Abstract
Aim: This pilot study assessed the association of BIM deletion polymorphism and BIM RNA isoform in patients with EGFR-positive non-small cell lung cancer (NSCLC). Patients and Methods: The study included 33 patients with EGFR-positive NSCLC treated with gefitinib. BIM deletion polymorphism and BIM RNA isoform (EL/L/S/γ) were determined by polymerase chain reaction (PCR). Results: BIM-γ expression was significantly higher in patients with BIM deletion polymorphism than among those without BIM deletion polymorphism inside tumors (p=0.038) and around tumors (p=0.0024). Relative BIM-γ expression was significantly higher in patients with BIM deletion polymorphism than among those without BIM deletion polymorphism (p=0.0017). Patients with BIM-γ had significantly shorter progression-free survival than those without BIM-γ (median: 304 vs. 732 days; p=0.023). Conclusion: Expression of BIM-γ mRNA and BIM deletion polymorphism were strongly associated. BIM-γ overexpression may have a role in apoptosis related to EGFR-tyrosine kinase inhibitor.
Keywords: BIM, non-small cell lung cancer, epidermal growth factor receptor tyrosine kinase inhibitor
AbbreviationsBIM, BCL2-like 11; BCL, B-cell CLL/lymphoma 2; BH3, BCL2 homology domain 3; NSCLC, non–small-cell lung cancer; EGFR, epidermal growth factor receptor; EGFR-TKI, epidermal growth factor receptor tyrosine kinase inhibitor; FFPE, formalin-fixed paraffin-embedded; PFS, progression-free survival; OS, overall survival; PCR, polymerase chain reaction; RR, response rate; DCR, disease control rate; CTC, National Cancer Institute Common Terminology Criteria.
Activating mutations in epidermal growth factor receptor (EGFR) are promising targets in the treatment of non-small cell lung cancer (NSCLC) (1,2). The frequency of EGFR mutations varies by population. In North America and Western Europe, approximately 5-10% of patients with adenocarcinoma harbor mutations, whereas in East Asia approximately 60-70% of never-smokers have EGFR mutations (3,4). EGFR tyrosine kinase inhibitors (EGFR-TKIs) induce marked radiographic and clinical improvement in patients with EGFR mutations. EGFR-TKIs such as gefitinib, erlotinib, and afatinib are recommended for treating EGFR-mutated NSCLC (5,6). NSCLC patients with such mutations who were treated with an EGFR-TKI as first-line therapy had longer progression-free survival (PFS) than those who received platinum-based chemotherapy (7-11). Therefore, detection of EGFR mutations in patients with metastatic NSCLC is important for selecting individualized therapies.
Treatment resistance invariably develops within 10 to 16 months after initial EGFR-TKI treatment (12). Approximately 60% of patients with acquired resistance to EGFR-TKIs had an EGFR T790M mutation (13,14). Other reported mechanisms underlying resistance are MET amplification, in 5-10% of cases (15,16), and small-cell cancer transformation, in fewer than 5% of cases (17). However, approximately 30% of patients with EGFR-active mutations do not exhibit an objective response to EGFR-TKI, which is known as primary resistance (18-22). Although the mechanisms of primary resistance have been investigated in several preclinical and retrospective studies, the clinical and molecular characteristics of such resistance remain poorly understood.
BCL2-like 11 (BIM) is a pro-apoptotic member of the B-cell CLL/lymphoma 2 (BCL2) family of proteins (23,24) and is a key modulator of apoptosis triggered by EGFR-TKIs (25,26). Faber et al. (27) used quantitative real-time polymerase chain reaction (PCR) and BIM immunohistochemistry to investigate BIM and β-actin RNA expression in pre-treatment tumors from 24 patients with EGFR-mutant lung cancer. The response rate to EGFR-TKIs was 44% in patients with low BIM expression and 77% in those with high BIM expression, although the difference was not significant. Recent data from the European Tarceva (EURTAC) trial showed that PFS and overall survival (OS) were shorter in patients with low/intermediate BIM mRNA levels in primary tumors than in those with high mRNA levels (PFS: 7.2 vs. 12.9 months, p=0.0003; OS: 22.1 vs. 28.6 months, p=0.0364) (28).
Ng et al. (29) reported a common intronic deletion polymorphism in the gene encoding BIM. This polymorphism switched BIM splicing from exon 4 to exon 3, which resulted in increased expression of BIM RNA isoforms lacking the proapoptotic BCL2-homology domain 3 (BH3), such as BIM-γ. The BIM isoforms with a BH3 domain were BIM-EL, L, and S. This BIM deletion polymorphism was absent in individuals from African and European populations but was present in 12% of an Asian population (29). After EGFR-TKI treatment, PFS was significantly shorter in patients with BIM deletion polymorphism than in those without this polymorphism, which suggests that reduced expression of BIM with a BH3 domain is associated with unfavorable response to EGFR-TKIs (29-33). However, few studies have examined the association between BIM polymorphism and expression of BIM RNA isoforms such as BIM-EL, L, S, and γ.
The present study investigated the association between BIM polymorphism and expression of the BIM RNA isoforms BIM-γ and BIM-EL/L/S in lung tissue from patients with EGFR-positive NSCLC.
Patients and Methods
Clinical samples. We studied 33 patients with EGFR mutation-positive NSCLC who were treated with EGFR-TKIs during the period from January 2008 to January 2016. BIM isoform and BIM deletion polymorphism were investigated by real-time PCR analysis of 33 formalin-fixed paraffin-embedded (FFPE) slides of surgical specimens of lung tissue.
Detection of BIM deletion polymorphism. To identify BIM deletion polymorphism, we performed 2 types of PCR analysis, using the method of Ng et al. (22). In brief, we used a single primer set that contained the deletion area in intron 2, as well as 2 separate primer sets designed for wild-type and deletion alleles. The DNA was subjected to PCR amplification using primers designed to detect the deletion site (2,903 bp) in intron 2 of the BCL2L11 gene. The resulting PCR products from the deletion (1,285 bp) and wild-type (4,188 bp) alleles were analyzed on agarose gels. In addition, the PCR products for the deletion (177 bp) and wild-type (174 bp) alleles were analyzed on agarose gels (30).
Detection of BIM-EL/L/S and BIM-γ. An miRNeasy FFPE Kit (Qiagen KK, Tokyo, Japan) was used to extract total RNA (including miRNA) from the FFPE sections of tumor tissue and non-tumor tissue. The extracted RNA was stored at −80˚C until use. cDNA was synthesized using PrimeScriptRT MasterMix (PerfectRealTime, Takara Bio Inc., Otsu, Japan). Quantitative real-time PCR was performed in a Thermal Cycler Dice Real Time System TP800 (Takara Bio Inc.), using SYBR Premix Ex Taq II (Tli RNaseH Plus, Takara Bio Inc.).
Quantification of BIM, BIM-EL/L/S, and BIM-γ. The quantitative real-time PCR primers (forward and reverse) used Perfect Real Time Primer (Takara Bio Inc.). To correct for differences in quality and quantity between samples, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a reference gene. The targets were obtained from the same mRNA preparations. Relative expression of BIM-EL/L/S and BIM-γ in mRNA from tissue sections inside and around tumors, as normalized to the reference gene (GAPDH mRNA), was calculated by using the KCL22 cell line for calibration.
Clinical outcomes. We retrospectively analyzed the clinical characteristics, response rate (RR), disease control rate (DCR), and toxicity of gefitinib in patients with and without BIM-γ. We then estimated PFS and overall survival (OS) in the same groups. The PFS of patients treated with EGFR-TKI was assessed from the date gefitinib therapy started to the first sign of disease progression, as determined by computed tomographic or magnetic resonance imaging, according to the Response Evaluation Criteria in Solid Tumors (RECIST) criteria. OS was defined as the interval from the date of diagnosis until death from any cause.
Statistical analysis. Statistical analyses were conducted using the SPSS software for Windows, version 12.0 (SPSS Inc., Tokyo, Japan). Differences in relative expressions of BIM, BIM-EL/L/S, and BIM-γ between patients with and without BIM-γ were compared with the Wilcoxon rank sum test. Differences in clinical characteristics, RR, and DCR, frequency of BIM deletion polymorphism, and BIM-γ expression between patients with and without BIM-γ were compared using the Fisher exact test. Survival curves were drawn by the Kaplan–Meier method, and statistical analysis was performed using the log-rank test. A p-value of less than 5% was considered statistically significant.
This single-center study was conducted at Toho University Omori Medical Center (Tokyo, Japan) and was approved by its Human Genome/Gene Analysis Research Ethical Committee (authorization number, 24-1).
Results
BIM deletion polymorphism in EGFR-positive NSCLC. We analyzed BIM deletion polymorphism in 33 patients with EGFR mutation-positive NSCLC who were treated with gefitinib. BIM deletion polymorphism was present in 4 of the 33 patients (12.1%); heterozygous deletion was noted in all 4 patients (Table I).
Table I. Patient characteristics (N=33).
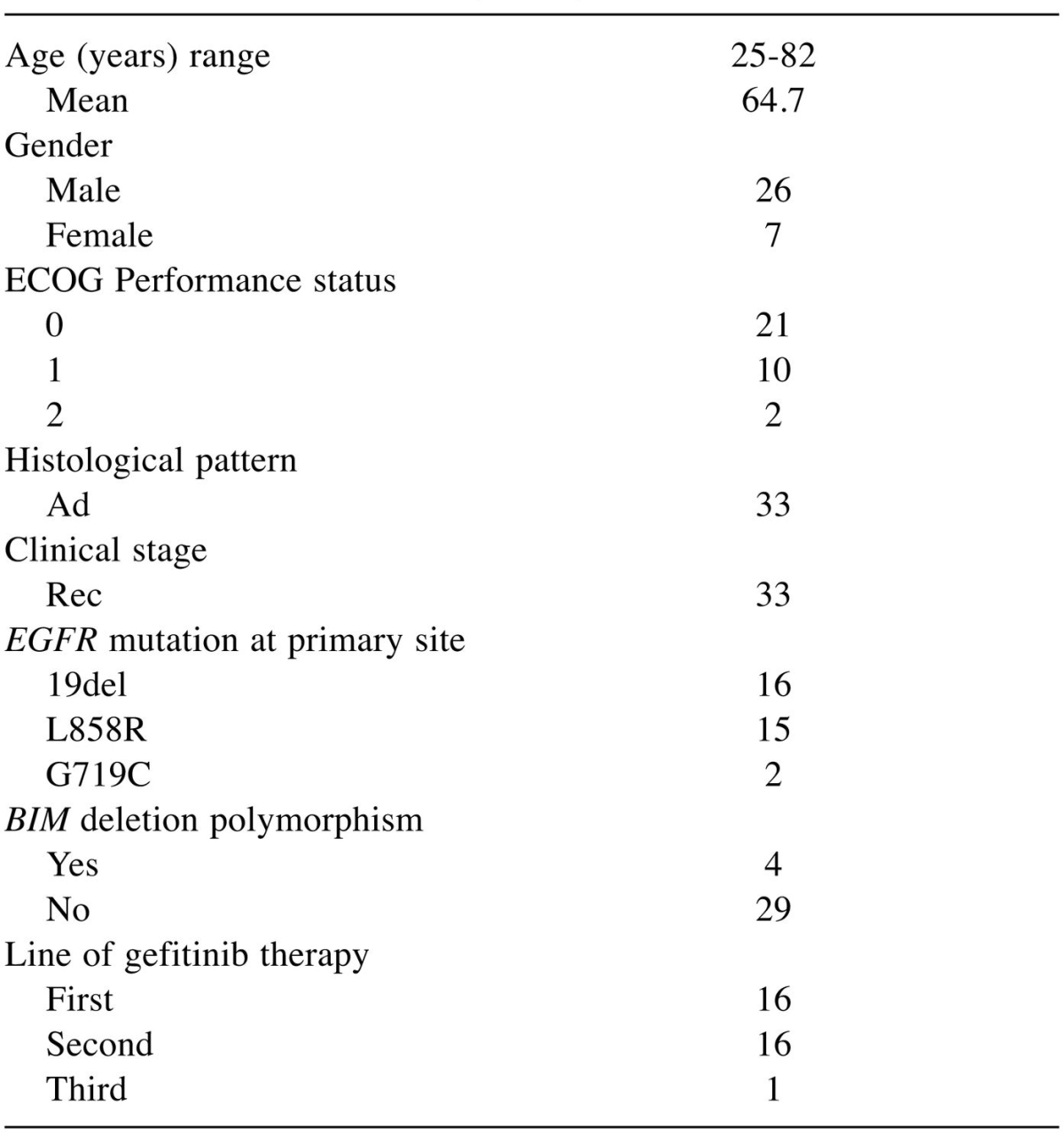
ECOG: Eastern Cooperative Oncology Group; Rec: recurrence after surgical resection; Ad: adenocarcinoma; EGFR: epidermal growth factor receptor; L858R: exon 21 L858R; 19del: exon 19 deletion; G719C: exon 18 G719C.
Clinical characteristics of patients with and without BIM deletion polymorphism. There was no significant difference in RR, DCR, or incidences of adverse events between patients with (n=8) and without (n=25) BIM-γ (Table II).
Table II. Clinical response and adverse events after EGFR-TKI therapy (N=33).
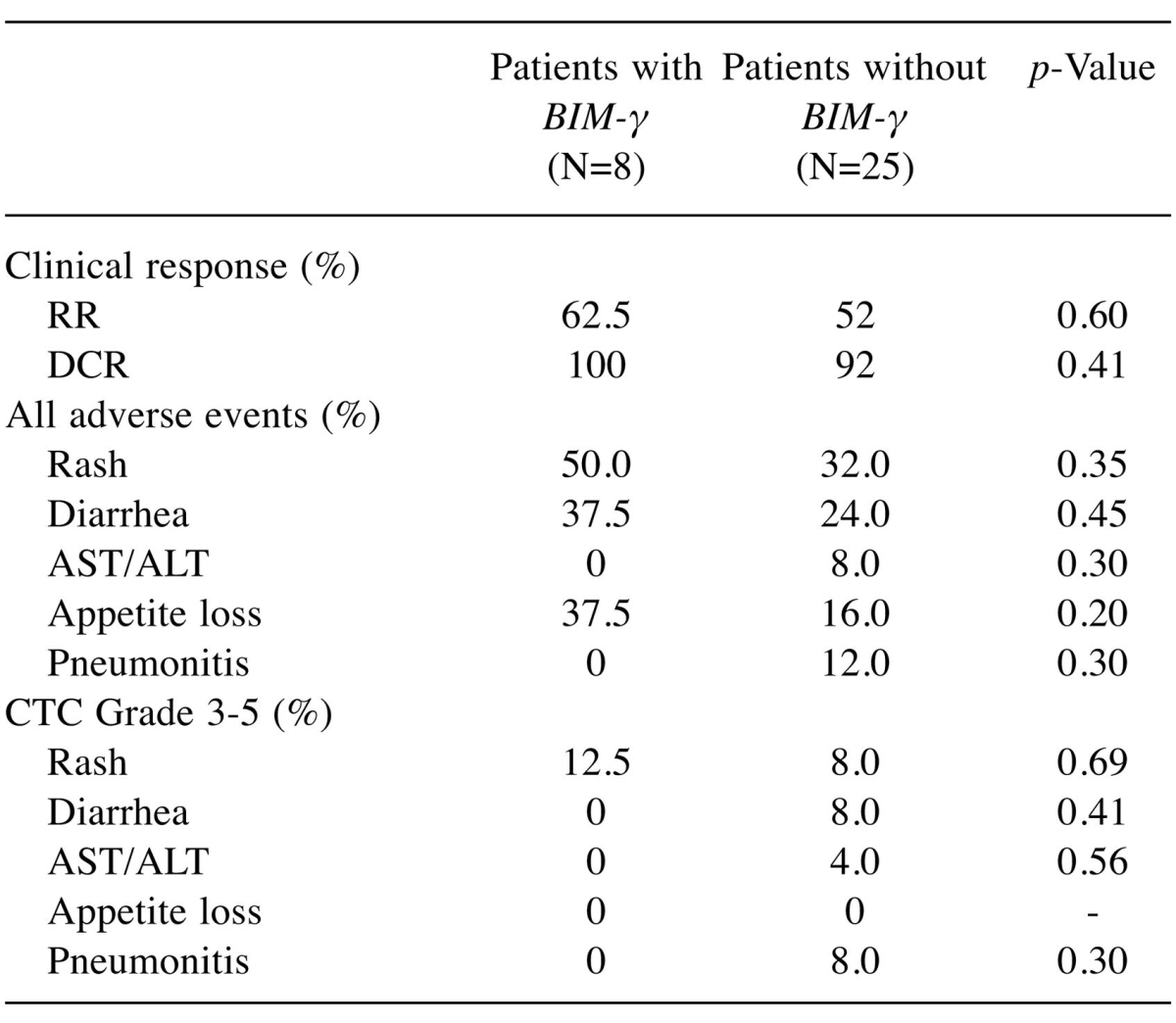
RR: Response rate, DCR: disease control rate, CTC: National Cancer Institute Common Terminology Criteria.
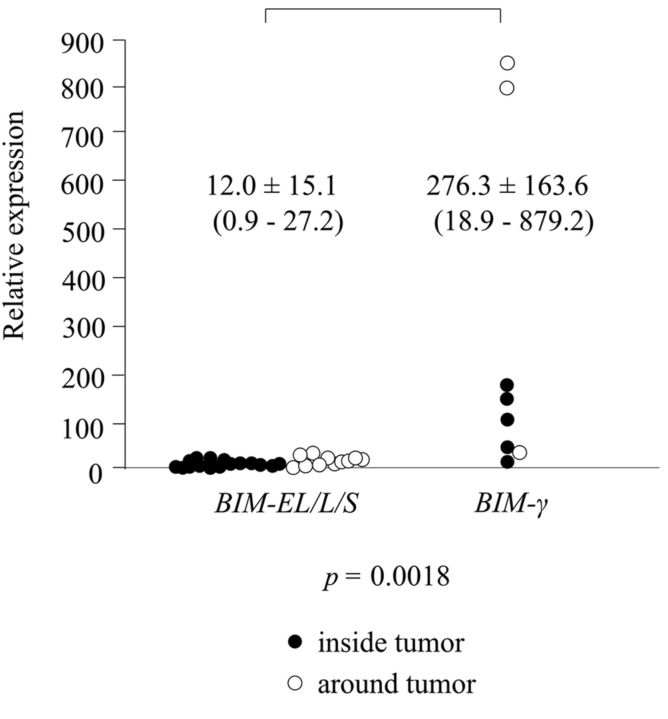
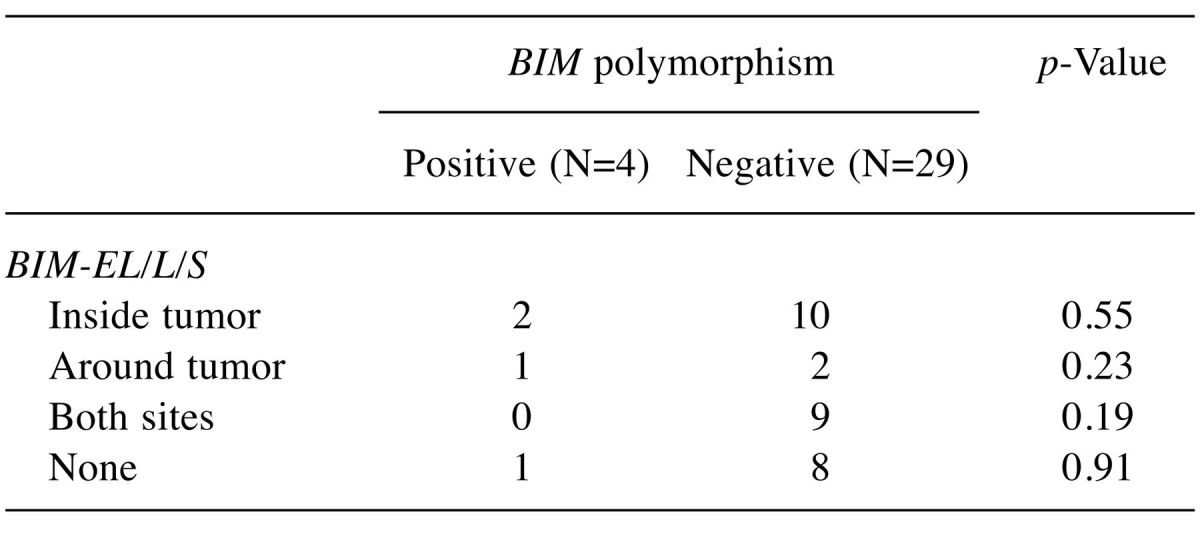
Association of BIM-EL/L/S and BIM-γ expression. Expression of BIM-EL/L/S mRNA was detected inside the tumor in 12 patients, around the tumor in 3 patients, and at both sites in 9 patients; 9 patients had no such expression. Expression of BIM-γ mRNA was detected inside the tumor in 5 patients and around the tumor in 3 patients; 25 patients had no such expression. There was no association between BIM-EL/L/S and BIM-γ expression (Table III). Relative expression was significantly higher for BIM-γ than for BIM-EL/L/S (276±163.6 vs. 12±15.1, p=0.0018) (Figure 1).
Table III. Association of BIM-EL/L/S and BIM-γ mRNA expression (N=33).

Figure 1. Relative expression was significantly higher for BIM-γ than for BIM-EL/L/S (276.3±163.6 vs. 12.0±15.1, p=0.0018).
Association of BIM deletion polymorphism and BIM-EL/L/S expression. We compared BIM-EL/L/S expression in relation to the frequency of BIM polymorphism inside and/or around tumors. There was no significant difference in BIM-EL/L/S expression in any comparison (Table IV).
Table IV. Association of BIM deletion polymorphism and BIM-EL/L/S expression (N=33).
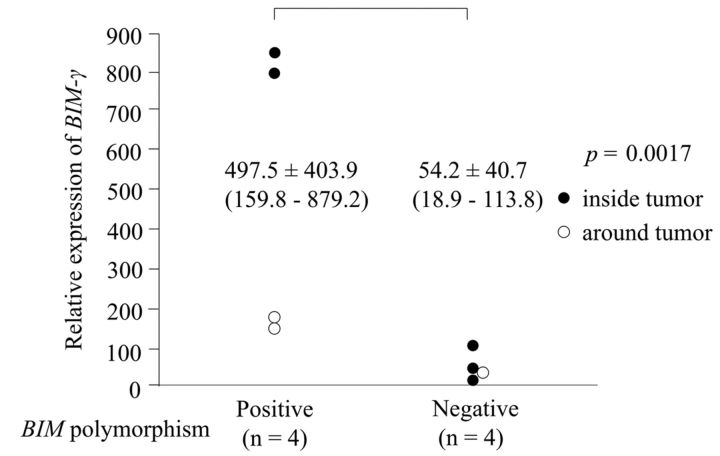
Association of BIM deletion polymorphism and BIM-γ expression. We compared the frequency of BIM deletion polymorphism and BIM-γ expression inside and/or around tumors. BIM-γ expression was significantly more frequent in patients with BIM deletion polymorphism than in those without BIM polymorphism inside tumors (p=0.038) and around tumors (p=0.0024). Absence of BIM-γ expression was significantly more frequent in patients without BIM polymorphism than in those with BIM polymorphism (p=0.00016) (Table V). Relative BIM-γ expression was significantly higher in patients with BIM deletion polymorphism than in those without BIM deletion polymorphism (p=0.0017; Figure 2).
Table V. Association of BIM deletion polymorphism and BIM-γ expression (n=33).
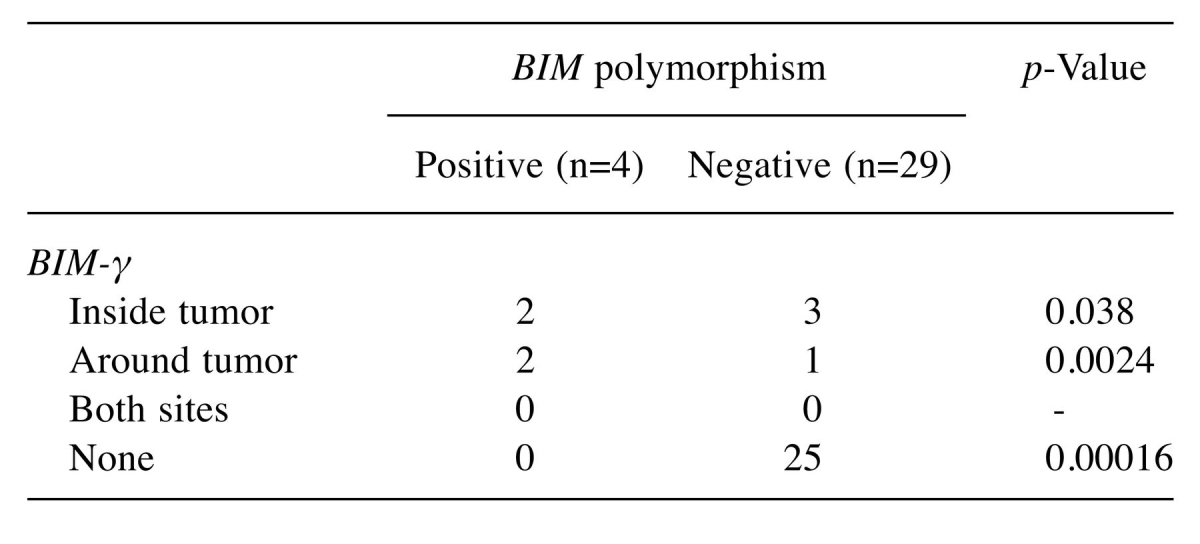
Figure 2. Frequency of BIM-γ expression was significantly higher in patients with BIM polymorphism than in those without BIM polymorphism (p=0.0017).
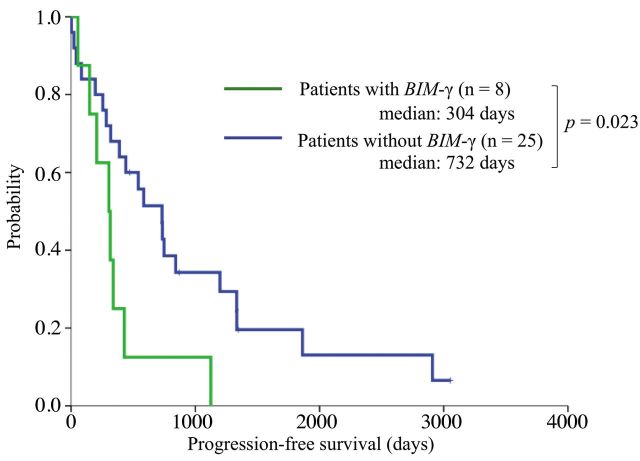
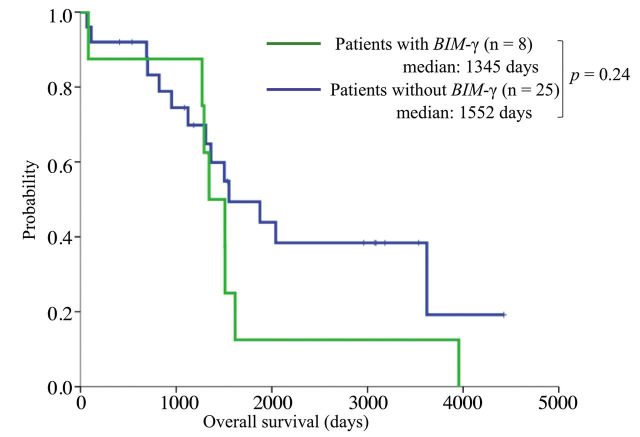
Survival and indicators of shorter PFS. We estimated PFS and OS in patients with and without BIM-γ. Patients with BIM-γ had significantly shorter PFS than those without BIM-γ (median: 304 vs. 732 days; p=0.023; Figure 3). There was no significant difference in OS (median: 1,345 vs. 1,552 days, p=0.24; Figure 4).
Figure 3. Kaplan-Meier curves for progression-free survival. Patients with BIM-γ had significantly shorter progression-free survival than those without BIM-γ (median: 304 vs. 732 days; p=0.023).
Figure 4. Kaplan-Meier curves for overall survival. Overall survival did not significantly differ between patients with and without BIM-γ (median: 1,345 vs. 1,552 days; p=0.24).
Discussion
The BIM deletion polymorphism is located in intron 2 of the BIM gene and results in expression of BIM isoforms lacking the BH3 domain, such as BIM-γ. However, we detected both mRNA BIM-γ and BIM-EL/L/S expression in and around tumors in patients with and without BIM deletion polymorphism. We found no association between BIM-EL/L/S and BIM-γ expression, regardless of the status of BIM deletion polymorphism. Furthermore, relative expression was significantly higher for BIM-γ than for BIM-EL/L/S (276±163.6 vs. 12±15.1, p=0.0018). Faber et al. (24) reported that BIM levels were important in determining response to targeted therapies in patients with solid tumors. This finding is consistent with research showing that cancer cells are sensitive to small changes in BIM protein concentration.
BIM-γ, a BIM isoform that lacks the BH3 domain, is up-regulated in most prostate cancer cell lines (34). BIM-γ inhibits clonal growth in prostate cancer and promotes apoptosis. Interestingly, BIM-γ was found in 13.7% (4 out of 29) of the present specimens without BIM deletion polymorphism. Relative BIM-γ expression in patients without BIM polymorphism was significantly lower than in those with BIM deletion polymorphism (p=0.0017). This suggests that, among the BIM isoforms, overexpression of BIM-γ suppresses TKI-related apoptosis. Further study of the mechanism of BIM-γ expression is warranted.
One hypothesis is that BIM deletion polymorphism itself results in relative resistance to EGFR-TKIs. Kuroda et al. (35) showed that cancer cells were sensitive to small changes in BIM protein concentrations and that BIM protein concentration had a dose-dependent effect on apoptosis and the degree of TKI resistance (35). We compared the frequency of BIM deletion polymorphism and BIM-γ inside and/or around tumors. Patients with BIM deletion polymorphism had significantly higher BIM-γ expression inside tumors (p=0.038) and around tumors (p=0.0024) than those without BIM deletion polymorphism. Absence of BIM-γ expression was significantly more frequent in patients without BIM polymorphism than among those with BIM polymorphism (p=0.00016). These findings suggest a strong association between an imbalance in BIM isoforms and BIM deletion polymorphism.
Clinical characteristics, response to EGFR-TKIs, and incidences of adverse events due to EGFR-TKI did not significantly differ among patients with and without BIM-γ. Thus, clinical characteristics are not sufficient to identify patients with and without BIM-γ. However, our analysis of PFS and OS in patients with and without BIM-γ showed that PFS was significantly shorter in patients with BIM-γ than in those without BIM-γ (median: 304 vs. 732 days; p=0.023). Future studies should attempt to clarify the association between BIM-γ and PFS in patients receiving gefitinib.
The major limitation of this study is that it was a retrospective single-center study with a small sample size. A large-scale multicenter study is thus needed in order to statistically confirm the validity of our results. Clinical application of our results would require a prospective study of patients receiving gefitinib for EGFR mutation-positive NSCLC with or without BIM-γ overexpression. Bean et al. (36) reported that BIM act as sentinels that interconnect kinase signaling networks and the mitochondria-dependent apoptotic program. Karachaliou et al. (37) reported that BIM and mTOR mRNA expression levels predict the outcome of erlotinib therapy in EGFR-mutant NSCLC. Future studies should examine the associations of BIM-γ with PUMA, mTOR, and other apoptosis markers.
In conclusion, the present study is the first to show that BIM-γ expression was strongly associated with BIM deletion polymorphism and that BIM-γ overexpression was associated with TKI-related apoptosis. These findings may be useful in developing treatment strategies for patients receiving EGFR-TKIs for EGFR mutation-positive NSCLC.
Conflicts of Interest
The Authors declare no conflicts of interest.
Acknowledgements
The Authors thank Yoshihisa Otsuka of SRL Inc. (Tokyo, Japan) and Yusuke Hashizawa and Akimitsu Iwama of Astra Zeneca Inc. (Tokyo Japan). They are also grateful to David Kipler for his review of the language of this article. This study was supported by JSPS KAKENHI Grant Numbers JP15K09195, JP2642140, and a Research Promotion Grant from Toho University Graduate School of Medicine (No. 16-3, to K.I.).
References
- 1.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Paez JG, Jänne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 3.Tan DS, Mok TS, Rebbeck TR. Cancer genomics: diversity and disparity across ethnicity and geography. J Clin Oncol. 2016;34:91–101. doi: 10.1200/JCO.2015.62.0096. [DOI] [PubMed] [Google Scholar]
- 4.Tan DS, Yom SS, Tsao MS, Pass HI, Kelly K, Peled N, Yung RC, Wistuba II, Yatabe Y, Unger M, Mack PC, Wynes MW, Mitsudomi T, Weder W, Yankelevitz D, Herbst RS, Gandara DR, Carbone DP, Bunn PA Jr., Mok TS, Hirsch FR. The international association for the study of lung cancer consensus statement on optimizing management of EGFR mutation-positive non-small cell lung cancer: status in 2016. J Thorac Oncol. 2016;11:946–963. doi: 10.1016/j.jtho.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Masters GA, Temin S, Azzoli CG, Giaccone G, Baker S Jr, Brahmer JR, Ellis PM, Gajra A, Rackear N, Schiller JH, Smith TJ, Strawn JR, Trent D, Johnson DH. American society of clinical oncology clinical practice. systemic therapy for stage IV non-small-cell lung cancer: American society of clinical oncology clinical practice guideline update. J Clin Oncol. 2015;33:3488–3515. doi: 10.1200/JCO.2015.62.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuan FC, Kuo LT, Chen MC, Yang CT, Shi CS, Teng D, Lee KD. Overall survival benefits of first-line EGFR tyrosine kinase inhibitors in EGFR-mutated non-small-cell lung cancers: a systematic review and meta-analysis. Br J Cancer. 2015;113:1519–1528. doi: 10.1038/bjc.2015.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mok TS, Wu YL, Thongprasert S, Yang CH, Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 8.Maemondo M, Inoue A, Kobayashi K, Sugawara S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I, Fujita Y, Okinaga S, Hirano H, Yoshimori K, Harada T, Ogura T, Ando M, Miyazawa H, Tanaka T, Saijo Y, Hagiwara K, Morita S, Nukiwa T. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 9.Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, Asami K, Katakami N, Takada M, Yoshioka H, Shibata K, Kudoh S, Shimizu E, Saito H, Toyooka S, Nakagawa K, Fukuoka M. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 10.Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, Zhang S, Wang J, Zhou S, Ren S, Lu S, Zhang L, Hu C, Hu C, Luo Y, Chen L, Ye M, Huang J, Zhi X, Zhang Y, Xiu Q, Ma J, Zhang L, You C. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011;12:735–742. doi: 10.1016/S1470-2045(11)70184-X. [DOI] [PubMed] [Google Scholar]
- 11.Yang JC, Wu YL, Schuler M, Sebastian M, Popat S, Yamamoto N, Zhou C, Hu CP, O'Byrne K, Feng J, Lu S, Huang Y, Geater SL, Lee KY, Tsai CM, Gorbunova V, Hirsh V, Bennouna J, Orlov S, Mok T, Boyer M, Su WC, Lee KH, Kato T, Massey D, Shahidi M, Zazulina V, Sequist LV. Afatinib versus cisplatin-based chemotherapy for EGFR mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6): analysis of overall survival data from two randomised, phase 3 trials. Lancet Oncol. 2015;16:141–151. doi: 10.1016/S1470-2045(14)71173-8. [DOI] [PubMed] [Google Scholar]
- 12.Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol. 2013;31:1070–1080. doi: 10.1200/JCO.2012.43.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 14.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adeno-carcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Jänne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 16.Bean J, Brennan C, Shih JY, Riely G, Viale A, Wang L, Chitale D, Motoi N, Szoke J, Broderick S, Balak M, Chang WC, Yu CJ, Gazdar A, Pass H, Rusch V, Gerald W, Huang SF, Yang PC, Miller V, Ladanyi M, Yang CH, Pao W. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932–20937. doi: 10.1073/pnas.0710370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, Akhavanfard S, Heist RS, Temel J, Christensen JG, Wain JC, Lynch TJ, Vernovsky K, Mark EJ, Lanuti M, Iafrate AJ, Mino-Kenudson M, Engelman JA. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeda M, Okamoto I, Fujita Y, Arao T, Ito H, Fukuoka M, Nishio K, Nakagawa K. De novo resistance to epidermal growth factor receptor-tyrosine kinase inhibitors in EGFR mutation-positive patients with non-small cell lung cancer. J Thorac Oncol. 2010;5:399–400. doi: 10.1097/JTO.0b013e3181cee47e. [DOI] [PubMed] [Google Scholar]
- 20.Cappuzzo F, Jänne PA, Skokan M, Finocchiaro G, Rossi E, Ligorio C, Zucali PA, Terracciano L, Toschi L, Roncalli M, Destro A, Incarbone M, Alloisio M, Santoro A, Varella-Garcia M. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol. 2009;20:298–304. doi: 10.1093/annonc/mdn635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanaka A, Sueoka-Aragane N, Nakamura T, Takeda Y, Mitsuoka M, Yamasaki F, Hayashi S, Sueoka E, Kimura S. Co-existence of positive MET FISH status with EGFR mutations signifies poor prognosis in lung adenocarcinoma patients. Lung Cancer. 2012;75:89–94. doi: 10.1016/j.lungcan.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 22.Sos ML, Koker M, Weir BA, Heynck S, Rabinovsky R, Zander T, Seeger JM, Weiss J, Fischer F, Frommolt P, Michel K, Peifer M, Mermel C, Girard L, Peyton M, Gazdar AF, Minna JD, Garraway LA, Kashkar H, Pao W, Meyerson M, Thomas RK. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res. 2009;69:3256–3261. doi: 10.1158/0008-5472.CAN-08-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Youle RJ, Strasser A. The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol. 2008;9:47–59. doi: 10.1038/nrm2308. [DOI] [PubMed] [Google Scholar]
- 24.Faber AC, Ebi H, Costa C, Engelman JA. Apoptosis in targeted therapy responses: the role of BIM. Adv Pharmacol. 2012;65:519–542. doi: 10.1016/B978-0-12-397927-8.00016-6. [DOI] [PubMed] [Google Scholar]
- 25.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, Tenen DG, Kobayashi S. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med. 2007;4:1669–1779. doi: 10.1371/journal.pmed.0040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, Pao W. Induction of BIM is essential for apoptosis triggered by EGFR kinase inhibitors in mutant EGFR-dependent lung adenocarcinomas. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faber AC, Corcoran RB, Ebi H, Sequist LV, Waltman BA, Chung E, Incio J, Digumarthy SR, Pollack SF, Song Y, Muzikansky A, Lifshits E, Roberge S, Coffman EJ, Benes CH, Gómez HL, Baselga J, Arteaga CL, Rivera MN, Dias-Santagata D, Jain RK, Engelman JA. BIM expression in treatment-naive cancers predicts responsiveness to kinase inhibitors. Cancer Discov. 2011;1:352–365. doi: 10.1158/2159-8290.CD-11-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costa C, Molina MA, Drozdowskyj A, Giménez-Capitán A, Bertran-Alamillo J, Karachaliou N, Gervais R, Massuti B, Wei J, Moran T, Majem M, Felip E, Carcereny E, Garcia-Campelo R, Viteri S, Taron M, Ono M, Giannikopoulos P, Bivona T, Rosell R. The impact of EGFR T790M mutations and BIM mRNA expression on outcome in patients with EGFR-mutant NSCLC treated with erlotinib or chemotherapy in the randomized phase III EURTAC trial. Clin Cancer Res. 2014;20:2001–2010. doi: 10.1158/1078-0432.CCR-13-2233. [DOI] [PubMed] [Google Scholar]
- 29.Ng KP, Hillmer AM, Chuah CT, Juan WC, Ko TK, Teo AS, Ariyaratne PN, Takahashi N, Sawada K, Fei Y, Soh S, Lee WH, Huang JW, Allen JC Jr, Woo XY, Nagarajan N, Kumar V, Thalamuthu A, Poh WT, Ang AL, Mya HT, How GF, Yang LY, Koh LP, Chowbay B, Chang CT, Nadarajan VS, Chng WJ, Than H, Lim LC, Goh YT, Zhang S, Poh D, Tan P, Seet JE, Ang MK, Chau NM, Ng QS, Tan DS, Soda M, Isobe K, Nöthen MM, Wong TY, Shahab A, Ruan X, Cacheux-Rataboul V, Sung WK, Tan EH, Yatabe Y, Mano H, Soo RA, Chin TM, Lim WT, Ruan Y, Ong ST. A common BIM deletion polymorphism mediates intrinsic resistance and inferior responses to tyrosine kinase inhibitors in cancer. Nat Med. 2012;18:521–528. doi: 10.1038/nm.2713. [DOI] [PubMed] [Google Scholar]
- 30.Isobe K, Hata Y, Tochigi N, Kaburaki K, Kobayashi H, Makino T, Otsuka H, Sato F, Ishida F, Kikuchi N, Hirota N, Sato K, Sano G, Sugino K, Sakamoto S, Takai Y, Shibuya K, Iyoda A, Homma S. Clinical significance of BIM deletion polymorphism in non-small-cell lung cancer with epidermal growth factor receptor mutation. J Thorac Oncol. 2014;9:483–487. doi: 10.1097/JTO.0000000000000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao M, Zhang Y, Cai W, Li J, Zhou F, Cheng N, Ren R, Zhao C, Li X, Ren S, Zhou C, Hirsch FR. The Bim deletion polymorphism clinical profile and its relation with tyrosine kinase inhibitor resistance in Chinese patients with non-small cell lung cancer. Cancer. 2014;120:2299–2307. doi: 10.1002/cncr.28725. [DOI] [PubMed] [Google Scholar]
- 32.Lee JH, Lin YL, Hsu WH, Chen HY, Chang YC, Yu CJ, Shih JY, Lin CC, Chen KY, Ho CC, Laio WY, Yang PC, Yang JC. Bcl-2-like protein 11 deletion polymorphism predicts survival in advanced non-small-cell lung cancer. J Thorac Oncol. 2014;9:1385–1392. doi: 10.1097/JTO.0000000000000238. [DOI] [PubMed] [Google Scholar]
- 33.Nie W, Tao X, Wei H, Chen WS, Li B. The BIM deletion polymorphism is a prognostic biomarker of EGFR-TKIs response in NSCLC: A systematic review and meta-analysis. Oncotarget. 2015;6:25696–25700. doi: 10.18632/oncotarget.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu JW, Chandra D, Tang SH, Chopra D, Tang DG. Identification and characterization of Bimgamma, a novel proapoptotic BH3-only splice variant of Bim. Cancer Res. 2002;62:2976–2981. [PubMed] [Google Scholar]
- 35.Kuroda J, Puthalakath H, Cragg MS, Kelly PN, Bouillet P, Huang DC, Kimura S, Ottmann OG, Druker BJ, Villunger A, Roberts AW, Strasser A. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci USA. 2006;103:14907–14912. doi: 10.1073/pnas.0606176103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bean GR, Ganesan YT, Dong Y, Takeda S, Liu H, Chan PM, Huang Y, Chodosh LA, Zambetti GP, Hsieh JJ, Cheng EH. PUMA and BIM are required for oncogene inactivation-induced apoptosis. Sci Signal. 2013;6:ra20. doi: 10.1126/scisignal.2003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karachaliou N, Codony-Servat J, Teixidó C, Pilotto S, Drozdowskyj A, Codony-Servat C, Giménez-Capitán A, Molina-Vila MA, Bertrán-Alamillo J, Gervais R, Massuti B, Morán T, Majem M, Felip E, Carcereny E, García-Campelo R, Viteri S, González-Cao M, Morales-Espinosa D, Verlicchi A, Crisetti E, Chaib I, Santarpia M, Luis Ramírez J, Bosch-Barrera J, Felipe Cardona A, de Marinis F, López-Vivanco G, Miguel Sánchez J, Vergnenegre A, Sánchez Hernández JJ, Sperduti I, Bria E, Rosell R. BIM and mTOR expression levels predict outcome to erlotinib in EGFR-mutant non-small-cell lung cancer. Sci Rep. 2015;5:17499. doi: 10.1038/srep17499. [DOI] [PMC free article] [PubMed] [Google Scholar]