Abstract
Background: Previous work characterized variants of the EL4 murine lymphoma cell line. Some are non-metastatic, and others metastatic, in syngenic mice. In addition, metastatic EL4 cells were stably transfected with phospholipase D2 (PLD2), which further enhanced metastasis. Materials and Methods: Microarray analyses of mRNA expression was performed for non-metastatic, metastatic, and PLD2-expressing metastatic EL4 cells. Results: Many differences were observed between non-metastatic and metastatic cell lines. One of the most striking new findings was up-regulation of mRNA for the matricellular protein WNT1-inducible signaling pathway protein 1 (CCN4) in metastatic cells; increased protein expression was verified by immunoblotting and immunocytochemistry. Other differentially expressed genes included those for reproductive homeobox 5 (Rhox5; increased in metastatic) and cystatin 7 (Cst7; decreased in metastatic). Differences between PLD2-expressing and parental cell lines were limited but included the signaling proteins Ras guanyl releasing protein 1 (RGS18; increased with PLD2) and suppressor of cytokine signaling 2 (SOCS2; decreased with PLD2). Conclusion: The results provide insights into signaling pathways potentially involved in conferring metastatic ability on lymphoma cells.
Keywords: Metastasis, lymphoma, differential gene expression, phospholipase D
EL4 is a murine lymphoma cell line developed in 1945 by treating C57 black mice with 9:10-dimethyl-1:2-benzanthra-cene (1). The cells were originally propagated in animal hosts, prior to adaptation to cell culture. EL4 cells were used for many years as a source of interleukin-2 (IL2), which they secrete when treated with phorbol 12-myristate 13-acetate (PMA).
The EL4 cell line originally provided to us, by a colleague at the University of Washington, was heterogeneous with respect to PMA response. We, therefore, developed and characterized EL4 sub-lines. Wild-type (WT)-derived cell lines, which are PMA responsive, grow readily in suspension culture as did the original strain. Variant (V)-derived (PMA-resistant) sub-lines were selected for enhanced adhesion to tissue culture plastic (2). Clonal lines were developed from both cell types by limiting dilution (3). The PMA sensitivity of WT-derived cells, as reflected by PMA-induced IL2 production and mitogen-activated protein kinase (ERK1/2) activation, has been attributed to expression of Ras guanyl nucleotide releasing protein 1 (RasGRP1), which binds PMA and directly activates Ras (4); V-derived cells do not express RasGRP1.
Clonal EL4 cell lines were used for experimental metastasis studies in syngeneic mice, with WT2 and V7 as prototypes (5,6). WT2 cells do not form tumors (non-metastatic), while V7 cells form liver tumors (metastatic) after tail vein injection. ‘Metastasis’ more strictly refers to tumorigenesis in this model, since circulating EL4 cells home to the liver to form tumors (5). The clonal C5 cell line was developed after stably overexpressing human hemagllutinin (HA)-tagged phospholipase D2 (PLD2) in V7 cells in order to characterize the signaling role of PLD2 using a cell line expressing little or no endogenous PLD2 (5). As compared to V7, C5 cells exhibit increased cell migration (7) and tumor growth (5), providing an early example of the positive role of PLD2 in tumorigenesis.
This report presents data from microarray analyses comparing transcripts expressed by these three EL4 cell types, with the goal of further defining their phenotypes.
Materials and Methods
Cell culture. Clonally-derived EL4 cells were maintained in RPMI-1640 (Gibco/Thermo Fisher Scientific, Waltham, MA, USA) with 10% fetal bovine serum (Atlanta Biologicals, Atlanta, GA, USA) as previously described (3,5). All cells were grown on standard tissue culture dishes, except for WT2, which was grown on suspension culture plastic (Corning Incorporated, Tewksbury, MA, USA). C5 was maintained with 0.25 mg/ml G418 (Calbiochem/EMD Millipore, Billerica, MA, USA), and switched to G418-free medium 24 hours prior to experiments.
Cell migration. For migration assays, 5×105 cells were seeded into 8.0 μm pore Falcon cell culture inserts (BD Biosciences, San Jose, CA, USA) in a 24-well plate. Cells were allowed to migrate for 8 or 12 hours at 37˚C with 5% CO2. Non-migrated cells were swabbed from the insert. Migrated cells were fixed with methanol at −20˚C for 10 minutes, stained with crystal violet at room temperature for 10 minutes, washed with distilled water, and counted using microscopy.
RNA preparation. Total RNA was extracted from cells (serum-starved overnight) using the RNeasy Mini kit (Qiagen, Hilden, Germany). Genomic DNA was removed by treating with DNase 1 from Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA). Concentration and purity of total RNA were determined by absorbance at 260 nm and by A260/280 ratio, respectively. RNA integrity was determined by separating 2 μg total RNA on a denaturing ethidium bromide-containing agarose gel, visualized by Gel Doc imaging system (BioRad Laboratories, Hercules, CA, USA). The 28s band was two-fold that of 18s rRNA for all three cell lines.
Microarray. Differential gene expression was assessed using an Affymetrix (Santa Clara, CA, USA) mouse 430 2.0 array containing 39,000 mouse transcripts as previously described (6). Two duplicate arrays, from separate aliquots of cells, were analyzed for each cell line. Normalized and log-transformed microarray data were analyzed using Genesifter software (PerkinElmer, Waltham, MA, USA). Differential expression of genes was determined by using pairwise analysis. Statistical significance was determined using Student’s t-test and corrected with Benjamini and Hochberg method. Genes differentially expressed by 2-fold or more were included in the analysis. The microarray data have been submitted to the GEO archive (GSE79635).
Immunoblotting. Cells were rinsed with phosphate-buffered saline (PBS); attached cells were harvested by scraping. Cells were collected by centrifugation at 1,200 × g. Whole cell lysates were prepared as previously described (3). Coomassie blue reagent (Pierce Chemical, Rockford, IL, USA) was used to determine protein concentrations. Equal amounts of protein (100 μg) were loaded in each lane of a 10% sodium dodecyl sulfate-polyacrylamide gel, electophoretically separated, transferred to polyvinylidene fluoride membranes, incubated with primary antibodies, and developed using enhanced chemiluminescence reagents (Amersham Pharmacia Biotech, Piscataway, NJ, USA). Anti-actin was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-HA was from Babco (Richmond, CA, USA). Anti-CCN4 was from R&D Systems (Minneapolis, MN, USA).
Immunocytochemistry. Cells were pelleted, fixed in formalin, embedded in paraffin, sectioned, and stained with anti-CCN4 (1:50 dilution; sc-25441; Santa Cruz Biotechnology, Santa Cruz, CA, USA) followed by fluorescent secondary antibody (1:100 dilution; Cy5-conjugated donkey anti-rabbit; Jackon ImmunoResearch, West Grove, PA, USA); controls lacked primary antibody. Nuclei were stained with 4’,6-diamidino-2-phenylindole (DAPI). Images were captured using an Olympus IX81 automated Inverted fluorescence microscope and processed using Image-Pro software (MediaCybernetics, Rockville, MD, USA).
Results
Differential expression of genes in WT2 (non-metastatic) vs. V7 (metastatic) EL4 cells. Over 2800 genes were found to be differentially expressed above the threshold (2-fold change) between WT2 and V7 cells in the microarrays. Genes of particular interest are presented in Table I. Several results confirmed published findings. Specifically, the Ptk2 transcript for focal adhesion kinase (FAK) was 12-fold higher in V7 than in WT2 cells, while the Pyk2 transcript for the related tyrosine kinase Pyk2 was 3-fold lower; both results are consistent with mRNA and protein differences reported previously (6). The Prkch transcript encoding protein kinase C η was 3-fold lower in V7 than in WT2 cells, consistent with protein levels (2). Rasgrp1 mRNA was 5-fold lower in V7 than in WT2 cells, consistent with the previous finding that RasGRP1 protein is greatly reduced in V7 as compared to WT2 cells (4). Thus, the results for previously examined genes validate the microarray method. Although the differences in mRNA expression noted above are only of the order of several-fold, they are consequential since some of the encoded proteins [e.g. FAK (6)] were previously noted to be essentially absent from the lower-expressing cell line.
Table I. Selected transcripts differentially expressed between WT2 and V7 EL4 cells.
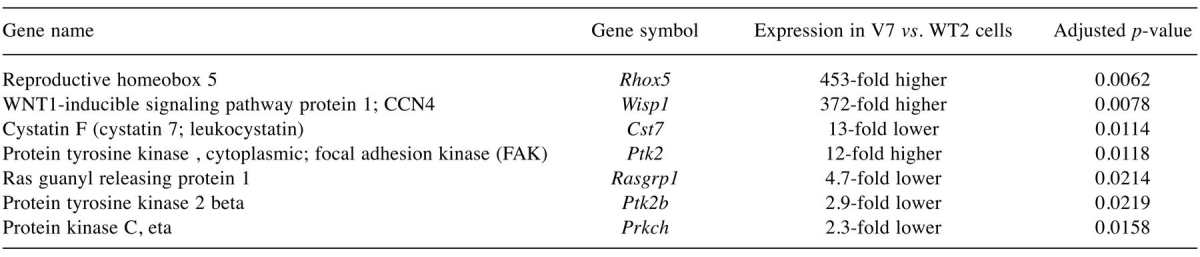
All of the listed genes are discussed in the text
The microarray analysis also identified many other interesting transcripts that had not been studied previously in the EL4 cell lines. The greatest difference in expression (453-fold higher in V7) was observed for reproductive homeobox 5 (Rhox5), a member of the homeobox gene family.
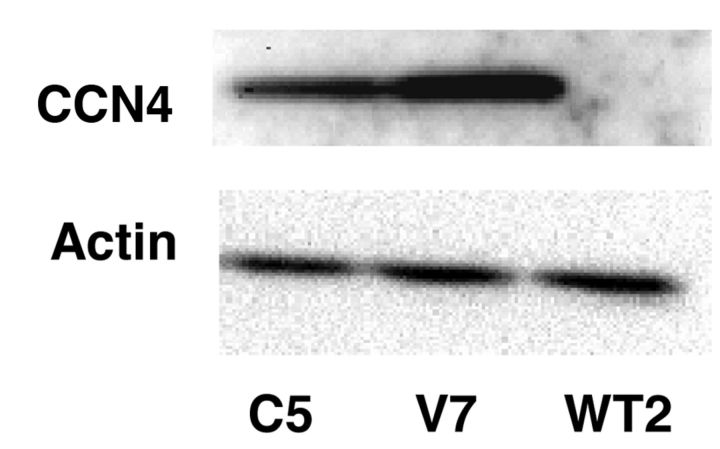
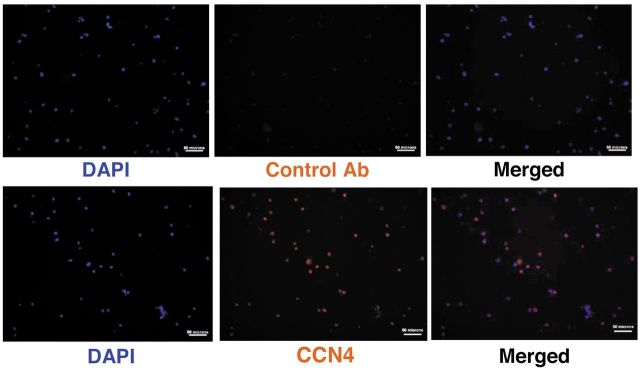
Of particular interest with respect to metastasis, transcript for CCN4 was 372-fold higher in V7 than in WT2. CCN4 is a member of the CCN family of matricellular proteins involved in cell signaling and adhesion. The expression of CCN4 protein was examined. CCN4 protein was found to be expressed in V7 cells, but was undetectable in WT2 cells (Figure 1). Immunocytochemistry showed that CCN4 was dispersed throughout the cytosol of V7 cells (Figure 2).
Figure 1. Expression of WNT1-inducible signaling pathway protein 1 (CCN4) in EL4 cell lines. Whole-cell extracts from WT2, V7 and C5 cells, equalized for protein, were immunoblotted for CCN4 and actin.
Figure 2. Localization of WNT1-inducible signaling pathway protein 1 (CCN4) in EL4 V7 cells. V7 cells were fixed and stained with 4’,5-diamidino-2-phenylindole (DAPI) and anti- CCN4 (merged in right panels). The control used secondary antibody only. Bars=50 μm.
Transcript for the protease cystatin F (leukocystatin; Cst7) was expressed at 13-fold higher levels in WT2 than in V7 cells; protein levels were not examined.
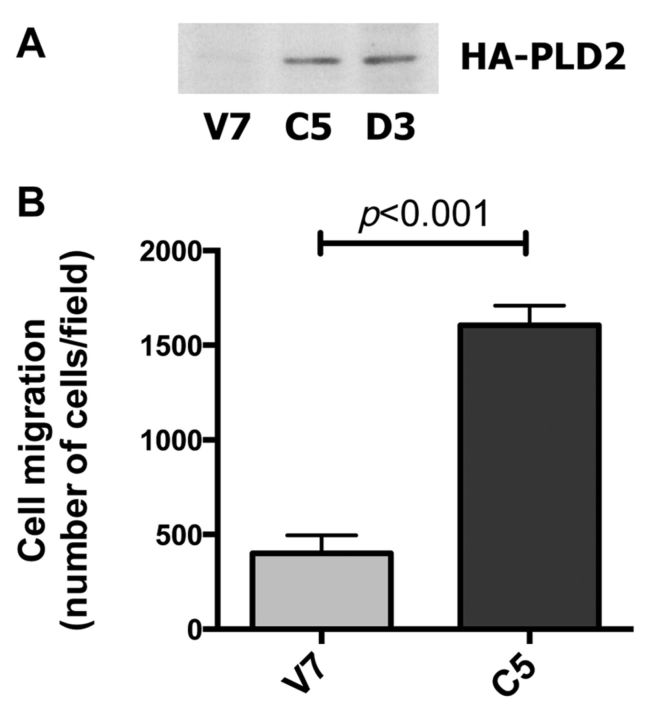
Differential expression of genes in V7 (metastatic) vs. C5 (PLD2-expressing, highly metastatic) EL4 cells. We first confirmed expression of transfected PLD2 at the time when the microarray analyses were performed, since the C5 cell line had been developed approximately 5 years earlier. C5 cells expressed human HA-PLD2, detected by immunoblotting for HA (Figure 3A), as reported previously (5). Human PLD2 was detected in C5, but not V7, cells by reverse transcription polymerase chain reaction (data not shown). The migration of C5 cells was enhanced as compared to that of V7 cells (Figure 3B), also as reported previously (5-7). Thus, any differences in gene expression between C5 and V7 cells are likely attributable to continued PLD2 overexpression.
Figure 3. Comparison of V7 and C5 EL4 cell lines. Panel A: Whole-cell extracts from the indicated cell lines, equalized for protein, were immunoblotted for hemagllutinin (HA) to detect transfected HA-tagged phospholipase D2 (PLD2). V7 is the non-transfected parental cell line, C5 was transfected with catalytically active human HA-PLD2. D3 (not otherwise discussed herein) was transfected with catalytically-inactive HA-PLD2 (5). Panel B: V7 and C5 cells were incubated for 6 hours in a Boyden chamber in serum-containing medium. Cells that migrated through the insert were counted. Each data point represents the mean±SEM of triplicate wells.
Microarray analysis was utilized to determine differential expression of genes in PLD2-transfected C5 as compared to V7 cells. There were 102 genes differentially expressed between V7 and C5 cells with greater than 2-fold change. The magnitude of the differential expression ranged up to 16.5-fold. Epidermal growth factor receptor (mRNA and protein), encoded by Egfr, was more highly expressed in C5 than V7 cells in a previous analysis using cells growing in serum (7), but was not noted in the current study, which used serum-starved cells. Table II lists selected genes differentially expressed between V7 and C5 cells.
Table II. Selected transcripts differentially expressed between V7 and C5 EL4 cells.
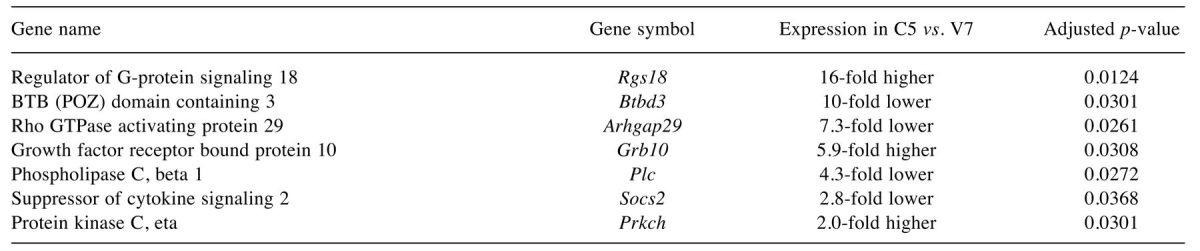
Some, but not all, of the listed genes are discussed in the text
Many of the differentially expressed genes are signal transduction proteins. One transcript of interest was that for suppressor of cytokine signaling 2 (Socs2), which negatively regulates signal transduction (9). Socs2 expression was lower in C5 cells by 2.8-fold at the mRNA level (Table I). Immunoblotting for SOCS2 protein suggested that SOCS2 expression is decreased in C5 as compared to V7 cells (10).
Regulator of G-protein signaling 18 (Rgs18) showed the greatest difference in expression between V7 and C5 cells, with 16-fold higher levels in C5. Growth factor receptor-bound protein 10 (Grb10) expression was increased 6-fold in C5 cells. The potential significance of these and other differences is discussed below.
Discussion
This report highlights results from two separate but concurrent microarray analyses of gene expression of clonal EL4 lymphoma cell lines. The first analysis, comparing PMA-sensitive, non-metastatic WT2 cells with PMA-resistant, metastatic V7 cells, identified nearly 3,000 differences (≥2-fold). The second analysis compared V7 cells to V7 stably transfected with human PLD2 (C5). In this case, only ~100 transcripts were differentially expressed.
Some differences between WT2 and V7 cells (Prkch, Ptk2, Rasgrp1) were discovered previously. Prkch transcript encoding protein kinase C η was decreased in V7 cells, consistent with protein expression (2), However, since Prkch transcript was higher in C5 than in V7 cells, there is no apparent correlation with metastasis. The potential roles of hundreds of other differentially expressed genes remain to be determined. The many differences may reflect that the cell line was heterogeneous when first developed in 1945, and that this heterogeneity has persisted and likely expanded over time. In the original report characterizing EL4 cells, the author commented that the phenotype of the cells, as seen by gross morbid anatomy, shifted from "lymphatic leukemia" to "lymphosarcoma" when they were propagated by intraperitoneal injection (1). Thus, different cell types in the original line could have been subject to selection under different growth conditions.
EL4 was characterized as an "NK T-cell tumor" by Gays and colleagues (11), NK referring to natural killer cells. They noted that NK-related markers are expressed in a fluctuating manner, even in clonal EL4 cell lines, ascribing this to an epigenetic process. Thus, another explanation for the heterogeneity is that variant phenotypes are continuously generated as the cells are maintained in culture. In our hands, clonal EL4 cell lines were relatively stable with respect to phenotype, although we discouraged selection for V-type cells by maintaining WT-derived cells in suspension culture.
From the WT2 vs. V7 microarray, Wisp1 was selected for verification at the protein level. CCN4 is a member of the CCN family of secreted matricellular proteins (12). CCN proteins modulate interactions between cells and extracellular matrix, largely via binding to integrins (13). A review of CCN4 in all cancer types emphasized its up-regulation in non-hematological neoplasms (14); an example is prostate cancer (15). In contrast, CCN4 is a potential biomarker for multiple myeloma, and is overexpressed in peripheral T-cell lymphomas (16). We detected cysteine-rich angiogenic inducer 61 (CYR61, CCN1) in V7 by immunoblotting (unpublished data), but Ccn4 transcript was not differentially expressed between WT2 and V7 cells in the microarray data. Wells and colleagues note little redundancy between the functions of CCN proteins (16). The authors commented that the same CCN protein can be involved in normal differentiation and in neoplasia, suggesting that differential expression of CCNs may provide a growth advantage to tumor cells through pathways that remain to be elucidated. These authors also noted that CCN4 is the least studied of the family with respect to human neoplasms of hematological origin. Thus, it is important to further explore the role of CCN4 in hematological neoplasms.
Rhox5, which encodes a homeobox transcription factor, was the transcript with greatest difference between WT2 and V7 cells, and was expressed primarily (if not exclusively) in metastatic V7 cells. RHOX5 is particularly important in development of reproductive tissues (17). It is expressed in immortalized and neoplastic cell lines, but not in normal adult tissues. The gene was originally identified from a murine T-lymphoma library as a gene differentially expressed between lymphomas at different stages of malignancy (18). It is tempting to hypothesize that expression of Rhox5 may be a driving force in many of the other differences in gene expression noted between V7 and WT2 cells. Further studies will be required to address this hypothesis.
Cystatin F, encoded by Cst7, is a protease inhibitor that may suppress apoptosis in hematopoietic cells, where it is selectively expressed (19). Its potential role in NK cells has been discussed (20). Although CST7 can be secreted, it localizes to lysosomes in a pro-monocyte cell line (21). Like other type II cystatins, CST7 is generally expressed at lower levels in tumors (19), consistent with its lower expression in metastatic V7 EL4 cells. However, CST7 was found to be overexpressed in colorectal tumor cells metastasized to liver (22). Since expression of CST7 protein was not tested in EL4 cells, the significance of its lower expression in metastatic V7 cells remains to be determined.
The changes in gene expression observed between C5 (PLD2-expressing) and V7 were generally modest in magnitude. SOCS2, which is decreased following PLD2 transfection, is an inducible protein of interest in neoplasia. SOCS2 expression is generally associated with increased body growth in rodents (9). Expression of SOCS2 is associated with positive prognosis in human breast cancer (23,24), but with malignancy in prostate (25) and colorectal (26) cancer. Its role in hematological malignancies is not yet clear, although SOCS2 is involved in T-cell differentiation (27), and stability (28). SOCS2 inhibits Janus kinase/signal transducer and activator of transcription signaling, but can also influence EGFR signaling. Since EGF response is increased in EL4 cells after PLD2 transfection (7), the observed decrease in SOCS2 could potentially play a role. However, additional studies will be needed to determine whether SOCS2 is downstream of PLD2.
Rgs18, which is up-regulated in C5 (PLD2-expressing) cells, is expressed in a myeloerythroid lineage-specific manner (29). RGS18 negatively modulates the function of G-protein-coupled receptors (GPCRs). RGS18 makes platelets less sensitive to activation (30,31). Since PLD2 can be activated downstream of GPCRs (32), it is conceivable that the increase in RGS18 is a compensatory response to some of the effects of PLD2 overexpression. The adapter protein GRB10, which is increased in PLD2-transfected cells, suppresses activation of protein kinase G (AKT) by growth factors (33); this could also be compensatory since C5 cells exhibit enhanced AKT activation (7).
In conclusion, our studies of mouse EL4 cell lines with different phenotypes have answered some questions, and raised many new ones. The microarray data provide the basis for further studies into the factors that contribute to the progression of hematologic malignancies.
Acknowledgements
This work was supported by National Institutes of Health CA094144. The Authors thank Derek J. Pouchnik (WSU Molecular Biology and Genomics Core) for performing the microarray analysis, and Hui Zhang (WSU) for helpful discussions. Patrick Hyunh and Jenna McKay assisted in early phases of this project.
References
- 1.Gorer PA. Studies in antibody response of mice to tumor inoculation. Br J Cancer. 1950;4:372–379. doi: 10.1038/bjc.1950.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sansbury HM, Fulwood S, Wisehart-Johnson AE, Qi C, Meier KE. Effects of protein kinase C activators on phorbol ester-sensitive and -resistant EL4 thymoma cells. Carcinogenesis. 1997;18:1817–1824. doi: 10.1093/carcin/18.9.1817. [DOI] [PubMed] [Google Scholar]
- 3.Ku H, Meier KE. Phosphorylation of paxillin via the ERK mitogen-activated protein kinase cascade in EL4 thymoma cells. J Biol Chem. 2000;275:11333–11340. doi: 10.1074/jbc.275.15.11333. [DOI] [PubMed] [Google Scholar]
- 4.Han S, Knoepp SM, Hallman M, Meier KE. RasGRP confers the phorbol ester-sensitive phenotype to EL4 lymphoma cells. Mol Pharm. 2007;71:314–322. doi: 10.1124/mol.106.028639. [DOI] [PubMed] [Google Scholar]
- 5.Knoepp SM, Chahal MS, Xie Y, Zhang Z, Brauner DJ, Hallman MA, Robinson SA, Han S, Imai M, Tomlinson S, Meier KE. Effects of phospholipase D2 expression on adhesion and tumori-genesis in EL4 thymoma cells. Mol Pharm. 2008;74:574–584. doi: 10.1124/mol.107.040105. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Z, Knoepp SM, Sansbury HM, Han S, Ku H, Xie Y, Hallman M, Meier KE. Differential expression of FAK and PYK2 in phorbol ester-sensitive and resistant EL4 thymoma cells. Clin Expt Metastasis. 2011;28:551–565. doi: 10.1007/s10585-011-9391-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chahal MS, Brauner D, Meier KE. Phospholipase D2 enhances epidermal growth factor-induced AKT activation in EL4 lymphoma cells. Pharmaceuticals. 2010;3:2045–2058. doi: 10.3390/ph3072045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gays F, Koh ASC, Mickiewicz KM, Aust JG, Brooks CG. Comprehensive analysis of transcript start sites in Ly49 genes reveals an unexpected relationship with gene function and a lack of upstream promoters. PLoS One. 2011;6:e18475. doi: 10.1371/journal.pone.0018475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trengove MC, Ward AC. SOCS proteins in development and disease. Am J Clin Exp Immunol. 2013;2:1–29. [PMC free article] [PubMed] [Google Scholar]
- 10.Chahal MS. Roles for PLD2 in growth factor-mediated signal transduction in EL4 lymphoma cells. Washington State University, 2008 Ph.D. Dissertation. [Google Scholar]
- 11.Gays F, Unnikrishnan M, Shrestha S, Fraser KP, Brown AR, Tristram CMG, Chrzanowska-Lightowlers ZMA, Brooks CG. The mouse tumor cell lines EL4 and RMA display mosaic expression of NK-related and certain other surface molecules and appear to have a common origin. J Immunol. 2000;164:5094–5102. doi: 10.4049/jimmunol.164.10.5094. [DOI] [PubMed] [Google Scholar]
- 12.Leask A, Abraham DJ. All in the CCN family: essential matricellular signaling modulators emerge from the bunker. J Cell Sci. 2006;119:4803–4810. doi: 10.1242/jcs.03270. [DOI] [PubMed] [Google Scholar]
- 13.Dhar A, Ray A. The CCN family proteins in carcinogenesis. Exp Oncol. 2010;32:2–9. [PubMed] [Google Scholar]
- 14.Gurbuz I, Chiquet-Ehrismann R. CCN4/WISP1 (WNT1-inducible signaling pathway protein 1): a focus on its role in cancer. Int J Biochem Cell Biol. 2015;62:142–146. doi: 10.1016/j.biocel.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 15.Ono M, Inkson CA, Sonn R, Kilts TM, de Castro LF, Maeda A, Fisher LW, Robey PG, Brendsen AD, Li L, McCartney-Francis N, Brown AC, Crawford NP, Molinolo A, Jain A, Fedarko NS, Young MF. WISP1/CCN4: a potential target for inhibiting prostate cancer growth and spread to bone. PLoS One. 2013;8:371709. doi: 10.1371/journal.pone.0071709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wells JE, Howlett M, Cheung LC, Kees UR. The role of CCN family genes in haematological malignancies. J Cell Commun Signal. 2015;9:267–278. doi: 10.1007/s12079-015-0296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacLean JA, Wilkinson MF. The Rhox genes. Reproduction. 2010;140:195–213. doi: 10.1530/REP-10-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson MF, Kleeman J, Richards J, MacLeod CL. A novel oncofetal gene is expressed in a stage-specific manner in murine embryonic development. Dev Biol. 1990;141:451–455. doi: 10.1016/0012-1606(90)90400-d. [DOI] [PubMed] [Google Scholar]
- 19.Magister S, Los J. Cystatins in immune system. J Cancer. 2013;4:45–56. doi: 10.7150/jca.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopitar-Jerala N. The role of cystatins in cells of the immune system. FEBS Lett. 2006;580:6295–6301. doi: 10.1016/j.febslet.2006.10.055. [DOI] [PubMed] [Google Scholar]
- 21.Langerholc T, Zavasnik-Bergant V, Turk B, Turk V, Abrahamson M, Kos J. Inhibitory properties of cystatin F and its localization in U937 promonocyte cells. FEBS J. 2005;6:1535–1545. doi: 10.1111/j.1742-4658.2005.04594.x. [DOI] [PubMed] [Google Scholar]
- 22.Utsonomoiya T, Hara Y, Kataoka A, Morita M, Arakawa H, Mori M, Nishimura S. Cystatin-like metastasis-associated protein mRNA expression in human colorectal cancer is associated with both liver metastasis and patient survival. Clin Cancer Res. 2002;8:2591–2594. [PubMed] [Google Scholar]
- 23.Farabegoli F, Ceccarelli C, Santini D, Taffurelli M. Suppressor of cytokine signalling 2 (SCOS2) expression in breast carcinoma. J Clin Pathol. 2005;58:1046–1050. doi: 10.1136/jcp.2004.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haffner MC, Petridou B, Peyrat PJ, Revillion F, Muller-Holzner E, Daxenbichler G, Marth C, Doppler W. Favorable prognostic value of SOCS2 and IGF-I in breast cancer. BMC Cancer. 2007;7:136. doi: 10.1186/1471-2407-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoefer J, Kern J, Ofer P, Eder IE, Schafer G, Dietrich D, Kirstiansen G, Geley S, Rainer J, Gunsilius E, Klocker H, Culig Z, Puhr M. SOCS2 correlates with malignancy and exerts growth-promoting effects in prostate cancer. Endocr Relat Cancer. 2014;21:175–187. doi: 10.1530/ERC-13-0446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letellier E, Schmitz M, Baig K, Beaume N, Schwartz C, Frasquilho S, Antunes L, Marcon N, Nazarov PV, Vallar L, Even J, Haan S. Identification of SOCS2 and SOCS6 as biomarkers in human colorectal cancer. Br J Cancer. 2014;111:726–735. doi: 10.1038/bjc.2014.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Letellier E, Haan S. SOCS2: physiological and pathological functions. Front Biosci Elite. 2016;8:189–204. doi: 10.2741/E760. [DOI] [PubMed] [Google Scholar]
- 28.Knosp CA, Schiering C, Spence S, Carroll HP, Hel HJ, Osbourn M, Jackson R, Lyubomska O, Malissen B Ingram R, Fitzgerald DC, Powrie F, Fallon PG, Johnston JA, Kissenpfennig A. Regulation of FOXP3+ inducible regulatory T-cell stability by SOCS2. J Immunol. 2013;190:3235–3245. doi: 10.4049/jimmunol.1201396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yowe D, Weih N, Prabhudas M, Poisson L, Errada P, Kapeller R, Yu K, Faron L, Shen M, Cleary J, Wilkie TM, Guitierrez-Ramos C, Hodge MR. RGS18 is a myeloerythroid lineage-specific regulator of G-protein-signalling molecule highly expressed in megakaryocytes. Biochem J. 2001;359:109–118. doi: 10.1042/0264-6021:3590109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delesque-Touchard N, Pendaries C, Volle-Challier C, Millet L, Salel V, Herve C, Pflieger A-M, Berthou-Soulie L, Prades C, Sorg T, Herbert J-M, Savi P, Bono F. Regulator of G-protein signaling 18 controls both platelet generation and function. PLoS One. 2014;9:3113215. doi: 10.1371/journal.pone.0113215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma P, Ou K, Sinnamon AJ, Jiang H, Siderovski DP, Brass LF. Modulating platelet reactivity through control of RGS18 availability. Blood. 2015;126:2611–2620. doi: 10.1182/blood-2015-04-640037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbs TC, Meier KE. Expression and regulation of phospholipase D isoforms in mammalian cell lines. J Cell Physiol. 2000;182:77–87. doi: 10.1002/(SICI)1097-4652(200001)182:1<77::AID-JCP9>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 33.Dufresne AM, Smith RJ. The adapter protein GRB10 is an endogenous negative regulator of insulin-like growth factor signaling. Endocrin. 2005;146:4399–4409. doi: 10.1210/en.2005-0150. [DOI] [PubMed] [Google Scholar]