (22). This has not only deleterious consequences for the envelope protein itself, but deeply impacts the complex splicing and protein expression. A proportion of type 2 transcripts is spliced in a regulated way to create mRNAs for the translation of Env and the accessory protein Rec (regulator of expression encoded by cORF; ref. 23). The nucleotide stretch missing in type 1 sequences harbors a splice donor site for the generation of doubly spliced transcripts coding for the two exons of the Rec protein (23, 24). Because of the absence of this splice donor site, type 1 viruses lack the 1.8-kb RNA message for the translation of this protein (Fig. 3A). To date, Rec is the only known auxiliary factor encoded by HERVs. It has a striking functional homology to the RNA-binding nuclear export proteins of lentiviruses, including the HIV protein Rev and HTLV protein Rex. Similar to Rev and Rex, the 14.7-kDa Rec protein binds to unspliced or partially spliced viral transcripts, stabilizes them, and facilitates their transport out of the nucleus to escape the cellular splicing machinery (25).
Fig. 3.
Genomic organization and transcript pattern of HERV-K. (A) The structure of a type 2 HERV-K is shown. (Right) A Northern blot with mRNA of a teratocarcinoma cell line, probed with a labeled HERV-specific cDNA. (B) Schematic representation of the splicing events resulting in rec (HERV-K type 2) and np9 (HERV-K type 1) specific mRNAs. The proviruses differ in a 292-bp deletion (Δ292 bp) that contains the splice donor site used to generate the rec transcripts (SD “b”). An alternative upstream splice donor site (SD “a”) is used for np9 messages. The numbers within the boxes refer to the coding reading frame. (This part of the figure has been redrawn with modifications from ref. 24.)
The Rec binding site or Rec Responsive Element (RcRE) is a highly structured RNA region that has been identified in the U3R segment of the 3′ LTR. Interestingly, Rex and even Rev interact with RcRE and both can partially substitute for Rec function, but Rec does not bind to the RRE element of HIV (25, 26). Because of the presence of the Rec/RcRE regulatory mechanism, HERV-K can no longer be regarded as a simple retrovirus but may represent an intermediate in the evolution from simple retroviruses, containing a constitutive transport element, to complex retroviruses. The possibility has been raised that the more simple exogenous retroviruses might have picked up Rec/RcRE sequences from HERVs to gain an evolutionary advantage (25).
Subcellular localization studies using Rec specific antisera have shown a highly specific staining of the nucleoli (23). Although the presence of Rec transcripts has been documented in healthy testicular tissues, it is expressed at much higher levels in germ cell tumors and cell lines derived therefrom. With respect to these observations, it has been suspected that a dysregulation of Rec expression in this tissue could have an effect on the onset of testicular cancers. The suspicion was substantiated by the observation that Rec supports tumor growth in nude mice and associates with the nuclear promyelocytic leukemia zinc finger protein (PLZF) that is implicated in leukemogenesis and spermatogenesis (27).
Recently, a HERV-K type 1-specific transcript coding for a 9-kDa protein, designated Np9, has been identified (24). This transcript represents a kind of type 1 substitute for the Rec sequence of type 2 proviruses. An alternative splice donor located just upstream of the type 1-specific 292-nt deletion is used for the splice event joining the two exons of the np9 mRNA (Fig. 3B). Np9 shares only the N-terminal 15 aa with Rec and Env. The C-terminal 59 aa are derived from the third (non-env, non-rec) reading frame. It is noteworthy that, similar to Rec, Np9 accumulates in the nucleus.
Armbruester et al. (24) found np9 transcripts in most transformed human cell lines by using an RT-PCR method. Interestingly, nearly all healthy and malignant tissues from humans express full-length HERV-K type 1 mRNAs, but no np9 transcripts are generated in normal human tissues. A remarkable difference between the rate of np9 and rec transcripts was observed in mammary carcinomas. Whereas np9 transcripts were present in 11 of 21 samples, rec mRNA was found in just 2 of 21 breast cancer tissues. The predominance of type 1 over type 2 HERV-K transcripts in breast cancer cells and cell lines had also been reported (28). It will be interesting to determine how many proviral loci actually contribute to these transcripts and whether differences in the LTR promoters, splice sites, or position in the genome are responsible for the preponderance of type 1 transcripts and the tumor associated expression of np9. However, as with Rec, a tumorigenic potential or a tumor promoting activity of Np9 in mammary epithelial or any other cell type has yet to be shown.
HERV-K Protein Expression
As with all betaretroviruses, two -1 frameshifts at the ribosome are needed to translate the 160-kDa HERV-K Gag-Prot-Pol precursor protein. The precursor is processed by the virus-encoded protease and the mature proteins are released. Active recombinant protease and reverse transcriptase enzymes have been successfully expressed in vitro (29, 30). Without the ribosomal frameshifts, only a Gag protein of 76 kDa is translated. The Gag protein itself is further cleaved by the protease and myristoylated matrix, and core and nucleocapsid components are released. These cleavages occur during the maturation process of the particle after budding from the producer cell. It results in a condensed core morphology, more typical of mature C-type retroviruses (31), and is an absolute requirement for the particle to become infectious. Analogous to the adjustment of transcript splicing, a preservation of the regulatory mechanisms leading to a balanced ratio of nascent Gag, Gag-Prot, and Gag-Prot-Pol proteins is a prerequisite for particle formation and viral infectivity. Gag-encoding transcripts have been detected in many cells and tissues from diseased and healthy individuals. They are easily detectable in teratocarcinoma cell lines as a typical staining pattern at the cellular membrane as well as in testicular tumor cells by using immunoperoxidase staining or immunogold labeling in electron micrographs (31–33).
Several lines of evidence indicate that the exogenous ancestors of contemporary HERVs used their envelope proteins to infect or reinfect human germ cells via a cellular receptor (7). In HERV-K genomes, the pol and env reading frames partially overlap, and the env protein is translated from a singly spliced transcript (Fig. 3A). The env reading frames of at least 16 HERV proviruses (among them six HERV-K) remained open and appear to encode functional proteins (34). Env transcripts comprise only a minor mRNA species in most of the tested cells, but their presence can be demonstrated in many cells and tissues including placenta, prostate, testis, trachea, ovarial carcinomas, kidney carcinomas, testicular tumors, and melanomas (ref. 34 and J. Denner, personal communication). The reason for the apparent scarcity of env transcripts in most cells is presumably the strong splice donor sites at the beginning of the reading frame provoking a second splice event which generates rec or np9 transcripts, depending on the HERV-K type. The Env precursor protein contains a characteristic long signal peptide and is slightly glycosylated (35). For unknown reasons, it is very inefficiently cleaved in most cells, and its transport to the cell membrane appears disorganized. However, fusion activity has been demonstrated for Env proteins of several HERV classes in syncytia formation assays as well as in infectivity assays by using pseudotyped retroviruses (36, 37).
Most HERVs encode a dUTPase domain in the protease ORF (38, 39). Its enzymatic capacity protects against toxic misincorporation of dUTP into cDNA during reverse transcription. The primate lentiviruses HIV and simian immunodeficiency virus (SIV) do not express their own dUTPase, and it is believed that a host cell enzyme provides this activity during reverse transcription.
Production of HERV Particles
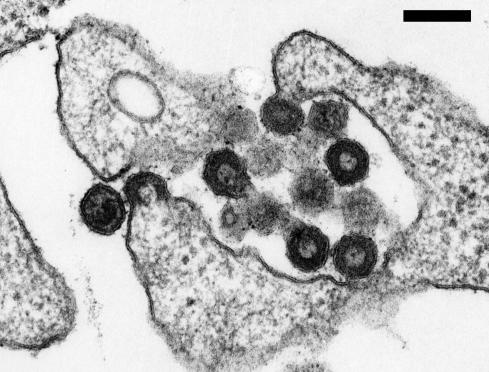
The HERV family first shown to produce viral particles was HERV-K. Retrovirus-like structures of endogenous origin were observed in the syncytial layer of full-term human placentas (40, 41) and cell lines derived from human teratocarcinomas (5, 32, 42). Today, there is no doubt that at least the human teratocarcinoma-derived virus (HTDV) particles are encoded by HERV-K proviruses (43). Using the RU5-PCR technique, it has been demonstrated that HERV-K is highly expressed in HTDV producing teratocarcinoma cell lines (Fig. 4 and ref. 22). In addition, antibodies generated with recombinant HERV-K proteins recognize HDTV elements as well as virus-like particles (VLPs) produced with compiled full-length molecular clones (31, 44). These VLPs display similar morphological characteristics as HTDV: (i) they lack an electron lucent space between the core and the viral membrane, (ii) mature condensed cores resembling those of C-type viruses are very rare, and (iii) spikes are virtually absent (44).
Fig. 4.
Electron microscopic analysis of HERV-K/HTDV particles produced by teratocarcinoma cell lines. Micrographs courtesy of K. Boller, Paul-Ehrlich-Institut (Langen, Germany). (Scale bar, 200 nm.)
HTDV particles are rarely seen separated from the plasma membrane, apparently being arrested at a late stage of the budding process (31, 43, 44). These obvious phenotypic deficiencies might account for the lack of detectable infectivity. The viruses seem to be crippled by disabilities resulting in the observed phenotypes. In addition to the handicaps already mentioned, negative transdominant effects of incorporated mutated proteins encoded by multiple proviruses (transcomplementation) could further obliterate the infectivity of the virions and might even prevent a hypothetical productive replication of those rare preserved complete elements such as HERV-K113. So far, it is not known whether this provirus is actively transcribed and can form particles. A molecular clone would be instrumental in shedding some light on this issue. This is also true for other known particles of endogenous retroviral origin belonging mostly to the HERV-H and HERV-W families (45, 46). Meanwhile, the formation of infectious HERV particles and their potential for transmission remain controversial open questions.
The Antibody Response to HERV-K
Initial reports indicating the presence of antibodies specific for proteins of endogenous retroviral origin were published >20 years ago (42, 47). A humoral response to these proteins has now been demonstrated in animals and in humans (48, 49). The existence of such a response remains difficult to explain because these proteins are regarded as self-antigens and should not provoke an immune response. How the tolerance is broken remains unknown. To test sera or other body fluids for the presence of HERV-reactive antibodies, most authors have used recombinant proteins and peptides in various immunoassays, including immunocytochemistry and electron microscopy (9, 33, 50). The exact origin of the antigen that elicited the antibody responses or the epitopes involved remained unclear in most studies. It therefore remains controversial whether many of these reactivities were indeed induced by endogenous retroviral antigens or whether they represent cross-reactions elicited by exogenous retroviruses or by nonviral antigens such as the cellular ribonucleoproteins, known to resemble some regions of Gag (51). However, in a recent study, antibodies recognizing multiple Env epitopes of two HERV-K proviruses were found in ≈30% of healthy individuals (52). Very solid data are also available documenting a significant increase in frequency and titer of antibodies directed against HERV-K proteins in patients suffering from testicular tumors. In an extensive study, Boller et al. (33) detected such reactivities in 60% of sera from males with germ cell tumors, in contrast to ≈4% of positive samples from healthy individuals. Interestingly, shortly after the elimination of the tumor, the antibody titers dropped and became undetectable by 5 years after surgery (33). Thus, the HERV-K-specific antibody titer might be useful as a testicular tumor marker for diagnosis, prognosis, and therapy. However, there is, as yet, no strong evidence that HERV-K proteins or proteins of any other endogenous retrovirus are the cause of tumor induction in germ cells or other cells. Reminiscent of reports of HERV-K expression in placentas, a doubled prevalence rate of HERV-K antibodies has been noted in sera from full-term pregnant woman (33, 53). This finding is in line with reports showing some dependence of several HERV LTRs on pregnancy hormones and their up-regulation in embryonic tissues (15, 54). There is also circumstantial evidence that HERV expression is elevated by proinflammatory cytokines (55, 56). A number of studies have, indeed, shown that HERV-encoded proteins are expressed in inflamed tissues, and antibodies to HERVs in patients with autoimmune diseases such as systemic lupus erythematodes (SLE), Sjogren's syndrome, multiple sclerosis, and related diseases were frequently detected (46, 52, 57–60). These studies are often cited to support a role for HERVs in the etiology or progression of these disorders. It has been speculated in this regard that molecular mimicry by HERV proteins of cellular components displaying similar epitopes could trigger events leading to autoimmune diseases (51, 61). There is, however, no definitive proof of this concept.
Biological Implications of Retroelement Inheritance
Contribution to the Evolution of the Host. It is obvious that the considerable number of retroelements inserted in mammalian chromosomes has had a profound impact on the shaping and plasticity of the genomes. Genomic rearrangements caused by scattered homologous proviral sequences gave rise to countless genetic variations on which the evolutionary powers of selection and adaptation could work (62). Analysis of human genes reveals that mobile elements, including retroelements, are overrepresented in the mRNAs of rapidly evolving mammalian genes with a high ratio of nonsynonymous to synonymous mutations, indicative of an increased diversifying selection (2). Such genes are mostly involved in immunity, stress responses, and responses to external stimuli. These findings point toward an active role for transposable elements in the diversification and expansion of gene families, increasing the speed of evolution in humans and other mammals. An alternative explanation for this observation, however, may be the higher redundancy of rapidly diversifying genes with an increased tolerance level for insertions (2).
Retroelements are also useful tools for phylogenetic studies and serve as genomic markers. A specific retroviral integration site shared by two species is indicative of a common ancestor because the likelihood of independent integrations at exactly the same locus (insertional homoplasy) is negligible (63). Endogenous retroviruses have proven to be especially instrumental for investigating the evolutionary separation of humans from other African great apes because some of the proviral loci appear only in distinct ape species, whereas others are exclusively human (12). However, care must be taken in the interpretation or generalization of single findings. Scenarios such as allelic segregation or loss of the proviral locus must be taken into account (63).
The integration process of a retroviral element per se is irreversible. A principal elimination of a provirus fixed in the genome of a population is not possible unless insert free alleles are recreated by recombination processes and intrachromosomal deletions (8). The most significant impacts on fixed retroviruses are exerted by recombination events. This is clearly demonstrated by a dramatic accumulation of class I and II elements on chromosome Y due to recombination deficits caused by its genomic singularity (64). The recombination mechanisms that could replace integrated proviruses, their probability, as well as the population dynamics of retroelements and other mobile elements are largely unknown. The advent of genome sequencing together with highly sophisticated statistical and phylogenetic tools will help to prove or lay to rest some of the present theories concerning forces and mechanisms of dispersion, fixation, maintenance, and clearance of endogenous retroviral sequences.
Consequences for the Expression of Cellular Genes. The influence of the transposable elements present in the human germ line on gene expression can be envisaged by the fact that roughly one quarter of all analyzed human promoter regions harbor sequences derived from these elements (65). In addition, a similar proportion of human genes code for at least one mRNA species with a retroelement in its untranslated region (2, 65). The majority of intragenic or near-gene insertions of retroelements were not favorable for the host and were either not fixed in the population or were removed from the lineage in the long term (8, 66). A general age dependent shift of retroelements from GC-rich coding regions to AT-rich isochores of low gene density corroborates this assumption. Although the younger class II HERV-K elements show a preference for integration in GC-rich areas near genes, older LTR-classes and L1 elements are underrepresented in these regions (8). More recent insertional mutagenesis events leading to alleles associated with diseases in humans are mostly caused by transpositionally active LINE and SINE. Examples include hemophilia A caused by the disruption of the factor VIII gene on the X-chromosome by an L1 sequence (67) and insertion of an L1 sequence into the dystrophin gene in some muscular dystrophy patients (68) (Table 2). The de novo insertion of an Alu repeat into an intron of the neurofibromatosis type 1 gene (NF1) resulted in deletion of the downstream exon during splicing and a shift of the reading frame causing neurofibromatosis (69). An Alu element is also suspected to have inactivated the GLO gene, leading to our inability to synthesize vitamin C (70). Insertion of mobile elements into somatic cells is mostly irrelevant unless proto-oncogenes, tumor suppressor genes, or cancer promoting genes are the targets. Insertion of an L1 element into c-myc was shown to be implicated in a breast carcinoma case (71) or to cause colon cancer if inserted into the adenomatous polyposis coli (APC) gene (72).
Table 2. Cases of insertional mutagenesis and recombinations caused by retroelements.
| Element | Gene | Functional role | Ref. |
|---|---|---|---|
| LINE-1 | Factor VIII | Hemophilia A | 67 |
| LINE-1 | Dystrophin | Muscular dystrophy | 68 |
| SINE | Fukutin | Muscular dystrophy | 102 |
| Alu | NF1 | Neurofibromatosis | 69 |
| LINE-1 | myc* | Breast carcinoma | 71 |
| LINE-1 | APC* | Colon cancer | 72 |
| LINE-1 | Attractin | Soluble protein form | 75 |
| HERV-E | Amylase | Activation of a promoter | 77 |
| HERV-K | FGFR1 kinase | Myeloproliferative disorder | 103 |
| HERVs | AZFa region | Male infertility | 104 |
FGFR1, fibroblast growth hormone receptor 1, chromosomal translocation; AZFa, azoospermia factor a, nonallelic homologous recombination between HERVs; APC, adenomatous polyposis carcinoma.
Somatic insertion
Interestingly, the largest proportion of older retroelements show an orientation bias opposite to that of the nearby gene (8, 73). This is another strong indication that LTR elements, predominantly these in the sense orientation, have already been excluded from genes or were not fixed because of their negative influence on transcription regulation. In particular, the provision of a premature polyadenylation site can result in truncated transcripts and eventually in genetic disorders. However, examples of adopted beneficial retroelement-derived polyadenylation sites have also been reported (74). One such example of an “exaptation,” a process in which a retroelement has taken on new functions for a genome, is the generation of the secreted form of the human transmembrane protein attractin. An L1 retrotransposon element that provides a premature stop codon and the polyadenylation sites is responsible for the truncated soluble attractin. Both forms, the transmembrane and the soluble protein, are involved in cell interactions during inflammatory processes. Thus, a retroelement insertion has provided an evolutionary mechanism for regulating an inflammatory response (75). Apart from polyadenylation sites, retroelements may also provide splice sites and other transcriptional processing signals that are often used in a cell-type-specific manner. One exception to the generally favored antisense orientation of retroelements in the vicinity of coding sequences (mentioned above) are LTR elements overlapping the 5′ and 3′ termini of human and mouse untranslated regions. These elements are more likely to have a sense orientation, suggesting a favorable impact on the transcriptional regulation of the gene (8). The number of documented cases in which retroviral sequences have been assimilated for promoter or enhancer functions has rapidly increased in recent years (Table 3). Many of these cases were identified by screening databases of expressed sequences for transcripts containing transposable elements within their 5′ UTRs. In general the LTRs act as minor complementary promoters driving considerable expression in selective tissues only, primarily in the placenta. However, for a few cellular genes, retroelements appear to be the sole promoter (2). Interestingly, although not the most abundant group, many of the retroelements contributing promoter or enhancer sequences are members of the HERV-E family. The explanation for this phenomenon may be a bias toward integration adjacent to coding sequences or successful sequence preservation and prevention of shutdown by methylation and other mechanisms (76). The most striking examples of HERV-E mediated expression of cellular genes are (i) the human salivary amylase gene into which an HERV-E integrated after divergence of Old World monkeys from the primate ancestral tree (77), (ii) the apolipoprotein-C1 gene, where ≈15% of the liver transcripts are derived from the LTR, and (iii) the endothelin-B receptor and pleiotrophin genes with LTR-driven transcripts limited to the placenta (78). Also worth mentioning in this context is the Opitz Syndrome gene Mid1, which is involved in a genetic disorder that primarily affects the development of midline structures. The gene encodes a microtubule-associated protein involved in phosphatase 2A turnover. In the placenta and in embryonic kidneys, HERV-E chimeric mRNAs make up to 25% of Mid1 transcription products. It is, however, not yet clear whether the LTR induced expression contributes to the pathology of Opitz syndrome (76). Recently, van de Lagemaat et al. (2) have published 15 previously unrecognized transcripts of human genes from the RefSeq database beginning with a transposable element, some of which are associated with disease.
Table 3. Examples of retroviral sequences involved in the regulation of cellular proteins.
| Element | Gene (promoter) | Functional role | Ref. |
|---|---|---|---|
| HERV-E | Mid1 | Opitz syndrome | 76 |
| HERV-E | Apolipoprotein C1 | Liver and other tissues | 78 |
| HERV-E | Endothelin-B receptor | Placenta | 78 |
| HERV-E | Pleiotrophin | Trophoblast | 105 |
| HERV-L | β1,3-galactosyltransferase | Colon, mammary gland | 106 |
| ERV II | BAAT (transferase) | Bile metabolism | 2 |
| ERV I | Aromatase | Placental estrogen synthesis | 2 |
| ERV III | Carbonic anhydrase 1 | Erythroid carbon metabolism | 2 |
| LTR + LINE-2 | Chaperonin | McKausick–Kaufman syndrome | 2 |
| HERV | INSL4 (insulin family) | Placenta | 107 |
Significance of the Expression of HERV Proteins. The consequences of a de novo incorporation, reinfection, or retrotransposition of a retroelement can vary considerably. Pathologic, irrelevant, and, in some cases, even beneficial outcomes have been reported for the expression of retroviral proteins and particles. The causative or disease-promoting association of HERV protein expression with germ cell tumors, mammary carcinomas, various autoimmune diseases, and neurological disorders, such as schizophrenia, has yet to be conclusively demonstrated (27, 59, 71, 79). Gag protein expression may induce massive T cell stimulation, apoptosis, and the generation of autoantibodies. Rec and Np9 are suspected to perturb nuclear cytoplasmic transport mechanisms or corrupt the function of nuclear factors (27, 51, 80). A significant pathological potential resides in the Env protein. The ectodomain of the transmembrane subunits of all known retroviruses contains an immunosuppressive domain that inhibits T and B cell activation and modulates cytokine expression (49, 81, 82). The effect has been demonstrated in an elegant way by using a murine in vivo model. Allogeneic tumor cells expressing the immunosuppressive ectodomains of exogenous and endogenous retroviruses were able to escape immune rejection and established tumors in recipient mice, whereas untransfected control tumor cells were eliminated. This experiment therefore provides extra evidence for the possible involvement of a HERV protein in tumoral processes (83). A deleterious effect might also result from the potential for fusing envelope-expressing cells with bystander cells carrying the receptor protein. On the other hand, this fusogenicity, as well as the immunosuppression, may serve an important function for the physiology of primate hosts under some circumstances.
Syncytin, the envelope protein of one of the defective HERV-W proviruses is obviously essential for the formation and function of syncytiotrophoblasts (84–87). These multinucleated cells originate from fetal trophoblasts and constitute the boundary layer between maternal and fetal tissue. The major functions of this layer include maternal–fetal exchange and the maintenance of immunologic tolerance toward the developing fetus. The persistence of the ORF coding for syncytin in the genomes of Old World primates for >25 million years, together with its restricted expression colinearly with primary cytotrophoblast differentiation, strongly suggests an active role of this envelope protein in hominoid placental physiology (87). It is conceivable that syncytin is not the only fusogenic Env that has been sequestered and put to work for the benefit of the host. Recently, the placental expression of syncytin 2, an even older fusogenic envelope protein belonging to the HERV-FRD family, has been reported (88).
Of some concern is the notion that the fusogenic envelopes of HERV-W and HERV-FRD family members could be incorporated into lentiviruses, conferring a dramatic change in cell tropism (37, 89). It is, however, likely that pseudotyping or other reported or suspected forms of transcomplementation involving ERVs are very rare, and their significance in vivo remains to be determined.
Another plausible option for a favorable effect of retroviral endogenization is a partial resistance to infection by pathogenic exogenous counterparts or their related viruses by receptor interference (90–92). This protection could endow the carrier of a less pathogenic or apathogenic germ line provirus with a selective advantage that might result in a population-wide allelic fixation and in the eradication of the exogenous competitor in the long term. Of note in this respect is the observation that endogenous Jaagsiekte sheep retrovirus (JSRV) blocks the entry of the corresponding exogenous virus. Both forms use hyaluronidase-2 as a receptor for entry, implying an interference between endogenous JSRV Env and exogenous viruses that could prevent infection (91).
Endogenization might also prove advantageous to an exogenous retrovirus, primarily by relieving the evolutionary pressure exerted on a virus during horizontal transmission. Because of the mutual benefits, one could even consider the relationship between some of the endogenous viruses (or their remnants) and the host as a kind of a symbiosis.
Env is not the only protein of endogenous retrovirus origin shown to confer resistance to exogenous viruses. The expression of Fv1, the gag-sequence of an endogenous murine retrovirus with homology to HERV-L family members, blocks certain strains of mouse leukemia virus (MLV) soon after entry (93). How Fv1 acts remains unclear, but a direct encounter with the incoming viral capsid is most likely. A restriction of this kind is not limited to mouse viruses. Many primates express Fv1-like activities toward a variety of retroviruses (94, 95), and it has been speculated that some of the responsible inhibitors are expressed by endogenous retroviral sequences analogous to Fv1. Recently, however, the activity that blocks HIV-1 in rhesus monkey cells at the postentry, preintegration stage has been identified as the cellular protein TRIM5α (96).
The expression of HERV sequences is not only suspected to cause or promote some types of cancer, but might also help to regress tumors by serving as tumor-specific antigens. The first reports of endogenous retroviral genes coding for peptides recognized by tumor-specific CTLs came from studies in mice (97–100). Subsequently, Schiavetti et al. (101) identified a HERV-K pseudogene (HERV-K-MEL) with a very short ORF. Surprisingly, the encoded peptide is expressed in most melanomas. The identification of CTL responses directed against epitopes of this peptide in a melanoma patient indicates presentation by MHC I complexes and suggests a potential use of tumor expressed, HERV-derived peptides for therapeutic vaccination (101).
Concluding Remarks
During the course of evolution, the human genome has accumulated a considerable number of random DNA insertions. The largest proportion of this genetic material consists of retrotranscribed and reinserted elements. A remarkable fraction is derived from ancient exogenous retroviruses that found their way into the germ line that became for most, if not all, a graveyard for the following millions of years. A clear proof for the existence of a HERV capable of productive replication remains elusive, but the recently identified youngest and most complete provirus, HERV-K113, provides the most promising candidate (7). It remains to be shown whether this virus or a theoretical transcomplementation between different retroviral elements might produce infectious particles.
Initially, HERVs and other retroelements were studied almost exclusively as a potential cause of disease. Initial efforts in this direction were based on the tumorigenic and immunomodulatory potential of related exogenous animal retroviruses. The detection of elevated HERV transcription, protein expression, and antibody titers associated with some tumor types, autoimmune diseases, and neurologic disorders increased the activities in this line of research. More decisive experiments are needed in the future to clarify the role of HERVs in disease.
The large-scale analysis of mammalian genomes and the development of powerful algorithms for the identification and phylogenetic analysis of retroelements have significantly broadened the research focus on these elements. As we learn more about the position, orientation, conservation, expression, and phylogeny of retroelements, we gain knowledge of our own evolution and uncover new examples of pathogenic and beneficial implications of this intimate relation.
Acknowledgments
We thank Drs. S. Norley, J. Denner, and O. Hohn for critical discussions of this review. The work in the laboratory of the authors was supported in part by a donation from the Heinz Kuthe de Mouson Legacy (to R.K.).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Therapeutic Vaccines: Realities of Today and Hopes for Tomorrow,” held April 1–3, 2004, at the National Academy of Sciences in Washington, DC.
Abbreviations: ERV, endogenous retrovirus; HERV, human ERV; SINE, short interspersed element; LINE, long-terminal interspersed element; HTDV, human tetratocarcinoma-derived virus.
References
- 1.Deininger, P. L. & Batzer, M. A. (2002) Genome Res. 12, 1455-1465. [DOI] [PubMed] [Google Scholar]
- 2.van de Lagemaat, L. N., Landry, J. R., Mager, D. L. & Medstrand, P. (2003) Trends Genet. 19, 530-536. [DOI] [PubMed] [Google Scholar]
- 3.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 4.McClintock, B. (1956) Cold Spring Harbor Symp. Quant. Biol. 21, 197-216. [DOI] [PubMed] [Google Scholar]
- 5.Bronson, D. L., Ritzi, D. M., Fraley, E. E. & Dalton, A. J. (1978) J. Natl. Cancer Inst. 60, 1305-1308. [DOI] [PubMed] [Google Scholar]
- 6.Lower, R., Lower, J., Frank, H., Harzmann, R. & Kurth, R. (1984) J. Gen. Virol. 65, 887-898. [DOI] [PubMed] [Google Scholar]
- 7.Turner, G., Barbulescu, M., Su, M., Jensen-Seaman, M. I., Kidd, K. K. & Lenz, J. (2001) Curr. Biol. 11, 1531-1535. [DOI] [PubMed] [Google Scholar]
- 8.Medstrand, P., van de Lagemaat, L. N. & Mager, D. L. (2002) Genome Res. 12, 1483-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lower, R., Lower, J. & Kurth, R. (1996) Proc. Natl. Acad. Sci. USA 93, 5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jurka, J. (2000) Trends Genet. 16, 418-420. [DOI] [PubMed] [Google Scholar]
- 11.Boeke, M. & Stoye, J. P. (1997) in Retroviruses, ed. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 343-436. [PubMed]
- 12.Medstrand, P. & Mager, D. L. (1998) J. Virol. 72, 9782-9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson, E., Kato, N. & Cohen, M. (1989) Curr. Top. Microbiol. Immunol. 148, 115-132. [DOI] [PubMed] [Google Scholar]
- 14.Repaske, R., O'Neill, R. R., Steele, P. E. & Martin, M. A. (1983) Proc. Natl. Acad. Sci. USA 80, 678-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ono, M., Kawakami, M. & Ushikubo, H. (1987) J. Virol. 61, 2059-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harada, F., Tsukada, N. & Kato, N. (1987) Nucleic Acids Res. 15, 9153-9162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeda, N. (1985) J. Biol. Chem. 260, 6698-6709. [PubMed] [Google Scholar]
- 18.Mayer, J., Sauter, M., Racz, A., Scherer, D., Mueller-Lantzsch, N. & Meese, E. (1999) Nat. Genet. 21, 257-258. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths, D. J. (2001) Genome Biol. 2, REVIEWS1017. [DOI] [PMC free article] [PubMed]
- 20.Buzdin, A., Ustyugova, S., Khodosevich, K., Mamedov, I., Lebedev, Y., Hunsmann, G. & Sverdlov, E. (2003) Genomics 81, 149-156. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, W. E. & Coffin, J. M. (1999) Proc. Natl. Acad. Sci. USA 96, 10254-10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lower, R., Lower, J., Tondera-Koch, C. & Kurth, R. (1993) Virology 192, 501-511. [DOI] [PubMed] [Google Scholar]
- 23.Lower, R., Tonjes, R. R., Korbmacher, C., Kurth, R. & Lower, J. (1995) J. Virol. 69, 141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Armbruester, V., Sauter, M., Krautkraemer, E., Meese, E., Kleiman, A., Best, B., Roemer, K. & Mueller-Lantzsch, N. (2002) Clin. Cancer Res. 8, 1800-1807. [PubMed] [Google Scholar]
- 25.Magin, C., Lower, R. & Lower, J. (1999) J. Virol. 73, 9496-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magin-Lachmann, C., Hahn, S., Strobel, H., Held, U., Lower, J. & Lower, R. (2001) J. Virol. 75, 10359-10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boese, A., Sauter, M., Galli, U., Best, B., Herbst, H., Mayer, J., Kremmer, E., Roemer, K. & Mueller-Lantzsch, N. (2000) Oncogene 19, 4328-4336. [DOI] [PubMed] [Google Scholar]
- 28.Wang-Johanning, F., Frost, A. R., Johanning, G. L., Khazaeli, M. B., LoBuglio, A. F., Shaw, D. R. & Strong, T. V. (2001) Clin. Cancer Res. 7, 1553-1560. [PubMed] [Google Scholar]
- 29.Schommer, S., Sauter, M., Krausslich, H. G., Best, B. & Mueller-Lantzsch, N. (1996) J. Gen. Virol. 77, 375-379. [DOI] [PubMed] [Google Scholar]
- 30.Berkhout, B., Jebbink, M. & Zsiros, J. (1999) J. Virol. 73, 2365-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boller, K., Konig, H., Sauter, M., Mueller-Lantzsch, N., Lower, R., Lower, J. & Kurth, R. (1993) Virology 196, 349-353. [DOI] [PubMed] [Google Scholar]
- 32.Bronson, D. L., Fraley, E. E., Fogh, J. & Kalter, S. S. (1979) J. Natl. Cancer Inst. 63, 337-339. [PubMed] [Google Scholar]
- 33.Boller, K., Janssen, O., Schuldes, H., Tonjes, R. R. & Kurth, R. (1997) J. Virol. 71, 4581-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Parseval, N., Lazar, V., Casella, J. F., Benit, L. & Heidmann, T. (2003) J. Virol. 77, 10414-10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tonjes, R. R., Limbach, C., Lower, R. & Kurth, R. (1997) J. Virol. 71, 2747-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.An, D. S., Kung, S. K., Bonifacino, A., Wersto, R. P., Metzger, M. E., Agricola, B. A., Mao, S. H., Chen, I. S. & Donahue, R. E. (2001) J. Virol. 75, 3547-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blaise, S., Ruggieri, A., Dewannieux, M., Cosset, F. L. & Heidmann, T. (2004) J. Virol. 78, 1050-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harris, J. M., McIntosh, E. M. & Muscat, G. E. (2000) Arch. Virol. 145, 353-363. [DOI] [PubMed] [Google Scholar]
- 39.Mayer, J. & Meese, E. U. (2003) J. Mol. Evol. 57, 642-649. [DOI] [PubMed] [Google Scholar]
- 40.Kalter, S. S., Helmke, R. J., Heberling, R. L., Panigel, M., Fowler, A. K., Strickland, J. E. & Hellman, A. (1973) J. Natl. Cancer Inst. 50, 1081-1084. [DOI] [PubMed] [Google Scholar]
- 41.Vernon, M. L., McMahon, J. M. & Hackett, J. J. (1974) J. Natl. Cancer Inst. 52, 987-989. [DOI] [PubMed] [Google Scholar]
- 42.Kurth, R., Löwer, R., Löwer, J., Harzmann, R., Pfeiffer, R., Schmitt, C.G., Fogh, J. & Frank, H. (1980) in Viruses in Naturally Occurring Cancer, ed. Todaro, G. Essex, M. E. & zur Hausen, H. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 835-846.
- 43.Bieda, K., Hoffmann, A. & Boller, K. (2001) J. Gen. Virol. 82, 591-596. [DOI] [PubMed] [Google Scholar]
- 44.Tonjes, R. R., Boller, K., Limbach, C., Lugert, R. & Kurth, R. (1997) Virology 233, 280-291. [DOI] [PubMed] [Google Scholar]
- 45.Perron, H., Jouvin-Marche, E., Michel, M., Ounanian-Paraz, A., Camelo, S., Dumon, A., Jolivet-Reynaud, C., Marcel, F., Souillet, Y., Borel, E., et al. (2001) Virology 287, 321-332. [DOI] [PubMed] [Google Scholar]
- 46.Christensen, T., Sorensen, P. D., Hansen, H. J. & Moller-Larsen, A. (2003) Mult. Scler. 9, 6-15. [DOI] [PubMed] [Google Scholar]
- 47.Toth, F. D., Vaczi, L., Szabo, B., Kiss, J., Rethy, A., Kiss, A., Telek, B., Kovacs, I., Kiss, C. & Rak, K. (1985) Acta Microbiol. Hung. 32, 267-273. [PubMed] [Google Scholar]
- 48.Charman, H. P., Kim, N., White, M., Marquardt, H., Gilden, R. V. & Kawakami, T. (1975) J. Natl. Cancer Inst. 55, 1419-1424. [DOI] [PubMed] [Google Scholar]
- 49.Denner, J., Persin, C., Vogel, T., Haustein, D., Norley, S. & Kurth, R. (1996) J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 12, 442-450. [DOI] [PubMed] [Google Scholar]
- 50.Sauter, M., Schommer, S., Kremmer, E., Remberger, K., Dolken, G., Lemm, I., Buck, M., Best, B., Neumann-Haefelin, D. & Mueller-Lantzsch, N. (1995) J. Virol. 69, 414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tenenbaum, S. A., Voss, T. G., Garry, R. F. & Gallaher, W. R. (1994) AIDS Res. Hum. Retroviruses 10, 521-522. [DOI] [PubMed] [Google Scholar]
- 52.Herve, C. A., Lugli, E. B., Brand, A., Griffiths, D. J. & Venables, P. J. (2002) Clin. Exp. Immunol. 128, 75-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simpson, G. R., Patience, C., Lower, R., Tonjes, R. R., Moore, H. D., Weiss, R. A. & Boyd, M. T. (1996) Virology 222, 451-456. [DOI] [PubMed] [Google Scholar]
- 54.Andersson, A. C., Venables, P. J., Tonjes, R. R., Scherer, J., Eriksson, L. & Larsson, E. (2002) Virology 297, 220-225. [DOI] [PubMed] [Google Scholar]
- 55.Sibata, M., Ikeda, H., Katumata, K., Takeuchi, K., Wakisaka, A. & Yoshoki, T. (1997) Leukemia 11, Suppl. 3, 145-146. [PubMed] [Google Scholar]
- 56.Katsumata, K., Ikeda, H., Sato, M., Ishizu, A., Kawarada, Y., Kato, H., Wakisaka, A., Koike, T. & Yoshiki, T. (1999) Clin. Immunol. 93, 75-80. [DOI] [PubMed] [Google Scholar]
- 57.Bengtsson, A., Blomberg, J., Nived, O., Pipkorn, R., Toth, L. & Sturfelt, G. (1996) Arthritis Rheum. 39, 1654-1663. [DOI] [PubMed] [Google Scholar]
- 58.Talal, N., Dauphinee, M. J., Dang, H., Alexander, S. S., Hart, D. J. & Garry, R. F. (1990) Arthritis Rheum. 33, 774-781. [DOI] [PubMed] [Google Scholar]
- 59.Hishikawa, T., Ogasawara, H., Kaneko, H., Shirasawa, T., Matsuura, Y., Sekigawa, I., Takasaki, Y., Hashimoto, H., Hirose, S., Handa, S., et al. (1997) Viral Immunol. 10, 137-147. [DOI] [PubMed] [Google Scholar]
- 60.Clerici, M., Fusi, M. L., Caputo, D., Guerini, F. R., Trabattoni, D., Salvaggio, A., Cazzullo, C. L., Arienti, D., Villa, M. L., Urnovitz, H. B., et al. (1999) J. Neuroimmunol. 99, 173-182. [DOI] [PubMed] [Google Scholar]
- 61.Perl, A. (2003) Rheum. Dis. Clin. North Am. 29, 123-143. [DOI] [PubMed] [Google Scholar]
- 62.Hughes, J. F. & Coffin, J. M. (2001) Nat. Genet. 29, 487-489. [DOI] [PubMed] [Google Scholar]
- 63.Barbulescu, M., Turner, G., Su, M., Kim, R., Jensen-Seaman, M. I., Deinard, A. S., Kidd, K. K. & Lenz, J. (2001) Curr. Biol. 11, 779-783. [DOI] [PubMed] [Google Scholar]
- 64.Kjellman, C., Sjogren, H. O. & Widegren, B. (1995) Gene 161, 163-170. [DOI] [PubMed] [Google Scholar]
- 65.Jordan, I. K., Rogozin, I. B., Glazko, G. V. & Koonin, E. V. (2003) Trends Genet. 19, 68-72. [DOI] [PubMed] [Google Scholar]
- 66.Kidwell, M. G. & Lisch, D. (1997) Proc. Natl. Acad. Sci. USA 94, 7704-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kazazian, H. H., Jr., Wong, C., Youssoufian, H., Scott, A. F., Phillips, D. G. & Antonarakis, S. E. (1988) Nature 332, 164-166. [DOI] [PubMed] [Google Scholar]
- 68.Holmes, S. E., Dombroski, B. A., Krebs, C. M., Boehm, C. D. & Kazazian, H. H., Jr. (1994) Nat. Genet. 7, 143-148. [DOI] [PubMed] [Google Scholar]
- 69.Wallace, M. R., Andersen, L. B., Saulino, A. M., Gregory, P. E., Glover, T. W. & Collins, F. S. (1991) Nature 353, 864-866. [DOI] [PubMed] [Google Scholar]
- 70.Challem, J. J. & Taylor, E. W. (1998) Free Radical Biol. Med. 25, 130-132. [DOI] [PubMed] [Google Scholar]
- 71.Morse, B., Rotherg, P. G., South, V. J., Spandorfer, J. M. & Astrin, S. M. (1988) Nature 333, 87-90. [DOI] [PubMed] [Google Scholar]
- 72.Miki, Y., Nishisho, I., Horii, A., Miyoshi, Y., Utsunomiya, J., Kinzler, K. W., Vogelstein, B. & Nakamura, Y. (1992) Cancer Res. 52, 643-645. [PubMed] [Google Scholar]
- 73.Smit, A. F. (1999) Curr. Opin. Genet. Dev. 9, 657-663. [DOI] [PubMed] [Google Scholar]
- 74.Baust, C., Seifarth, W., Germaier, H., Hehlmann, R. & Leib-Mosch, C. (2000) Genomics 66, 98-103. [DOI] [PubMed] [Google Scholar]
- 75.Tang, W., Gunn, T. M., McLaughlin, D. F., Barsh, G. S., Schlossman, S. F. & Duke-Cohan, J. S. (2000) Proc. Natl. Acad. Sci. USA 97, 6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Landry, J. R., Rouhi, A., Medstrand, P. & Mager, D. L. (2002) Mol. Biol. Evol. 19, 1934-1942. [DOI] [PubMed] [Google Scholar]
- 77.Samuelson, L. C., Phillips, R. S. & Swanberg, L. J. (1996) Mol. Biol. Evol. 13, 767-779. [DOI] [PubMed] [Google Scholar]
- 78.Medstrand, P., Landry, J. R. & Mager, D. L. (2001) J. Biol. Chem. 276, 1896-1903. [DOI] [PubMed] [Google Scholar]
- 79.Karlsson, H., Bachmann, S., Schroder, J., McArthur, J., Torrey, E. F. & Yolken, R. H. (2001) Proc. Natl. Acad. Sci. USA 98, 4634-4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hugin, A. W., Vacchio, M. S. & Morse, H. C., III (1991) Science 252, 424-427. [DOI] [PubMed] [Google Scholar]
- 81.Cianciolo, G. J., Copeland, T. D., Oroszlan, S. & Snyderman, R. (1985) Science 230, 453-455. [DOI] [PubMed] [Google Scholar]
- 82.Mangeney, M., de Parseval, N., Thomas, G. & Heidmann, T. (2001) J. Gen. Virol. 82, 2515-2518. [DOI] [PubMed] [Google Scholar]
- 83.Mangeney, M. & Heidmann, T. (1998) Proc. Natl. Acad. Sci. USA 95, 14920-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mi, S., Lee, X., Li, X., Veldman, G. M., Finnerty, H., Racie, L., LaVallie, E., Tang, X. Y., Edouard, P., Howes, S., et al. (2000) Nature 403, 785-789. [DOI] [PubMed] [Google Scholar]
- 85.Lavillette, D., Marin, M., Ruggieri, A., Mallet, F., Cosset, F. L. & Kabat, D. (2002) J. Virol. 76, 6442-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Blond, J. L., Lavillette, D., Cheynet, V., Bouton, O., Oriol, G., Chapel-Fernandes, S., Mandrand, B., Mallet, F. & Cosset, F. L. (2000) J. Virol. 74, 3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frendo, J. L., Olivier, D., Cheynet, V., Blond, J. L., Bouton, O., Vidaud, M., Rabreau, M., Evain-Brion, D. & Mallet, F. (2003) Mol. Cell. Biol. 23, 3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blaise, S., de Parseval, N., Benit, L. & Heidmann, T. (2003) Proc. Natl. Acad. Sci. USA 100, 13013-13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.An, D. S., Xie, Y. & Chen, I. S. (2001) J. Virol. 75, 3488-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Melder, D. C., Pankratz, V. S. & Federspiel, M. J. (2003) J. Virol. 77, 10504-10514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spencer, T. E., Mura, M., Gray, C. A., Griebel, P. J. & Palmarini, M. (2003) J. Virol. 77, 749-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ponferrada, V. G., Mauck, B. S. & Wooley, D. P. (2003) Arch Virol. 148, 659-675. [DOI] [PubMed] [Google Scholar]
- 93.Best, S., Le Tissier, P., Towers, G. & Stoye, J. P. (1996) Nature 382, 826-829. [DOI] [PubMed] [Google Scholar]
- 94.Hatziioannou, T., Cowan, S., Goff, S. P., Bieniasz, P. D. & Towers, G. J. (2003) EMBO J. 22, 385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Schmitz, C., Marchant, D., Neil, S. J., Aubin, K., Reuter, S., Dittmar, M. T. & McKnight, A. (2004) J. Virol. 78, 2006-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stremlau, M., Owens, C. M., Perron, M. J., Kiessling, M., Autissier, P. & Sodroski, J. (2004) Nature 427, 848-853. [DOI] [PubMed] [Google Scholar]
- 97.Hayashi, H., Matsubara, H., Yokota, T., Kuwabara, I., Kanno, M., Koseki, H., Isono, K., Asano, T. & Taniguchi, M. (1992) J. Immunol. 149, 1223-1229. [PubMed] [Google Scholar]
- 98.Huang, A. Y., Gulden, P. H., Woods, A. S., Thomas, M. C., Tong, C. D., Wang, W., Engelhard, V. H., Pasternack, G., Cotter, R., Hunt, D., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 9730-9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.de Bergeyck, V., De Plaen, E., Chomez, P., Boon, T. & Van Pel, A. (1994) Eur. J. Immunol. 24, 2203-2212. [DOI] [PubMed] [Google Scholar]
- 100.Yang, J. C. & Perry-Lalley, D. (2000) J. Immunother. 23, 177-183. [DOI] [PubMed] [Google Scholar]
- 101.Schiavetti, F., Thonnard, J., Colau, D., Boon, T. & Coulie, P. G. (2002) Cancer Res. 62, 5510-5516. [PubMed] [Google Scholar]
- 102.Kobayashi, K., Nakahori, Y., Miyake, M., Matsumura, K., Kondo-Iida, E., Nomura, Y., Segawa, M., Yoshioka, M., Saito, K., Osawa, M., et al. (1998) Nature 394, 388-392. [DOI] [PubMed] [Google Scholar]
- 103.Guasch, G., Popovici, C., Mugneret, F., Chaffanet, M., Pontarotti, P., Birnbaum, D. & Pebusque, M. J. (2003) Blood 101, 286-288. [DOI] [PubMed] [Google Scholar]
- 104.Bosch, E. & Jobling, M. A. (2003) Hum. Mol. Genet. 12, 341-347. [DOI] [PubMed] [Google Scholar]
- 105.Schulte, A. M., Malerczyk, C., Cabal-Manzano, R., Gajarsa, J. J., List, H. J., Riegel, A. T. & Wellstein, A. (2000) Oncogene 19, 3988-3998. [DOI] [PubMed] [Google Scholar]
- 106.Dunn, C. A., Medstrand, P. & Mager, D. L. (2003) Proc. Natl. Acad. Sci. USA 100, 12841-12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bieche, I., Laurent, A., Laurendeau, I., Duret, L., Giovangrandi, Y., Frendo, J. L., Olivi, M., Fausser, J. L., Evain-Brion, D. & Vidaud, M. (2003) Biol. Reprod. 68, 1422-1429. [DOI] [PubMed] [Google Scholar]