Abstract
Background: Polyomaviruses (PyVs) are potential transforming viruses. Despite their involvement in human tumours still being debated, there is evidence to suggest a role for PyVs in bladder carcinoma (BC). Therefore, a possible association between PyVs and BC was investigated. Materials and Methods: Urine, blood and fresh bladder tissue specimens were collected from 29 patients with BC. PyV prevalence, non-coding control region (NCCR) organization and genotypic analysis were assessed. Results: Data showed a significant prevalence of John Cunningham (JC) PyV in BC tissues and in urine with respect to BKPyV, while simian virus 40 was not revealed. A BKPyV rearranged NCCR sequence was isolated, whereas a JCPyV archetypal structure was consistently retained. A prevalence of European genotypes was observed. Conclusion: Our data would suggest a JCPyV involvement in cancer progression and a BKPyV association with BC pathogenesis in immunocompetent patients. However, further work is necessary to better understand the exact role of PyVs in urothelial carcinogenesis.
Keywords: Polyomaviruses, PyVs, urothelial bladder carcinoma, BKPyV
During recent years, the potential role of the three polyomaviruses (PyVs) detected in human cancers, namely BKPyV, John Cunningham (JC) PyV and simian virus 40 (SV40), has been emphasized. Although evidence implicating PyVs in a variety of human tumours, including brain, pancreas, lung, liver, colon and urinary tract cancer, have been described over the years, there is no conclusive proof that either of these PyVs directly causes or acts as a co-factor in human cancer (1-3). The polyomavirus genome is a double-stranded circular DNA molecule of approximately 5 kb, functionally divided into an early region, which codes for the T antigens (large T and small t); a late region coding for the late proteins (agno and capsid proteins); and a non-coding control region (NCCR, also called regulatory region), which contains several cis-acting sequences important for viral DNA replication (promoter/enhancer elements, binding sites for T antigens, origin of replication) and the binding sites for cellular transcription factors (4). The NCCR of the proposed archetypal BKPyV strain WW has been arbitrarily divided into five transcriptional factor binding sequence blocks, called O (35 bp), P (68 bp), Q (39 bp), R (63 bp), and S (63 bp) (5), whereas the NCCR of the archetype CY, the naturally-occurring JCPyV variant shed in urine of healthy patients, is divided into six regions named box A (36 bp), B (23 bp), C (55 bp), D (66 bp), E (18 bp) and F (69 bp) (6). Different BKPyV and JCPyV strains display a marked heterogeneity in their NCCRs due to point mutations, deletions, duplications, and rearrangements in this region. These rearrangements may play a key role in viral replication, allowing the virus to adapt to ongoing changes within the host cell environment, by increasing or reducing the number or the affinities of host transcription factor binding sites (7,8). Similarly to BKPyV and JCPyV, SV40 possesses variations in the structure of its regulatory region and shares with these closest relative viruses the lowest similarity in the sequences of this region. In fact, the SV40 ‘archetypal’ regulatory region can be divided into the Ori, the G+C-rich domain, containing Sp1-binding sites and comprising part of the early promoter (referred to as the 21-bp repeat region), the enhancer area containing a segment referred to as the 72-bp element, and a region containing the late promoter/initiator (9).
BKPyV and JCPyV usually infect the human population during early childhood. After primary infection, which is usually unapparent, both viruses persist in a latent form in the kidney and possibly in the bone marrow, the hematopoietic system, the leukocytes and the central nervous system (CNS). Conditions of severe immunosuppression may lead to the reactivation of these viruses. JCPyV is the aetiological agent of progressive multifocal leukoencephalopathy (PML), a fatal CNS demyelinating disease, whereas BKPyV causes nephropathy and haemorragic cystitis in kidney or bone marrow transplants (10,11). Conversely, SV40 is a monkey virus that was introduced into the human population through contaminated poliovirus vaccine in the late 1950s; nevertheless, it has been recently associated with certain human tumours such as brain and bone cancer, mesotheliomas and non-Hodgkin’s lymphoma (3).
In 2015, an estimated 74,000 new diagnoses of bladder cancer (BC) were expected to occur in the United States, with approximately 16,000 deaths, 72% of which will be in men. BC incidence is about four times higher in men than in women and almost twice as high in White men than in Black men (12). Urothelial carcinoma, the most common BC type, is the fourth most common cancer in men and the twelfth most common in women in US and results in significant morbidity and mortality. Meanwhile, an estimated 26,000 new diagnoses of BC were expected to occur in Italy in 2015. A total of 5,727 deaths from BC occurred in 2012, 80% of which in men. In Italy, BC represents 4% of total cancer deaths, 5% among men, and 2% among women (13).
Despite the relatively low incidence of bladder carcinoma compared to other cancer types, the high local recurrence rate requires regular medical examination associated with a significant impact on healthcare resources and the patient’s quality of life. Smoking is the most well-established risk factor for BC being approximately 4-fold higher among smokers than non-smokers. Moreover, about half of all BC in both men and women are attributed to smoking. Risk is also increased among workers in the dye, rubber, leather, and aluminium industries, painters, people who live in communities with high levels of arsenic in the drinking water or in areas with a high prevalence of Schistosoma haematobium infection (a parasite encountered in Asia, Africa and South America) and people with certain birth bladder defects (12,14). Recently, viral infections have also been associated with this pathology. BC has been observed in SV40 T antigen (TAg) transgenic mice (15,16). Geetha and colleagues found a strong BKV DNA detection in neoplastic tissue but not in the surrounding non-dysplastic urothelium (17), whereas Weinreb et al. demonstrated a strong association between a prior PyV infection and the subsequent diagnosis of bladder carcinoma (18). The high percentage of BKPyV and JCPyV detection found by Fioriti et al. shows that PyVs may be an important co-factor in BC pathogenesis (14). Despite several reports suggesting PyV involvement in BC aetiology, a prospective sero-epidemiological study in individuals with malignant lesions of the genitourinary tract did not support a clear relationship between BKPyV infection and this type of cancer (19).
Therefore, taking into account these assumptions, the aim of this study was to evaluate the correlation between PyVs and BC and their possible role in the pathogenesis of this tumour. Moreover, the analysis of the genomic sequences of the PyV hypervariable NCCR region was performed in order to determine what type of NCCR configuration was mainly associated with a normal and cancerous bladder cellular environment.
Materials and Methods
Study population. Bladder tissue specimens, blood and urine samples were obtained from 29 immunocompetent patients (mean age=67.6±9.5 years, range=56-87 years) with bladder hyperplasia and carcinoma attending the Department of Obstretics, Gynecology and Urological Sciences of the Umberto I General Hospital (Rome, Italy) between December 2012 and April 2014. When biopsies were performed, both the neoplastic and the adjacent non-neoplastic tissue were considered. Participants gave their written informed consent to the study.
Clinical specimen collection and processing. Standard laboratory procedures for sterile DNA extraction and polymerase chain reaction (PCR) were practiced for all specimens, including the use of ultra-pure, ultraviolet (UV)-irradiated and autoclaved water, autoclaving of all tubes and equipment, tissue processing under a laminar flow, UV irradiated hood, the use of filter-barrier pipette tips, etc.
Fresh urine specimens, collected without preservatives, were centrifuged at 1,376 × g for 10 min at 4˚C. DNA for molecular analysis was extracted from the pellet of each specimen using the QIAamp® DNA Mini Kit (QIAGEN S.p.A, Milan, Italy) according to the manufacturer’s instructions.
Blood samples were collected in 7-ml Vacuntainer® tubes containing EDTA (BD Becton Dickinson, S.p.A, Milan, Italy) and centrifuged at 1,376 × g for 10 min. DNA was extracted from 200 μl of plasma by DNeasy® Blood & Tissue Kit (QIAGEN S.p.A) following manufacturer’s instructions.
Small pieces of fresh normal and cancerous bladder tissue specimens were processed for DNA extraction using QIAamp® DNA Mini Kit (QIAGEN S.p.A) and the recommended tissue protocol. All extracted DNA samples were stored at −20˚C until use.
Histological and cytological examinations. Bladder transurethral resection was used to collect samples for histological examination. Specimens were formalin fixed and paraffin embedded; serial 5-μm microtome sections were stained with haematoxylin-eosin and examined under light microscopy. The tumours were classified according to the World Health Organization diagnostic criteria and staged according to the 2010 American Joint Committee on Cancer consensus guidelines (20-22). Urinary samples were stained with Papanicolaou method and analysed for the presence of cytological abnormalities.
Real-time TaqMan PCR (Q-PCR). Total DNA extracted from each sample was tested for human PyV genome detection and quantification using the BKPyV Q-PCR Alert Kit and the JCPyV Q-PCR Alert Kit (ELITechGroup S.p.A, Trezzano S/N, Milan, Italy). These two commercial kits are specific for both a region of the gene that encodes BKPyV and JCPyV TAg and for the promoter region and 5’ untranslated region (5’ UTR) of the human beta globin (β-globin) gene. β-globin gene was amplified simultaneously with the target sequence to verify successful DNA isolation and exclude false-negative results. Conversely, primers and probes were designed to detect sequences in the conserved N-terminal region of the SV40 TAg gene (23). A real-time PCR for β-actin gene was performed to control for the suitability of DNA samples tested for SV40 following the manufacturer’s instructions (Applied Biosystems, Foster City, CA, USA).
All three quantitative assays, based on TaqMan-MGB® (Minor Groove Binder) technology, were performed using a 7300 Real-Time PCR System and SDS 1.4 software to analyze the amplification data (Applied Biosystems).
All samples were tested in triplicate and the number of viral copies in each sample was calculated from a standard curve. The standard curve for quantification of human PyVs was obtained using four stabilized serial dilutions at known titer of a plasmid containing part of the BKPyV and JCPyV TAg (range: 102-105 plasmid copies). Conversely, the standard curve for SV40 quantification was constructed using serial dilutions (ranging from 102-105 copies) of a plasmid containing the whole genome of SV40-B2E. Standard precautions designed to prevent contamination during Q-PCR were followed and a negative control with the reaction mixture without a DNA template was included in each run. The lower detection limit for the assays was 10 DNA copies of the target gene per amplification reaction, corresponding to 10 genome equivalents per reaction (10 gEq/reaction).
Data are expressed as genome equivalents of viral DNA (gEq)/ml of urine and plasma samples and as genome equivalents (gEq)/mg of DNA isolated from tissues.
PCR for NCCR and VP1 gene typing region. All samples resulted positive for viral DNA were subsequently screened for the NCCR of human BKPyV and JCPyV by nested-PCR using specific sets of primers (24,25), whereas a single PCR amplification was used to analyze the SV40 NCCR sequences (26) by means of a 9700 GeneAmp® PCR System (AB Applied Biosystems). In order to define the viral genotype of the isolated BKPyV and JCPyV strains, a PCR amplifying the VP1 typing region of both viruses was carried out with specific primers for these regions (27-29). All assays included positive (previously amplifiable DNA) and negative (pure water as template) controls to exclude false-positive and false-negative results. PCR products were detected by electrophoresis on a GelRed™ (Biotium)-stained 2% agarose gel and visualized under UV light.
Sequencing of NCCR and VP1 regions. PCR products corresponding to the NCCR and VP1 regions were purified with QIAquick® PCR purification kit, according to QIAGEN protocol (30). DNA sequencing was performed using a dedicated facility (Bio-Fab research s.r.l., Rome, Italy). All NCCR sequences were compared to the BKPyV prototypic sequence (31) and to the JCPyV archetype strain CY (32), in order to investigate the presence of possible rearrangements within these regions. SV40 sequence homology searches, instead, were performed using the Basic Local Alignment Search Tool (BLAST) at the National Center for Biotechnology Information (NCBI) website. Conversely, sequences obtained from amplification of the VP1 gene typing region of both viruses were analysed on the basis of single nucleotide polymorphisms (SNPs) used to classify the BKPyV subtypes/subgroups (33) and the JCPyV genotypes/subtypes (34). Sequence alignments were performed with ClustalW2 at the European Molecular Biology Laboratory–European Bioinformatics Institute (EMBL-EBI) website using default parameters (35).
Cloning and sequencing. PCR amplicons corresponding to the exact size of human PyV NCCR, were cloned into a pGEM-T Easy Vector (Promega Corporation, Madison, WI, USA). Plasmid DNA was purified from up to 25 clones of amplicons using the Wizard® Plus SV mini-preps DNA purification system (Promega Corporation), visualized in 1% agarose gel electrophoresis and sequenced (BioFab research s.r.l.). Homology between amplicons cloned and the human PyV archetypal NCCR sequences (31,32) was searched by ClustalW2 at the EMBL-EBI website (35).
Statistical analysis. Data are summarized as medians and ranges or as mean, as appropriate. If a Z test indicated a non-normal distribution, we used non-parametric test such as Mann–Whitney U-tests and Kruskal–Wallis tests. Categorical data were analysed by using χ2 test and Student’s t-test. p-Values less than 0.05 were considered statistically significant.
Results
PyV detection in biological specimens. This study was conducted on 29 patients with BC from whom we collected 58 fresh bladder specimens (29 benign and 29 cancerous tissue samples) at surgery and 29 urine samples. The histological examination showed high-grade urothelial carcinoma in 31% of patients, low-grade urothelial carcinoma in 58.6% and bladder hyperplasia in 10.3% of patients (Table I).
Table I. Summary of polyomavirus (PyV) detection and characteristics in all clinical specimens provided.
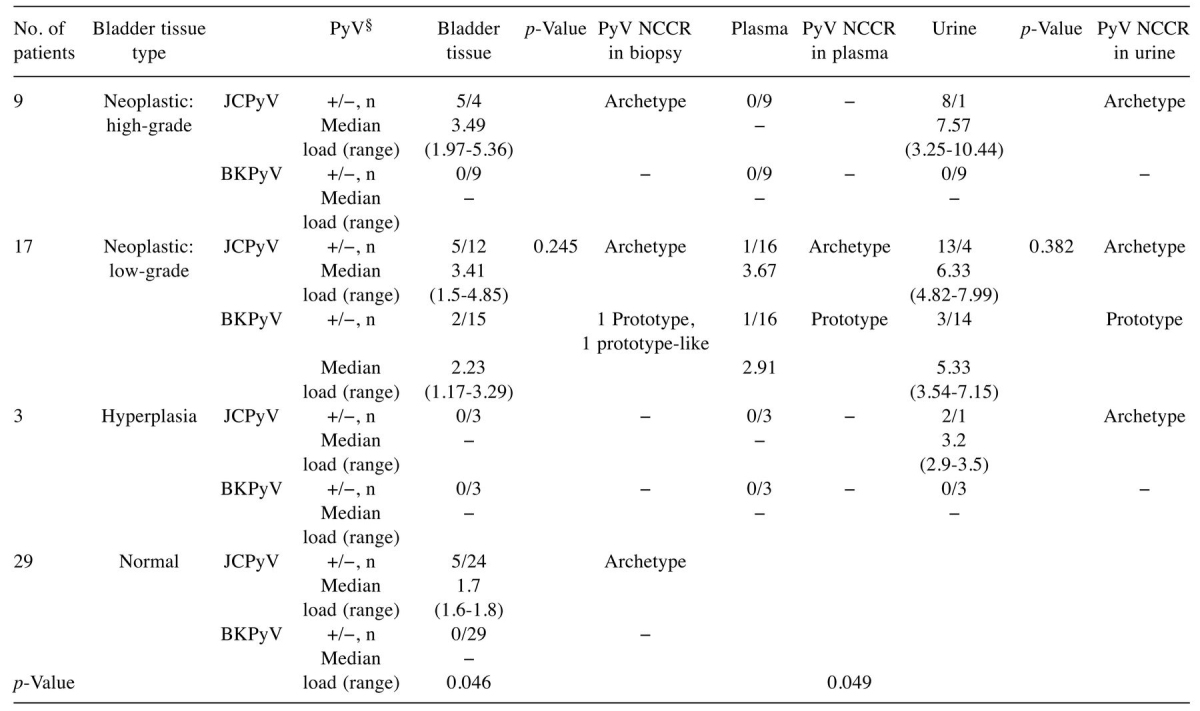
JCPyV: John Cunningham polyomavirus; BKPyV: BK polyomavirus; NCCR: non-coding control region.*By Mann–Whitney U-test; p<0.05 was considered statistically significant. §BKPyV and JCPyV loads are expressed as median (range) of log10 genome equivalent (gEq)/ml in urine and in plasma and as median (range) of log10 genome equivalent (gEq)/mg in tissues.
Q-PCR assays did not reveal the presence of SV40 DNA sequences in any of the recruited patients. Regarding human PyV DNA detection in urine sample, JCPyV was found in 88.9% of patients with high-grade urothelial carcinoma, in 76.5% with low-grade urothelial carcinoma, and in 66.7% of patients with bladder hyperplasia. Conversely, BK viruria was detected only in 17.6% of patients with bladder hyperplasia. Concerning blood samples, JCPyV DNA was detected in the plasma sample of one patient (5.9%) with low-grade urothelial carcinoma. Similarly, BK viremia was found in another patient with low-grade urothelial carcinoma (5.9%) (Table I). Considering bladder specimens, JCPyV was detected in 55.6% of patients with high-grade urothelial carcinoma and in 29.4% of patients with low-grade urothelial carcinoma. BKPyV DNA was detected only in 29.4% of patients with low-grade urothelial carcinoma. No viral DNA was found in hyperplastic tissue. Finally, JCPyV DNA was found in 17.2% of benign bladder tissue specimens, whereas BKPyV genome was not detected (Table I).
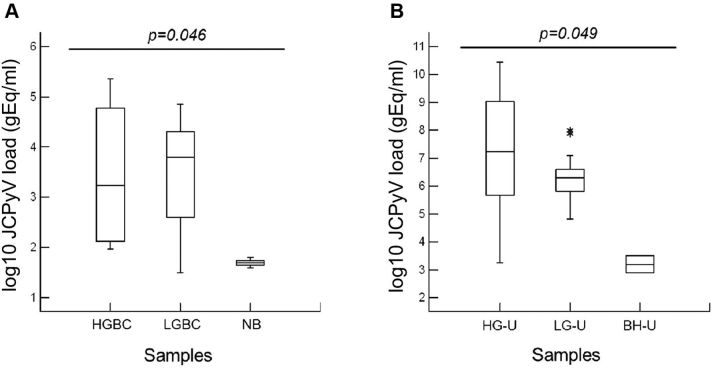
Statistical analysis of these data highlighted that there was no statistically significant difference between BKPyV and JCPyV loads detected in low-grade urothelial carcinoma tissues (p=0.245) nor in urine belonging to the same class of patients with BC (p=0.382) (Table I). Conversely, the comparison between JCPyV load in patients with neoplastic and benign bladder tissues was statistically significant (p=0.046) (Figure 1A). A statistically significant difference was also obtained comparing the JCPyV load found in urine specimens of patients with high- and low-grade urothelial carcinoma and bladder hyperplasia (p=0.049) (Figure 1B).
Figure 1. John Cunningham polyomavirus (JCPyV) load values found in cancerous and benign bladder tissue and in urine specimens of patients with bladder carcinoma (BC). A statistically significant difference was obtained comparing the JCV loads detected (A) in patients with neoplastic and benign bladder tissues (p=0.046) and (B) in urine specimens of patients with high- (HGBC) and low-grade (LGBC) urothelial carcinoma and bladder hyperplasia (p=0.049). NB: Normal bladder; HG-U: urine from patients with HGBC; LG-U: urine from patients with LGBC; BH-U: urine from patients with bladder hyperplasia.
Finally, categorizing the results according to the number of patients and type of virus, JCPyV was detected in 79.3% (23 out of 29) of the individuals and BKPyV in 10.3% (3 out of 29). In two patients, both human PyVs were detected, whereas five patients were negative for both PyVs. Therefore, JCPyV seems to be more common than BKPyV in the cohort of patients enrolled in this study.
Sequencing analysis of human PyV NCCR region. All specimens positive for viral DNA were further analyzed for the presence of rearrangements within the NCCR region of both PyVs. The NCCR sequences amplified by each BKPyV-positive clinical sample were compared to the prototypic NCCR sequence defined by Yogo et al. (31), which spans from the midpoint of the origin of replication and the start site of the agnoprotein gene. Specifically, the 268 nucleotides of the prototypic NCCR sequence were arbitrarily divided into five blocks (O, P, Q, R, and S) with lengths of 35, 68, 39, 63, and 63 nucleotides, respectively, in order to more easily describe the major rearrangements within this region.
Homology sequence analysis evidenced that the BKPyV NCCR sequences isolated from the urine of two patients were identical to the prototypic NCCR, consisting of a single complete copy of the five blocks arranged in the correct order, with the occurrence of a few point mutations at nucleotide positions 4 within R-block and 18 within S-block, and a single thymidine nucleotide deletion at nucleotide position 50 within S block. These point-mutations did not involve any cellular transcriptional factor binding sites, except for the single-nucleotide deletion at position 50 within the S block involving the estrogen response element (ERE) (Figure 2, isolate A). One of these two patients exhibited a rearranged NCCR structure in the malignant tissue, consisting of a whole R block deletion and of a single complete copy of the remaining P, Q and S blocks arranged in the correct order (Figure 1). This rearranged sequence was similar to the DDP strain of BKPyV isolated from peripheral blood mononuclear cells by Degener et al. (36), and it was also characterized by the same nucleotide changes within the R and S blocks found in isolate A (Figure 2, isolate B). Finally, for the three remaining sequences, obtained from the malignant tissue, plasma and urine samples of a single patient, a prototypic NCCR consisting of a single complete copy of the P, Q, R and S blocks organized in the correct order was isolated. However, all these sequences were characterized by various nucleotide substitutions at nucleotide positions 55 in the P block, 7 and 41 in the R block, 24 in the S block and by the deletion of a single adenine nucleotide at nucleotide position 58 within P block. The nucleotide substitutions at position 55 in the P block and 41 in the R block involved the binding sites for the cellular factors p53 and nuclear factor 1 (NF1), respectively, whereas the adenine deletion at position 58 within the P block invoIved the binding sites for the transcriptional factor c-myc (Figure 2, isolate C).
Figure 2. Alignment of BK polyomavirus non-coding control region (BKPyV NCCR) isolates from different clinical specimens of patients with bladder cancer the prototypic NCCR sequence. The prototypic NCCR defined by Yogo and colleagues (31) is shown at the top of the figure, while nucleotide sequences of three variants, determined directly from malignant (low-grade) bladder tissue, urine and blood samples, are shown below in relation to the prototypic NCCR, with nucleotide substitutions indicated by bold highlighted letters and deletions identified by bold highlighted hyphens. The genotype/subtype corresponding to each isolate is also indicated. Blocks (P, Q, R and S) commonly used to denote archetypal NCCRs (5) are indicated under the NCCR sequences, while proven and putative binding sites for transcriptional factors are illustrated above the prototypic NCCR sequence. The O-block, containing the origin of replication, was omitted.
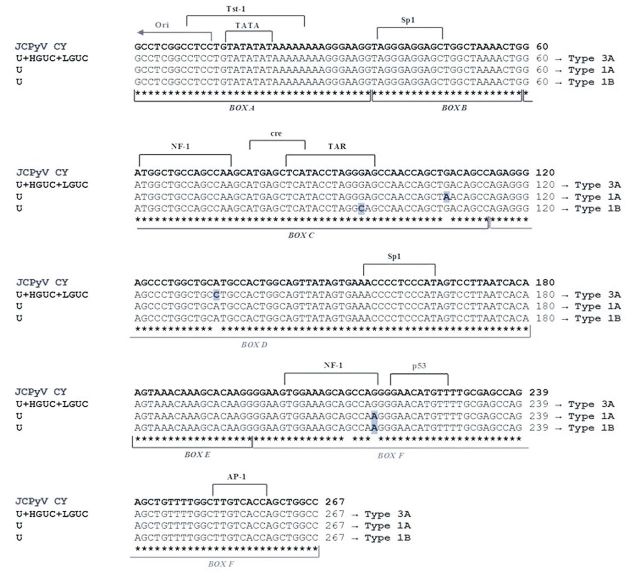
Concerning JCPyV, the structure of its NCCR allows the virus to be classified into two forms: the non-pathogenic archetype CY and the pathogenic Mad-1 variants. As mentioned above, the NCCR of the archetype CY is divided into six regions named box A (36 bp), B (23 bp), C (55 bp), D (66 bp), E (18 bp) and F (69 bp) (6). In contrast, the NCCR of the Mad-1 strain contains a 98-bp tandem repeats of boxes A, C and E, with the deletion of boxes B and D (37). The sequence analysis of the JCPyV NCCR performed on positive urine and bioptical tissue specimens always showed the presence of archetypal structures with a few single-nucleotide substitutions (G95C, G108A, A133C and G217A). In particular, the nucleotide variations G108A and A133C did not involve any cellular transcriptional factor binding sites, whereas the nucleotide substitutions G95C and G217A involved the binding sites for the transactivation response element (TAR) and NF1, respectively (Figure 3).
Figure 3. Alignment of John Cunningham polyomavirus non-coding control region (JCPyV NCCR) isolates from different clinical specimens of patients with bladder cancer the archetypal NCCR sequence. The NCCR sequence of the archetypal JCPyV strain CY, as proposed by Yogo et al. (32) is shown at the top of the figure and the consensus sequences of the NCCR showing variations isolated from malignant (high- and low-grade) bladder tissues (CPT and NPT, respectively) and urine specimens, with related genotype, are shown below. The four nucleotide changes (G95C, G108A, A133C and G217A), identified by comparing with CY isolate, are indicated by bold highlighted letters. The JCPyV NCCR isolated from specimens of the other patients always showed an archetypal CY-like structural organization and the nucleotide substitution G217A in those patients with a JCPyV European genotype (data not shown). The six boxes commonly used to divide the archetypal NCCR (6) are indicated under the NCCR sequences, while proven and putative binding sites for transcriptional factors are illustrated above the CY NCCR sequence. U: Urine; HGBC: high-grade urothelial bladder carcinoma; LGBC: low-grade urothelial bladder carcinoma.
PCR and sequencing analysis of human PyV VP1 region. An additional specific PCR was undertaken to detect the presence of VP1 region in all BKPyV- and JCPyV-positive samples. Regarding BKPyV, the 327 bp VP1 gene sequence was amplified in three urine, one plasma and two low-grade urothelial carcinoma samples. The amplified PCR products were then sequenced in order to classify each BKPyV strain into the corresponding subtype/subgroup, analysing the SNPs within the amplified VP1 region and aligning our 327 bp typing isolates with the consensus sequences generated for each BKPyV subtype/subgroup (33). Based on the SNPs and on the consensus sequences, subtype I/subgroup b-2 was detected in two out of three urine samples (66.7%) and in one out of two bioptical samples (50%), while subtype II was detected in urine, in plasma and in bioptical specimens belonged to the same patient.
The SNPs and the consensus sequence generated from the alignment of 32 complete JCPyV genomes (34), were used to determine the JCPyV genotype of our positive samples. In particular, among the 29 enrolled patients, a prevalence of the European genotypes 1A, 1B and 4 was observed, followed by the African genotype 3A. In particular, genotypes 1A and 1B were found in six (20.7%) and in 10 (34.5%) individuals, respectively, whereas type 4 was identified in two patients (6.9%). Finally, five patients (17.2%) carried the African genotype 3A.
Discussion
The potential oncogenicity of PyVs in both animal and in vitro models has been now thoroughly documented. The three PyVs detected in humans (BKPyV, JCPyV and SV40) exhibit oncogenic properties via the TAg, which can bind and block the tumor-suppressor proteins p53 and pRb (38,39). The profound PyV-induced effects on cell activation and on the cell cycle, i.e. the shift to proliferation and inhibition of apoptosis, in the context of marked immunosuppression, constitute a potentially ideal background for malignant transformation. However, no study has provided unambiguous evidence of a statistical or temporal link between PyV infection and subsequent cancer onset. Nevertheless, several lines of evidence have suggested a significant role of PyVs, mainly of BKPyV, in the development of urothelial carcinoma. After primary infection, both BKPyV and JCPyV remain latent lifelong in the epithelium of the urinary tract (40). Two reports showed that the urothelium-specific expression of polyomavirus SV40 TAg in transgenic mice produced tumors strongly resembling human carcinoma in situ of the bladder (15,16). Using PCR technology, two independent Italian studies reported that a high percentage of BKPyV sequences detected was significantly associated with urinary bladder carcinoma (14,41). Furthermore, a statistically significant association between urine cytological evidence of PyV infection (decoy cells) and BC was also demonstrated in immunocompetent patients (18). Despite this experimental and epidemiological evidence, clinical cases of PyV-associated urological malignancies are rare and presented mainly as isolated case reports, mostly in transplant recipients or rarely in immunocompetent patients (17,42-52).
The intriguing hypothesis that BKPyV might contribute to bladder carcinogenesis was not supported by the results of this study. In fact, the percentage of BKPyV DNA-positive cancerous bladder tissues (7.7%, 2/17) was found to be lower than those reported by two previous molecular studies, where BKV genome was found in 55% (14/32) and in 58% (15/26) of urinary bladder carcinoma cases, mainly in papillar urothelial carcinomas (14,41). Similarly to these two reports, we detected BKPyV sequences in the same tumour histotype. Nevertheless, our percentage of BKPyV detection (7.7%) is similar to that reported in a tissue-based study by Rollison and colleagues (45), who detected BKPyV DNA by PCR in only 5.5% (4/76) of urothelial carcinomas. Moreover, both our patients with BKPyV-positive cancerous bladder tissues exhibited viruria and one also viremia.
Since there are no known differences in the population prevalence of BKPyV infection, methodological differences in laboratory procedures most likely account for the wide range of BKPyV DNA prevalence reported in bladder tumour tissues.
Despite the high structural homology between BKPyV and JCPyV, the latency site in the genitourinary tract and their potential oncogenicity, only one study, to our knowledge, has investigated the relationship between JCPyV and BC (14). Therefore, we extended this study to investigate a possible causative role of this virus in BC development. Our results showed the presence of JCPyV DNA in 38.5% (10/26) of cancerous bladder tissue samples, with the highest positivity detected in high-grade carcinoma (55.6% vs. 29.4% of low-grade), whereas in benign bladder tissue, JCPyV was found in 17.2% (5/29) of patients. These data are consistent with those reported by Fioriti et al. in 2003, who detected JCPyV DNA in 25% (6/32) of urinary bladder carcinoma cases (14), with the same percentage of infection in both high- and low-grade urothelial carcinoma (50%). Moreover, Fioriti et al. reported co-infection with BKPyV and JCPyV in 72% (9/13) of infected bladder carcinoma specimens, whereas in the current study, only one patient showed a co-infection of BKPyV with JCPyV in the infected neoplastic tissues (1/12: 8.3%).
The comparison between the JCPyV loads detected in neoplastic (high- and low-grade) and benign bladder tissues was statistically significantly different (p=0.046) (Figure 1A), whereas there was no a statistically significant difference between BKPyV and JCPyV loads detected in low-grade urothelial carcinoma tissues (p=0.245).
Regarding JCPyV DNA detection in circulatory and urinary compartments, JC viremia was detected in only one patient with low-grade urothelial carcinoma. By contrast, JC viruria was found in 88.9% (8/9) and in 76.5% (13/17) of patients with high- and low-grade urinary bladder cancer, respectively, and in 66.7% (2/3) of bladder hyperplasia. Furthermore, a statistically significant difference in the rate of urinary JCPyV excretion among these patients was observed (p=0.049) (Figure 1B). Conversely, no significant difference between BKPyV and JCPyV loads was detected in the urine samples belonging to the low-grade carcinoma class (p=0.382). In particular, two individuals with low-grade carcinoma showed simultaneous urinary excretion of both viruses. Therefore, JCPyV reactivation from latency is by far more frequent than BKPyV in the cohort of patients enrolled in this study. This finding is not surprising since Kitamura and colleagues reported a high incidence of urinary JCPyV excretion in older immunocompetent patients (53).
The purpose of the histological examination of the bladder tissue specimens was to assess whether a specific BKPyV and JCPyV NCCR configuration may be associated with a normal or cancerous cellular environment. The NCCR of both human PyVs contains several cis-acting sequences crucial for viral DNA replication (promoter/enhancer elements, binding sites for T antigens, origin of replication) and the binding sites for cellular transcription factors (4). The main BKPyV and JCPyV NCCR characteristic is its being a highly variable region which may undergo sequence rearrangements yielding viral variants that could up-modulate viral expression in a specific anatomical site or acquire new tissue tropism and greater pathogenic potential (7). Monini and colleagues reported the detection of BKPyV sequences by PCR and Southern blotting in normal as well as malignant tissues from bladder, prostate and kidney, with a prevalence ranging from 50 to 83% (41). The sequence analysis of these BKPyV NCCRs showed that these tissues were infected by a novel BKPyV strain, URO1. This strain, mainly prevalent in the italian population, is characterized by NCCR rearrangements that may affect viral DNA replication. However, it remains to be determined whether the mutations in URO1 affect replication or transformation (1). Recently, Mischitelli and colleagues found two rearranged BKPyV variants, characterized by enhancement of the binding sites for c-myc and p53, in cancerous prostatic tissues of two patients affected by advanced prostate cancer (54). Moreover, Anzivino et al. isolated a BKPyV DDP-like NCCR sequence from the benign prostatic tissue of one patient with Gleason 7 prostate cancer (7).
In this study, a prototypic and a DDP-like NCCR sequence were isolated from the neoplastic (low-grade) bladder tissue of two BKPyV-positive patients. This rearranged NCCR differed from the prototypical one for a whole R-block deletion and for a perfectly conserved single copy of the remaining O, P, Q and S blocks arranged in the correct order (Figure 2). The simultaneous presence of a rearranged NCCR sequence in low-grade cancerous tissue and of a prototypical structural organization of this region in the excreted virus could likely be explained by the occurrence of a NCCR structural alteration during BKPyV persistence in the urogenital tract. We might speculate that alterations in the urinary bladder microenvironment allow the virus to actively replicate and select for one or more viral strains or NCCR variants able to transform the host tissue. Ongoing replication of the virus might also lead to viral and cellular modifications that could promote genomic instability upon which further hits could occur, contributing to tumourigenesis or cancer progression. This hypothesis is supported by Monini et al., who noticed that BKPyV DNA rearrangement occurred at an early stage of neoplastic initiation or progression (41). Moreover, our assumption might explain the finding of a BKPyV rearranged NCCR in normal prostatic tissue by Anzivino and colleagues (7). However, the possibility of ‘hit-and-run’ carcinogenic mechanisms induced by BKPyV cannot be excluded (55).
Interestingly, a BKPyV prototypic NCCR sequence was isolated from a patient whose malignant bladder tissue was also infected with JCPyV. The finding of a non-rearranged regulatory region could be explained assuming that cellular environment created by neoplastic-cell proliferation, in association with JCPyV infection, might cause favourable conditions for BKPyV replication and its subsequent rearrangement. Indeed, the BKPyV load detected in this bioptical tissue (3.29 gEq/mg) was higher than that found in the malignant tissue infected by the rearranged virus (1.17 gEq/mg) (Table I).
Concerning JCPyV, the NCCR of the archetype CY is the naturally occurring JCPyV variant shed in urine of immunocompetent individuals, whereas that of the pathogenic JCPyV Mad-1 strain is usually isolated from the brain or cerebrospinal fluid of patients with PML (11). In addition, the JCPyV Mad-1 strain has been described to be the only JCPyV variant found in the human colon, and a variant lacking one of the characteristic 98 bp repeats within the NCCR was only found in cancer tissue, suggesting that Mad-1-like strain might be implicated in development of chromosomal instability in colonic epithelium (56,57). The Mad-1 strain was also detected in the prostate gland associated with the urinary tract together with different NCCR sequences, suggesting that a single prostate may be inhabited by more than one JCPyV strain or NCCR variant (58). The archetype CY-like structural organization of NCCR in cancerous and benign urinary bladder tissues was found (Figure 3). This led us to speculate that the non-pathogenic JCPyV CY strain might be selected for replication and persistently infect the bladder urothelium. Moreover this could explain the higher replication rate in the urinary epithelium, with concomitant excretion of progeny virions into the urine, observed in patients with high-grade BC compared with individuals with low-grade BC and bladder hyperplasia (p=0.046) (Figure 1A).
As JCPyV is a virus closely related to BKPyV, it could also play a role in tumourigenesis or cancer progression of infected urinary bladder cells. Since the viral archetypal form was detected in high-grade urothelial bladder carcinoma, JCPyV might be involved in cancer progression rather than tumourigenesis. However, carcinogenic mechanisms induced by JCPyV, as the ‘hit-and-run’ model proposed for BKPyV, cannot be excluded.
The archetype CY-like NCCRs, isolated from JCPyV-positive patients, showed four nucleotide changes (G95C, G108A, A133C and G217A) by comparing with the archetypal JCPyV CY strain. All these changes corresponded to known sequence polymorphisms (28,59,60). Moreover, the nucleotide variations G108A and A133C did not involve any cellular transcriptional factor binding site, whereas the nucleotide substitutions G95C and G217A involved the binding sites for TAR and NF1, respectively (Figure 3). In particular, the 217-G to A nucleotide transition inside the box F was isolated within the NCCR sequences of patients with European genotypes. In fact, this point mutation seems to be a common feature of the European strains and was also found in European Americans (60). Indeed in our study, there was a prevalence of the European genotypes 1A, 1B and 4, followed by the East African genotype 3A. Similarly, the 133-A to C nucleotide transversion inside box D and the maintained nucleotide G at position 217, isolated from all patients with African genotype 3A, seem to be a common feature of the type 3 JCPyV strain (59).
Finally, the molecular analysis of the BKPyV VP1 coding sequence between nucleotides 1744 and 1812 (amino acids 61 to 83) allows the definition of BKPyV genotypes and their different distribution in human populations (27). In particular, subtype I (further divided into four subgroups, each of which has a unique geographical distribution pattern: I/a, I/b-1, I/b-2 and I/c) is widespread throughout the world, subtype IV (further divided into six subgroups with their own geographical distribution pattern: IV/a-1, IV/a-2, IV/b-1, IV/b-2, IV/c-1 and IV/c-2) is prevalent in East Asia and part of Europe, while subtypes II and III are rarely detected throughout the world (61). Furthermore, all BKPyV subtypes/subgroups are defined by the presence of specific nucleotide substitutions and deletions within the NCCR sequence (31). We found the European subtype I/b-2 and the subtype II. In fact, alignment of all NCCR sequences, isolated from each BKPyV-positive clinical sample of our patients, with the prototypic NCCR sequence proposed by Yogo and colleagues (31) revealed that the point mutations detected in these sequences, including the rearranged ones, were typical of the consensus NCCR sequences identified for the BKPyV subtype I/subgroup b-2 and subtype II.
In summary, our data support the hypothesis that alterations in the local urinary bladder environment may be crucial for promoting active PyV replication and consequently those NCCR rearrangements able to transform the host tissue. Moreover, human PyV infection could mechanistically contribute to oncogenesis in the bladder of immunocompetent patients, impeding activity of tumour-suppressors pRb and p53 and, therefore, leading to changes in cellular DNA. In conclusion, our results would suggest an involvement of JCPyV in cancer progression rather than in tumourigenesis in immunocompetent patients, even though carcinogenic mechanisms cannot be excluded. Concerning BKPyV, although only one patient showed a rearranged NCCR in BC tissue, this virus would seem mainly associated with the pathogenesis of urinary BC in non-immunocompromised conditions, supporting the experimental evidence and previous reports. However, further work on a larger samples size will be required to better understand the exact role of human PyVs in urothelial carcinogenesis and to prove the exact molecular steps of oncogenesis in this setting.
Competing Interests
The Authors declare that they have no competing interests.
Acknowledgements
This work was supported by MIUR grant.
References
- 1.Abend JR, Jiang M, Imperiale MJ. BK virus and human cancer: innocent until proven guilty. Semin Cancer Biol. 2009;19:252–260. doi: 10.1016/j.semcancer.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellizzi A, Nardis C, Anzivino E, Rodìo D, Fioriti D, Mischitelli M, Chiarini F, Pietropaolo V. Human polyomavirus JC reactivation and pathogenetic mechanisms of progressive multifocal leukoencephalopathy and cancer in the era of monoclonal antibody therapies. J Neurovirol. 2012;18(1):1–11. doi: 10.1007/s13365-012-0101-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah KV. SV40 and human cancer: a review of recent data. Int J Cancer. 2007;120(2):215–223. doi: 10.1002/ijc.22425. [DOI] [PubMed] [Google Scholar]
- 4.Imperiale MJ, Major EO. Polyomaviruses. In: Field’s Virology, Fifth Edition. Knipe DM and Howley PM (eds.) Philadelphia, Lippincott, Williams. 2007:pp. 2263–2298. [Google Scholar]
- 5.Moens U, Van Ghelue M. Polymorphism in the genome of non-passaged human polyomavirus BK: implications for cell tropism and the pathological role of the virus. Virology. 2005;331:209–231. doi: 10.1016/j.virol.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 6.Frisque RJ, Bream GL, Cannella MT. Human polyomavirus JC virus genome. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anzivino E, Rodio DM, Mischitelli M, Bellizzi A, Sciarra A, Salciccia S, Gentile V, Pietropaolo V. High frequency of JCV DNA detection in prostate cancer tissues. Cancer Genomics Proteomics. 2015;12(4):189–200. [PubMed] [Google Scholar]
- 8.Bellizzi A, Anzivino E, Ferrari F, Di Nardo G, Colosimo MT, Fioriti D, Mischitelli M, Chiarini F, Cucchiara S, Pietropaolo V. Polyomavirus JC reactivation and noncoding control region sequence analysis in pediatric Crohn’s disease patients treated with infliximab. J Neurovirol. 2011;17(4):303–313. doi: 10.1007/s13365-011-0036-3. [DOI] [PubMed] [Google Scholar]
- 9.Butel JS, Lednicky JA. Cell and molecular biology of simian virus 40: implications for human infections and disease. J Natl Cancer Inst. 1999;91(2):119–134. doi: 10.1093/jnci/91.2.119. [DOI] [PubMed] [Google Scholar]
- 10.Bennett SM, Broekema NM, Imperiale MJ. BK polyomavirus: emerging pathogen. Microbes Infect. 2012;14(9):672–683. doi: 10.1016/j.micinf.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fedele CG, Ciardi MR, Delia S, Contreras G, Perez JL, De Oña M, Vidal E, Tenorio A. Identical rearranged forms of JC polyomavirus transcriptional control region in plasma and cerebrospinal fluid of acquired immunodeficiency syndrome patients with progressive multifocal leukoencephalopathy. J Neurovirol. 2003;9(5):551–558. doi: 10.1080/13550280390241188. [DOI] [PubMed] [Google Scholar]
- 12.American Cancer Society. Surveillance Research Cancer Facts & Figures. Available at: http://www.cancer.org/acs/groups/content/@editorial/documents/document/acspc-044552.pdf. 2015.
- 13.Associazione Italiana Registri Tumori. I numeri del cancro in Italia 2015. Available at: http://www.registritumori.it/PDF/AIOM2015/I_numeri_del_cancro_2015.pdf. 2015.
- 14.Fioriti D, Pietropaolo V, Dal Forno S, Laurenti C, Chiarini F, Degener AM. Urothelial bladder carcinoma and viral infections: different association with human polyomaviruses and papillo-maviruses. Int J Immunopathol Pharmacol. 2003;16(3):283–288. doi: 10.1177/039463200301600315. [DOI] [PubMed] [Google Scholar]
- 15.Grippo PJ, Sandgren EP. Highly invasive transitional cell carcinoma of the bladder in a simian virus 40 T-antigen transgenic mouse model. Am J Pathol. 2000;157(3):805–813. doi: 10.1016/S0002-9440(10)64594-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Espana A, Salazar E, Sun TT, Wu XR, Pellicer A. Differential expression of cell cycle regulators in phenotypic variants of transgenically induced bladder tumors: implications for tumor behavior. Cancer Res. 2005;65:1150–1157. doi: 10.1158/0008-5472.CAN-04-2074. [DOI] [PubMed] [Google Scholar]
- 17.Geetha D, Tong BC, Racusen L, Markowitz JS, Westra WH. Bladder carcinoma in a transplant recipient: evidence to implicate the BK human polyomavirus as a causal transforming agent. Transplantation. 2002;73:1933–1936. doi: 10.1097/00007890-200206270-00015. [DOI] [PubMed] [Google Scholar]
- 18.Weinreb DB, Desman GT, Amolat-Apiado MJ, Burstein DE, Godbold JH Jr., Johnson EM. Polyoma virus infection is a prominent risk factor for bladder carcinoma in immunocompetent individuals. Diagn Cytopathol. 2006;34:201–203. doi: 10.1002/dc.20429. [DOI] [PubMed] [Google Scholar]
- 19.Newton R, Ribeiro T, Casabonne D, Alvarez E, Touze A, Key T, Coursaget P. Antibody levels against BK virus and prostate, kidney and bladder cancers in the EPIC-Oxford cohort. Br J Cancer. 2005;93:1305–1306. doi: 10.1038/sj.bjc.6602869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eble JN, Sauter G, Epstein JI, Sesterhenn IA (eds.) World Health Organization Classification of Tumours. Pathology and Genetics of Tumors of the Urinary System and Male Genital Organs. Lyon, IARC Press. 2004 [Google Scholar]
- 21.Lopez-Beltran A, Montironi R. Non-invasive urothelial neoplasms: according to the most recent WHO classification. Eur Urol. 2004;46(2):170–176. doi: 10.1016/j.eururo.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 22.Sobin LH, Gospodarowicz MK, Wittekind C (eds.) TNM Classification of Malignant Tumours, Seventh edition. New York, Wiley Liss. 2010 [Google Scholar]
- 23.McNees AL, White ZS, Zanwar P, Vilchez RA, Butel JS. Specific and quantitative detection of human polyomaviruses BKV, JCV, and SV40 by real-time PCR. J Clin Virol. 2005;34(1):52–62. doi: 10.1016/j.jcv.2004.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Flaegstad T, Sundsfjord A, Arthur RR, Pedersen M, Traavik T, Subramani S. Amplification and sequencing of the control regions of BK and JC virus from human urine by polymerase chain reaction. Virology. 1991;180:553–560. doi: 10.1016/0042-6822(91)90069-n. [DOI] [PubMed] [Google Scholar]
- 25.Markowitz RB, Thompson HC, Mueller JF, Cohen JA, Dynan WS. Incidence of BK virus and JC virus viruria in human immunodeficiency virus-infected and uninfected subjects. J Infect Dis. 1993;167:13–20. doi: 10.1093/infdis/167.1.13. [DOI] [PubMed] [Google Scholar]
- 26.Lednicky JA, Garcea RL, Bergsagel DJ, Butel JS. Natural simian virus 40 strains are present in human choroid plexus and ependymoma tumors. Virology. 1995;212:710–717. doi: 10.1006/viro.1995.1529. [DOI] [PubMed] [Google Scholar]
- 27.Jin L, Gibson PE, Knowles WA, Clewley JP. BK virus antigenic variants: sequence analysis within the capsid VP1 epitope. J Med Virol. 1993;39:50–56. doi: 10.1002/jmv.1890390110. [DOI] [PubMed] [Google Scholar]
- 28.Agostini HT, Yanagihara R, Davis V, Ryschkewitsch CF, Stoner GL. Asian genotypes of JC virus in Native Americans and in a Pacific Island population: markers of viral evolution and human migration. Proc Natl Acad Sci USA. 94;1997 (a):14542–14546. doi: 10.1073/pnas.94.26.14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietropaolo V, Di Taranto C, Degener AM, Jin L, Sinibaldi L, Baiocchini A, Melis M, Orsi N. Transplacental transmission of human polyomavirus BK. J Med Virol. 1998;56:372–376. doi: 10.1002/(sici)1096-9071(199812)56:4<372::aid-jmv14>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 30.Pietropaolo V, Fioriti D, Simeone P, Videtta M, Di Taranto C, Arancio A, Orsi N, Degener AM. Detection and sequence analysis of human polyomaviruses DNA from autoptic samples of HIV-1 positive and negative subjects. Int J Immunopathol Pharmacol. 2003;16:269–276. doi: 10.1177/039463200301600313. [DOI] [PubMed] [Google Scholar]
- 31.Yogo Y, Zhong S, Xu Y, Zhu M, Chao Y, Sugimoto C, Ikegaya H, Shibuya A, Kitamura T. Conserved archetypal configuration of the transcriptional control region during the course of BK polyomavirus evolution. J Gen Virol. 2008;89:1849–1856. doi: 10.1099/vir.0.2008/000836-0. [DOI] [PubMed] [Google Scholar]
- 32.Yogo Y, Kitamura T, Sugimoto C, Ueki T, Aso Y, Hara K, Taguchi F. Isolation of a possible archetypal JC virus DNA sequence from nonimmunocompromised individuals. J Virol. 1990;64:3139–3143. doi: 10.1128/jvi.64.6.3139-3143.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anzivino E, Bellizzi A, Mitterhofer AP, Tinti F, Barile M, Colosimo MT, Fioriti D, Mischitelli M, Chiarini F, Ferretti G, Taliani G, Pietropaolo V. Early monitoring of the human polyomavirus BK replication and sequencing analysis in a cohort of adult kidney transplant patients treated with basiliximab. Virol J. 2011;8:407–416. doi: 10.1186/1743-422X-8-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jobes DV, Friedlaender JS, Mgone CS, Agostini HT, Koki G, Yanagihara R, Ng TCN, Chima SC, Ryschkewitsch CF, Stoner GL. New JC virus (JCV) genotypes from Papua New Guinea and Micronesia (type 8 and type 2E) and evolutionary analysis of 32 complete JCV genomes. Arch Virol. 2001;146:2097–2113. doi: 10.1007/s007050170023. [DOI] [PubMed] [Google Scholar]
- 35.ClustalW2–Multiple Sequence Alignment. Available at: http://www.ebi.ac.uk/Tools/msa/clustalw2/ 2015.
- 36.Degener AM, Pietropaolo V, Di Taranto C, Jin L, Ameglio F, Cordiali-Fei P, Trento E, Sinibaldi L, Orsi N. Identification of a new control region in the genome of the DDP strain of BK virus isolated from PBMC. J Med Virol. 1999;58:413–419. doi: 10.1002/(sici)1096-9071(199908)58:4<413::aid-jmv15>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 37.Frisque RJ. Nucleotide sequence of the region encompassing the JC virus origin of DNA replication. J Virol. 1983;46(1):170–176. doi: 10.1128/jvi.46.1.170-176.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dyson N, Bernards R, Friend SH, Gooding LR, Hassell JA, Major EO, Pipas JM, Vandyke T, Harlow E. Large T antigens of many polyomaviruses are able to form complexes with the retinoblastoma protein. J Virol. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev. 2006;20:2373–2382. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drachenberg CB, Hirsch HH, Ramos E, Papadimitriou JC. Polyomavirus disease in renal transplantation: review of pathological findings and diagnostic methods. Hum Pathol. 2005;36:1245–1255. doi: 10.1016/j.humpath.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Monini P, Rotola A, Di Luca D, De Lellis L, Chiari E, Corallini A, Cassai E. DNA rearrangements impairing BK virus productive infection in urinary tract tumors. Virology. 1995;214:273–279. doi: 10.1006/viro.1995.9928. [DOI] [PubMed] [Google Scholar]
- 42.Monini P, de Lellis L, Rotola A, Di Luca D, Ravaioli T, Bigoni B, Cassai E. Chimeric BK virus DNA episomes in a papillary urothelial bladder carcinoma. Intervirology. 1995;38:304–308. doi: 10.1159/000150455. [DOI] [PubMed] [Google Scholar]
- 43.Tong BC, Racusen L, Markowitz JS, Westra WH. Bladder carcinoma in a transplant recipient: evidence to implicate the BK human polyomavirus as a causal transforming agent. Transplantation. 2002;73:1933–1936. doi: 10.1097/00007890-200206270-00015. [DOI] [PubMed] [Google Scholar]
- 44.Herawi M, Parwani AV, Chan T, Ali SZ, Epstein JI. Polyoma virusassociated cellular changes in the urine and bladder biopsy samples: a cytohistologic correlation. Am J Surg Pathol. 2006;30:345–350. doi: 10.1097/01.pas.0000179117.38787.57. [DOI] [PubMed] [Google Scholar]
- 45.Rollison DE, Sexton WJ, Rodriguez AR, Kang LC, Daniel R, Shah KV. Lack of BK virus DNA sequences in most transitional-cell carcinomas of the bladder. Int J Cancer. 2007;120:1248–1251. doi: 10.1002/ijc.22494. [DOI] [PubMed] [Google Scholar]
- 46.Roberts IS, Besarani D, Mason P, Turner G, Friend PJ, Newton R. Polyoma virus infection and urothelial carcinoma of the bladder following renal transplantation. Br J Cancer. 2008;99:1383–1386. doi: 10.1038/sj.bjc.6604711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill P, Slavin J, Goodman D. High-grade urothelial carcinoma in a kidney transplant recipient with BK virus infection. Nephrol Dial Transplant Plus. 2009;2:246–249. doi: 10.1093/ndtplus/sfp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang HH, Liu KL, Chu SH, Tian YC, Lai PC, Chiang YJ. BK virus infection in association with post-transplant urothelial carcinoma. Transplant Proc. 2009;41:165–166. doi: 10.1016/j.transproceed.2008.08.138. [DOI] [PubMed] [Google Scholar]
- 49.Chen CH, Wen MC, Wang M, Lian JD, Cheng CH, Wu MJ, Yu TM, Chuang YW, Chang D, Shu KH. High incidence of malignancy in polyomavirus-associated nephropathy in renal transplant recipients. Transplant Proc. 2010;42:817–818. doi: 10.1016/j.transproceed.2010.02.068. [DOI] [PubMed] [Google Scholar]
- 50.Alexiev BA, Randhawa P, Vazquez Martul E, Zeng G, Luo C, Ramos E, Drachenberg CB, Papadimitriou JC. BK virus-associated urinary bladder carcinoma in transplant recipients: report of two cases, review of the literature, and proposed pathogenetic model. Hum Pathol. 2013;44(5):908–917. doi: 10.1016/j.humpath.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 51.Alexiev BA, Papadimitriou JC, Chai TC, Ramos E, Staats PN, Drachenberg CB. Polyomavirus (BK)-associated pleomorphic giant cell carcinoma of the urinary bladder: a case report. Pathol Res Pract. 2013;209(4):255–259. doi: 10.1016/j.prp.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 52.van Aalderen MC1, Yapici Ü, van der Pol JA, de Reijke TM, van Donselaar-van der Pant KA, Florquin S, Bemelman FJ, Ten Berge IJ. Polyomavirus BK in the pathogenesis of bladder cancer. Neth J Med. 2013;71(1):26–28. [PubMed] [Google Scholar]
- 53.Kitamura T, Aso Y, Kuniyoshi N, Hara K, Yogo Y. High incidence of urinary JC virus excretion in nonimmunosuppressed older patients. J Infect Dis. 1990;161(6):1128–1133. doi: 10.1093/infdis/161.6.1128. [DOI] [PubMed] [Google Scholar]
- 54.Mischitelli M, Bellizzi A, Anzivino E, Rodio DM, Sciarra A, Gentile V, Pietropaolo V. Results, questions, perspectives of a study on human polyomavirus BK and molecular actors in prostate cancer development. Cancer Genomics Proteomics. 2015;12:57–65. [PubMed] [Google Scholar]
- 55.Das D, Wojno K, Imperiale MJ. BK virus as a cofactor in the etiology of prostate cancer in its early stages. J Virol. 2008;82:2705–2714. doi: 10.1128/JVI.02461-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laghi L, Randolph AE, Chauhan DP, Marra G, Major EO, Neel JV, Boland CR. JC virus DNA is present in the mucosa of the human colon and in colorectal cancers. Proc Natl Acad Sci USA. 1999;96:7484–7489. doi: 10.1073/pnas.96.13.7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ricciardiello L, Chang DK, Laghi L, Goel A, Chang CL, Boland CR. Mad-1 is the exclusive JC virus strain present in the human colon, and its transcriptional control region has a deleted 98-base-pair sequence in colon cancer tissues. J Virol. 2001;75:1996–2001. doi: 10.1128/JVI.75.4.1996-2001.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zambrano A, Kalantari M, Simoneau A, Jensen JL, Villarreal LP. Detection of human polyomaviruses and papillomaviruses in prostatic tissue reveals the prostate as a habitat for multiple viral infections. Prostate. 2002;53:263–276. doi: 10.1002/pros.10157. [DOI] [PubMed] [Google Scholar]
- 59.Agostini HT, Ryschkewitsch CF, Brubaker GR, Shao J, Stoner GI. Five complete genomes of JC virus type 3 from Africans and African Americans. Arch Virol. 1997;142(4):637–655. doi: 10.1007/s007050050108. [DOI] [PubMed] [Google Scholar]
- 60.Agostini HT, Deckhut A, Jobes DV, Girones R, Schlunck G, Prost MG, Frias C, Pérez-Trallero E, Ryschkewitsch CF, Stoner GL. Genotypes of JC virus in East, Central and Southwest Europe. J Gen Virol. 2001;82:1221–1331. doi: 10.1099/0022-1317-82-5-1221. [DOI] [PubMed] [Google Scholar]
- 61.Zhong S, Randhawa PS, Ikegaya H, Chen Q, Zheng HY, Suzuki M, Takeuchi T, Shibuya A, Kitamura T, Yogo Y. Distribution patterns of BK polyomavirus (BKV) subtypes and subgroups in American, European and Asian populations suggest co-migration of BKV and the human race. J Gen Virol. 2009;90:144–152. doi: 10.1099/vir.0.83611-0. [DOI] [PubMed] [Google Scholar]