Abstract
The monoamine neurotransmitter dopamine (DA) acts across phylogeny to modulate both simple and complex behaviors. The presynaptic DA transporter (DAT) is a major determinant of DA signaling capacity in ensuring efficient extracellular DA clearance. In humans, DAT is also a major target for prescribed and abused psychostimulants. Multiple structural determinants of DAT function and regulation have been defined, though largely these findings have arisen from heterologous expression or ex vivo cell culture studies. Loss of function mutations in the gene encoding the Caenhorhabditis elegans DAT (dat-1) produces rapid immobility when animals are placed in water, a phenotype termed Swimming-induced paralysis (Swip). The ability of a DA neuron-expressed, GFP-tagged DAT-1 fusion protein (GFP::DAT-1) to localize to synapses and rescue Swip in these animals provides a facile approach to define sequences supporting DAT somatic export and function in vivo. In prior studies, we found that truncation of the last 25 amino acids of the DAT-1 C-terminus (Δ25) precludes Swip rescue, supported by a deficit in GFP::DAT-1 synaptic localization. Here, we further defined the elements within Δ25 required for DAT-1 export and function in vivo. We identified two conserved motifs (584KW585 and 591PYRKR595) where mutation results in a failure of GFP::DAT-1 to be efficiently exported to synapses and restore DAT-1 function. The 584KW585 motif conforms to a sequence proposed to support SEC24 binding, ER export from the endoplasmic reticulum (ER), and surface expression of mammalian DAT proteins, whereas the 591PYRKR595 sequence conforms to a 3R motif identified as a SEC24 binding site in vertebrate G-protein coupled receptors. Consistent with a potential role of SEC24 orthologs in DAT-1 export, we demonstrated DA neuron-specific expression of a sec-24.2 transcriptional reporter. Mutations of the orthologous C-terminal sequences in human DAT (hDAT) significantly reduced transporter surface expression and DA uptake, despite normal hDAT protein expression. Although, hDAT mutants retained SEC24 interactions, as defined in co-immunoprecipitation studies. However, these mutations disrupted the ability of SEC24D to enhance hDAT surface expression. Our studies document an essential role of conserved DAT C-terminal sequences in transporter somatic export and synaptic localization in vivo, that add further support for important roles for SEC24 family members in efficient transporter trafficking.
Keywords: dopamine, transporter, nematode, synaptic trafficking, Sec proteins
INTRODUCTION
The catecholamine dopamine (DA) exerts powerful modulatory control of neurotransmission and behavior across phylogeny (Carlsson, 1987; McDonald et al., 2006). Not surprisingly, altered DA signaling is implicated in multiple brain disorders, ranging from Parkinson’s disease and dystonia (Hornykiewicz, 2006) to addiction (Keiflin and Janak, 2015; Ritz et al., 1987), major depression, bipolar disorder (Cousins et al., 2009; Heshmati and Russo, 2015; Horschitz et al., 2005), attention-deficit hyperactivity disorder (Del Campo et al., 2011; Mazei-Robison et al., 2005) and schizophrenia (Bonoldi and Howes, 2013; Seeman, 2010). The capacity for DA signaling is dictated by mechanisms supporting intraneuronal DA biosynthesis and metabolism, DA vesicular packaging and release, and transporter-mediated DA clearance. The presynaptic, Na+/Cl− coupled DA transporter (DAT, SLC6A3) (Torres et al., 2003) provides for efficient DA reuptake and pharmacological manipulation of DAT leads to significant behavioral effects, exemplified by the addictive and/or therapeutic impact of DAT interactions with cocaine, methylphenidate (Ritalin™) and amphetamine (AMPH)(Nestler, 2005; Ritz et al., 1987; Sulzer et al., 2005).
Decades of in vitro studies have shown that DAT localization and function is regulated through mechanisms that support trafficking of transporters between endosomal and cell surface membranes (Chen et al., 2010; Melikian, 2004), as well as plasma membrane recycling. We and others have demonstrated that such processes can be disrupted by disease-associated coding variation (Hansen et al., 2014; Kurian et al., 2009; Sakrikar et al., 2012; Wu et al., 2015). Recent studies from the Melikian lab (Wu et al., 2015) provide evidence that a basic understanding of structural determinants of DAT trafficking can lead to strategies to overcome the deleterious impact of DAT mutations.
DAT and other neurotransmitter transporters require post-translational processing (e.g. N-glycosylation and S-palmitoylation) in the endoplasmic reticulum (ER) and Golgi before mature proteins can be exported from the cell soma and trafficked to the synapse (Foster and Vaughan, 2011). Export of all proteins from the ER is predominantly mediated by the COPII complex, with SEC24 family members responsible for cargo recognition (Pagano et al., 1999). Cell culture experiments have identified binding sites for SEC24 family members to DAT, the serotonin transporter (SERT) and other SLC6 transporters, specifically involving an R/K-L/I/V motif in the cytoplasmic C-terminus (El-Kasaby et al., 2010; Sucic et al., 2011). Interactions with mammalian SEC24 members is isoform specific with SEC24C supporting ER export of SERT, whereas SEC24D supports the export of DAT and the γ-aminobutyric acid (GABA) transporter (GAT-1) (Sucic et al., 2011; Sucic et al., 2013). To date, however, in vivo evidence for SEC24-dependent export and synaptic localization of neurotransmitter transporters has not been reported.
Our lab has adopted the nematode C. elegans as a model system to identify regulatory networks sustaining DA signaling and, more specifically, to elucidate mechanisms supporting DAT function and regulation in vivo (Carvelli et al., 2008; Hardaway et al., 2012; Nass et al., 2005; Nass et al., 2002). Genes that dictate DA signaling capacity are well conserved between nematodes and humans, including cat-2 (tyrosine hydroxylase), cat-1 (vesicular monoamine transporter), dop1,2,3,4 (D1 and D2 type DA receptors) and dat-1 (DAT) (McDonald et al., 2006). Worms with loss of function mutations in dat-1 are insensitive to the neurotoxic actions of the DAT-1 substrate 6-hydroxydopamine (6-OHDA) (Nass et al., 2005; Nass et al., 2002) and exhibit immobility when placed in water termed Swimming-induced paralysis (Swip) (Hardaway et al., 2012; McDonald et al., 2007). Both loss of 6-OHDA sensitivity and Swip can be significantly rescued by transgenic expression of a N-terminally-tagged, green fluorescent protein (GFP) - DAT-1 fusion (GFP::DAT-1), as well as by cat-1, cat-2 mutations and dop-3 mutations (McDonald et al., 2007). Swip therefore represents a robust measure of constrained DA signaling and proper DAT-1 mediated DA clearance, and can be used to identify novel regulators of DA signaling as well as the functional impact of dat-1 mutations in vivo (Hardaway et al., 2012; Hardaway et al., 2015).
Previously, we reported that a 25 amino acid C-terminal deletion (Δ25) eliminates the ability of GFP::DAT-1 to rescue the Swip exhibited by a dat-1 deletion mutant (ok157)(McDonald et al., 2007). Imaging of GFP::DAT-1 Δ25 localization reveals a significantly diminished ability of the mutant transporter to localize to DA synapses (McDonald et al., 2007). Initially, we suspected that the impact of the Δ25 truncation might arise from a loss of the Type II PDZ-type motif (IML) present in the DAT-1 C-terminus, given evidence for DAT interactions with PDZ domain-containing proteins, such as PICK1, and their contribution to transporter surface trafficking (Bjerggaard et al., 2004; Torres et al., 2001). However, subsequent studies with a ΔIML truncation mutation of DAT-1 revealed rescue of both Swip and 6-OHDA sensitivity, as well as efficient localization of the ΔIML transporter to DA synapses (McDonald et al., 2007). We therefore pursued a more extensive analysis of the Δ25 mutation, studies that lead to the identification of two motifs previously linked to interactions of SEC24 proteins with membrane proteins. We also provide correlative data from studies of human DAT (hDAT) transfected mammalian cells that these sequences also dictate mammalian DAT surface localization and DAT functional capacity.
MATERIALS AND METHODS
Chemical reagents, worm strains and husbandry
All buffers, salts and miscellaneous chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise specified. C. elegans strains unless otherwise noted were provided by the Caenorhabditis Genetics Center (University of Minnesota, Twin Cities), which is funded by the NIH Office of Research Infrastructure Programs (P40-OD101440). The wild type strain is N2 Bristol. The dat-1(ok157) III strain was a gift of J. Duerr and J. Rand (Oklahoma Medical Research Foundation, Oklahoma City). VC20226 is a part of the million mutation project (Thompson et al., 2013). The latter line was outcrossed four times, tracking the presence of the dat-1 mutation encoding the missense variant P596S, prior to analysis. C. elegans strains were cultured on bacterial lines of OP50 or NA22 and maintained using standard methods and conditions (Brenner, 1974).
Creation of plasmids and transgenic animals Plasmids
Pdat-1:GFP::DAT-1, (Carvelli et al., 2004), Pdat-1::GFP (Nass et al., 2002), Pdat-1:GFP::DAT-1 (Δ25) (McDonald et al., 2007), HA::hDAT (Sakrikar et al., 2012) and HA::DAT-1 (Nass et al., 2005) were described previously. GFP tagged SEC24D was a kind gift from Guangyu Wu (Georgia Health Sciences University). GFP reporters for sec-24.1 and sec-24.2 were created by a PCR fusion approach as previously described with at least 1 kB of sequence upstream of the start (Hobert, 2002) using fosmids WRM061BG12 and WRM061BA06 for sec-24.2 and 1, respectively. Mutations in Pdat-1::GFP::DAT-1, HA::DAT-1 and HA::hDAT were all created using the Quick Change XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) and were verified by sequencing prior to use. Animals carrying extrachromosomal arrays were created by co-injecting transgenes using methods described previously (Mello et al., 1991). All transgenes were fully sequenced prior to injections. Plasmids containing Pdat-1::DAT-1::GFP deletions and Pdat-1::DAT-1::GFP 591PYRKR595-AAAAA were injected into dat-1(ok157); lin-15 (n765ts) background at 90 ng/μl to generate the following transgenic lines: BY636-640: Pdat-1::DAT-1::GFPΔ591-615; BY631-635: Pdat-1::DAT-1::GFPΔ596-615; BY622-625: Pdat-1::DAT-1::GFPΔ601-615; BY626-630: Pdat-1::DAT-1::GFP Δ606-615; BY622-625: Pdat-1::DAT-1::GFPΔ611-615; BY816-818: Pdat-1::DAT-1::GFP 591PYRKR595-AAAAA. Single point mutations were injected into dat-1(ok157); lin-15 (n765ts) background at 15 ng/μl with pJM23 (40ng/μl) to generate BY515-517 (Pdat-1::DAT-1::GFP P591A), BY518-520 (Y592A), BY5521-523 (R593A), BY524-526 (K594A), BY527-529 (R595A), BY5530-532 (P596A), BY1226 (P596S). Pdat-1::DAT-1::GFP 591PYR593-AAA (BY533-535) and Pdat-1::DAT-1::GFP 594KRP596-AAA (BY536-538) were injected into dat-1(ok157); lin-15 (n765ts) background at 20 ng/μl with pJM23 (40 ng/μl) and Punc-122:RFP (40 ng/μl). Pdat-1::DAT-1::GFP 591PYRKR595-AYAKA was injected into dat-1(ok157) with at 40 ng/μl with Punc-122::RFP (20ng/μl) and Pelt-2::GFP (40 ng/μl) to generate BY1173. Pdat-1::DAT-1::GFP 585KW585-AA was injected into dat-1(ok157); lin-15 (n765ts) background at 2 ng/μl with lin-15 (4 ng/μl) and Punc-122:RFP (4 ng/μl) to generate lines BY854 and BY855. PCR products Psec-24.1::GFP and Psec-24.2::GFP were injected onto N2 background at 50 ng/μl with Pdat-1::mCherry (30 ng/μl) and pcDNA3 (20 ng/μl) to generate BY1070-1072: Psec24.1::GFP and BY1073-1075: Psec24.2::GFP.
Swip assays
Swip assays were performed according to (McDonald et al., 2007). Briefly, synchronous nematode populations were generated by hypochlorite treatment and L1 animals were plated onto nematode growth medium (NGM) plates seeded with OP50 bacteria. Early- to mid-stage L4 animals were identified by characteristic morphology and ten to fifteen worms were placed into 100 μl water in a Pyrex Spot Plate with the number of worms paralyzed visually scored after ten minutes of incubation. In the case of transgenic lines, fluorescence-positive animals at L2 or L3 stage were picked onto plates and placed at 12°C to mature to the L4 stage. All Swip assays were performed blinded to genotype.
Confocal imaging and quantification
In vivo imaging of transgenic nematodes was performed as previously described (McDonald et al., 2007). Briefly, animals were placed on freshly prepared 2% agarose pads and immobilized using 225 mM 2,3 butanedione monoxime (Sigma-Aldrich, St. Louis, MO) and 2.5mM levamisole (Sigma-Aldrich) in 10mM HEPES (Smith et al., 2010). A Z-stack of images was obtained using a Zeiss LSM 510 or 710. By setting the gain of confocal illumination of synaptic regions just below the maximum intensity of fluorescence, we obtain the relative intensity of signal from cell soma and dendritic regions within the same animal. For imaging and quantification of GFP::DAT-1 localization, imaging stacks obtained using the LSM and containing cell body, dendrites and synaptic regions were thresholded, and pseudocolored using Metamorph (Molecular Devices, Sunnyvale, CA). Pixel density was calculated as the average fluorescence intensity in regions of interest, and these values were used to calculate the ratio of fluorescence in the cell body to the intensity determined over synapses or dendrites in the same animal. A minimum of 10 animals per genotype was used for analysis. Sec-24 promotor fusions were imaged using the LSM 710 and Z stacks of images containing individual fluorophores were merged using ImageJ (NIH). Final images were processed using identical brightness and contrast values.
Cell culture methods and transfections
HEK-293T cells were cultured for transient transfection as previously described (Mazei-Robison et al., 2005). Trans-IT LT-1 (Mirus, Madison, WI) in serum free media was used to transfect cells approximately 48 hr after plating HEK-293T cells using a 1:3 DNA:lipid ratio.
Dopamine transport assays
[3H]DA transport assays were performed in triplicate in 24-well plates at 37°C as described previously (Apparsundaram et al., 2001; Sakrikar et al., 2012). Briefly, cells were washed with Krebs–Ringers–HEPES (KRH) (130 mM NaCl, 1.3 mM KCl, 2.2 mM CaCl2, 1.2 mM MgSO4, 1.2 mM KH2PO4, 10 mM HEPES, pH 7.4) buffer and were incubated in the uptake assay buffer (KRH, 10 mM glucose, 100 M pargyline, ascorbic acid, and 10 M tropolone) before incubation with 50nM [3H]DA (Perkin Elmer, Waltham, MA) for 10 min followed by three KRH washes. MicroScint scintillation reagent (Perkin Elmer) was added to the wells at the end of the assay and DA uptake was quantified using a TopCount Scintillation Counter (Beckman Coulter, Indianapolis, IN). Mean± SEM values reported derive from three to six independent experiments performed in triplicate.
Cell surface biotinylation and co-immunoprecipitation experiments
Experiments were performed as previously described (Sakrikar et al., 2012). HEK-293T cells were seeded in six-well plates and incubated for 48 hr before experiments. Cells were washed twice with warm KRH buffer prior to incubation with Sulfo-NHS-SS-Biotin (Pierce, ThermoFisher Scientific, Waltham, MA) for 30 min at 4°C. Excess biotin was quenched by two washes with 0.1 M glycine in PBS, and cells were solubilized using radioimmunopreciptation assay (RIPA) buffer (100 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate) for 30 min at 4°C. Samples were extracted and particulate removed by brief centrifugation before co-immunoprecipitation and immunoblotting.
Co-IP experiments were performed as described previously (Sakrikar et al., 2012). Cells were lysed using co-immunoprecipitation buffer (50 mM Tris, pH 7.4, 150 mM NaCl, 1% Triton X-100) containing protease inhibitors 48 hr after transfection. Immunoprecipitation was performed overnight using goat anti-GFP antibody (BioLegend, San Diego, CA) prior to capture on Protein G beads (GE Life Sciences) and elution with laemmli buffer. Recovered DAT was visualized after SDS-PAGE and transfer to PVDF membrane (GE Healthcare Life Sciences, Pittsburgh, PA) using rat anti-hDAT antibody (Millipore; MAB 369; 1:5000). Bands were visualized using direct capture with ImageQuant LAS 4000 (GE Healthcare Life Sciences) of enhanced chemiluminescence signal (Clarity Western ECL substrate, Bio-Rad, Hercules, CA).
Graphical and statistical analyses
All graphical and statistical analyses were obtained using Prism 5.0 software (GraphPad, La Jolla, CA). Statistical tests are reported in the figure legends. For all analyses, a P value <0.05 was taken as statistically significant.
RESULTS
Identification of sequences within the DAT-1 C-terminus that support normal swimming behavior
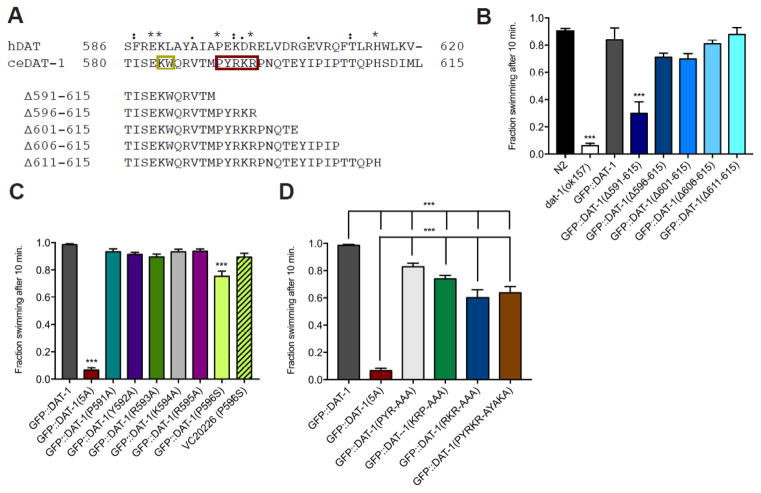
We previously reported that truncation of the final twenty-five amino acids of the GFP::DAT-1 C-terminus (GFP::DAT-1 Δ25, Δ591-615) (Figure 1A) fails to rescue Swip, or restore the 6-OHDA sensitivity of a DAT-1 loss the function mutant dat-1(ok157), nor does the mutant transporter localize efficiently to DA synapses (McDonald et al., 2007). Finding that the critical sequences within the Δ25 truncation were not those of the PDZ recognition motif (613IML615), we generated a series of mutant GFP::DAT-1 constructs bearing progressive deletions that shortened the C-terminus in steps of 5 amino acids (Figure 1A) and then generated dat-1(ok157) animals that transmitted these DNAs as extrachromosomal arrays, followed by Swip testing (Figure 1B). As expected, a construct bearing a deletion of the final five amino acids (GFP::DAT-1 Δ606-615) rescued the Swip of dat-1(ok157) animals, comparable to that seen with GFP::DAT-1 injections. Wildtype levels of Swip rescue were also evident when we expressed constructs bearing deletions of the terminal ten (GFP::DAT-1 Δ606-615), fifteen (GFP::DAT-1 Δ601-615) or twenty (GFP::DAT-1 Δ596-615) amino acids. These data indicated that the relevant contributor to the altered function and trafficking of GFP::DAT-1 Δ591-615 likely lay at or near amino acids 591-595 (shaded in Figure 1A).
Figure 1. The 591PYRKR595 C-terminal motif is required for DAT-1 function.
A. Alignment of amino acids 580-615 of the DAT-1 C-terminus with sequences of the hDAT C-terminus. Identical (*), conservative (:) and semi-conservative (.) changes are noted. Amino acids retained in each deletion construct are indicated below the full sequence. Red box, novel 5 amino acid motif (PRYKR). Gold box, SEC-24 binding motif (KW).
B–D. Swip behavior of GFP::DAT-1 wild-type and mutations, narrowing down the C-terminal motif to 5 amino acids, PYRKR. All GFP::DAT-1 lines were produced on a dat-1(ok157) background, which is not noted. The figures represent the mean swimming behavior assessed by manual scoring, as described in Material and Methods. Data were analyzed using one-way ANOVA with multiple Bonferroni post-tests, comparing Swip levels of mutants to GFP::DAT-1 (B–D) or N2, (B) and with ***P<0.001 and error bars represent SEM.
To determine whether a deletion encompassing amino acids 591-596 (PYRKR) lost the ability to rescue Swip due to a structural or sequence perturbation of the DAT-1 C-terminus, we generated animals expressing a construct in which amino acids 591PYRKR595 were substituted by alanine (GFP::DAT-1(5A)). Swip testing of these animals revealed a paralytic phenotype identical to that of dat-1(ok157), indicative of a failure of GFP::DAT(5A) to confer DAT activity at DA synapses (Figure 1C). Interestingly, animals produced to express constructs with individual alanine substitutions across 591PYRKR595 demonstrated efficient Swip rescue, comparable in efficiency to GFP::DAT-1 (Fig 1C). We also tested for rescue by a DAT-1 missense allele from the Million Mutation Project (Thompson et al., 2013) that converts a proline residue immediately adjacent to the 591PYRKR595 sequence to serine (P596S) (Figure 1C), as well as an alanine substitution at the same residue in GFP::DAT-1 (data not shown), and also found levels of Swip rescue slightly but significantly reduced relative to rescue found with GFP::DAT-1 (Figure 1C). These findings suggested that the 591PYRKR595 region harbors multiple residues that contribute to the proper synaptic localization and transport function of DAT-1. Animals produced to express constructs with either alanine substitutions at 591PYR593 (GFP::DAT-1 PYR-AAA) or at 593RKR595 (GFP::DAT-1 RKR-AAA) exhibited a rescue of Swip that was significantly, though modestly, lower than that seen with GFP::DAT-1 (Figure 1D). A similarly blunted rescue of Swip was detected in animals expressing constructs with either alanine substitutions of interior residues in the motif (594KRP596, GFP::DAT-1 PYRKRP-PYRAAA) or at residues conserved with hDAT (PYRKR-AYAKA) (Fig 1D). Together, these results point to a contribution of multiple residues within the 591PYRKR595 sequence in establishing the efficient synaptic targeting and/or intrinsic functional capacity of DAT-1.
Evaluation of somatic export and presynaptic localization of DAT-1 C-terminal mutants
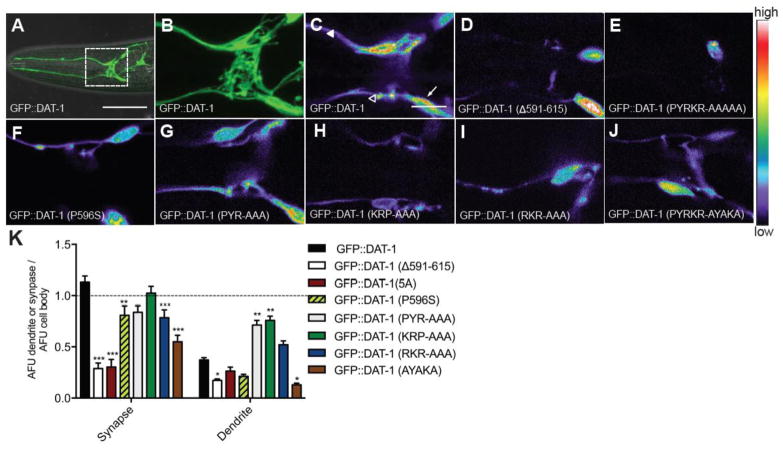
To determine whether the diminished in vivo function of the mutated GFP::DAT-1 constructs studied above arises from altered trafficking of transporters to synapses versus an intrinsic functional deficit, we imaged the distribution of the transporter fusion proteins in live, anesthetized transgenic animals using confocal microscopy. To quantify relative compartmental localization, we determined the ratio of GFP::DAT-1 fluorescence (in a dat-1(ok157) null background) at DA synapses and dendrites vs. that obtained in the cell body, with all elements captured in the same focal plane. As shown in our prior work (McDonald et al., 2007), synapses display the highest GFP::DAT-1 intensity, followed by the cell body and the dendrites (Figure 2C, K). The Δ591-615 deletion restricts GFP expression almost exclusively to the cell soma (Figure 2D, K). The five alanine substitution mutant (GFP::DAT-1 PYRKR-AAAAA, 5A) which reproduced the loss of Swip rescue seen in the GFP::DAT-1 Δ25 mutant also resulted in somatic retention of the transporter (Figure 2E, K). Consistent with their relatively normal swimming behavior, animals expressing PYR-AAA and RKR-AAA mutant transgenes displayed synaptic localization patterns that were more similar to wildtype GFP::DAT-1. Although the GFP signal in KRP-AAA mutant animals was considerably lower, quantification of relative transporter distribution revealed a more normal expression pattern than the 5A mutant (Figure 2G, H, I, K). Finally, as seen with the Swip phenotype, the synaptic localization of the GFP::DAT-1 AYAKA mutant was more affected, but not to the degree seen with the five-alanine mutant (Figure 2J, K). Together these results support the idea that the functional deficits observed after in vivo expression of the mutant GFP::DAT-1 constructs engineered to target the 591PYRKR595 motif derives from a disruption in transporter localization to synapses. The retention for many mutants of readily detectible cell body expression is consistent with a deficit in somatic export.
Figure 2. The 591PYRKR595 C-terminal motif is required for DAT-1 synaptic localization.
A. Wild-type animals injected with Pdat-1:GFP::DAT-1 on a dat-1(ok157) background display GFP expression in CEP and ADE soma and dendrites. Anterior is left. White box indicates zoomed in area for B–H. Scale bar represents 50 μm.
B. Zoomed in image of A. Scale bar represents 10 μm.
C–J. Zoomed in images of GFP::DAT-1 wildtype and mutants, all on a dat-1(ok157) background. Representative single plane images have been pseudocolored. In wildtype, GFP expression is highest in the synapse (open triangles), followed by the cell body (white arrows) and lowest in the dendrites (closed triangles). Δ591-615 (D) and 5A (E) GFP expression is localized mostly in the cell body. Other mutants (F–J) appear more similar to wildtype. Scale bar represents 10 μm. Each image is representative of three independent transgenic lines. All images were taken using the same confocal parameters.
K. Quantification of GFP::DAT-1 fluorescence, expressed as a ratio of synapse/cell body or dendrite/cell body. Single planes containing cell body, synapse and dendrites were imaged and quantified as described in Methods and Materials. Data were ratio normalized against fluorescence of the cell body and then analyzed using two-way ANOVA with post-test comparisons made to GFP::DAT-1 and where *P<0.05, **P<0.01 and ***P<0.001 and error bars represent SEM. Three independent transgenic lines were scored.
SEC-24 binding is required for DAT-1 export and function
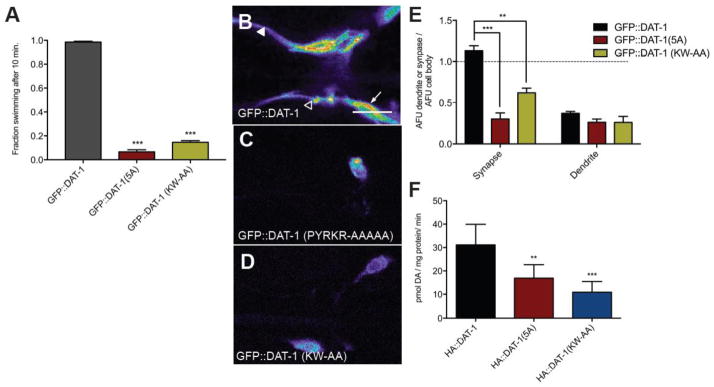
The 591PYRKR595 sequence we found to support somatic export of DAT-1 contains a “3R” motif that has been identified as a SEC24 binding site in G-protein coupled receptors (Dong et al., 2012). In close proximity to 591PYRKR595 is a 584KW585 sequence at a site previously suggested to bind the COPII export complex protein SEC24 (Lord et al., 2013). Recent experiments in cultured mammalian cells support for a role of the latter sequences in dictating ER export of human SERT by SEC24C and of hDAT by SEC24D (Sucic et al., 2011). To assess the role of 584KW585 for DAT-1 function in C. elegans DA neurons in vivo, we mutated the 584KW585 sequence in GFP::DAT-1 to 584AA585. Transgenic dat-1(ok157) animals expressing GFP::DAT-1 KW-AA displayed a robust Swip phenotype, similar to the behavior of nontransgenic dat-1(ok157) animals or dat-1(ok157) animals expressing the GFP::DAT-1 PYRKR-AAAAA mutant (Figure 3A). Confocal imaging of these lines revealed an almost exclusive localization of GFP-tagged DAT-1 proteins to the cell body (Figure 3D, E).
Figure 3. The nearby SEC24 binding motif is also required for DAT-1 export and function.
A. Swip behavior of GFP::DAT-1 bearing a mutation in a putative SEC-24 binding site (584KW585-AA) is similar to behavior of the 5A mutant. Data were analyzed using one-way ANOVA with multiple Bonferroni posttests comparing levels to GFP::DAT-1 animals, and where ***P<0.001 and error bars represent SEM. For all transgenic lines, three independent lines were scored.
B–D. Confocal Z stack images of GFP::DAT-1 fluorescence in CEP/ADE neurons. Anterior is left. In wildtype, GFP expression is highest in the synapse (open triangles), followed by the cell body (white arrows) and lowest in the dendrites (closed triangles). GFP expression in both 5A and KW-AA mutants is mainly observed in the cell body. Each image is representative of three independent transgenic lines. All images were taken at the same confocal parameters. Scale bar represents 10 μm.
E. Analysis of GFP::DAT-1 mutant fluorescence. AFU values were normalized to cell soma levels and analyzed using two-way ANOVA comparing levels against GFP::DAT-1 animals where *P<0.05, **P<0.01 and ***P<0.001 and error bars represent SEM. Three independent transgenic lines were scored for each data set.
F. Uptake of [3H]DA in HEK293T cells transfected with HA::DAT-1 or constructs bearing 5A and KL-AA mutations. Both mutations induced significantly reduced DA uptake compared to HA::DAT-1. Transport expressed as picomoles * milligram protein−1 *min−1. Values represent a mean of four separate experiments with error bars representing SEM. Data were analyzed using one-way ANOVA where ***p<0.0001.
We previously reported that functional DAT-1 protein could be expressed in mammalian cells, as assessed by [3H]DA uptake, with mutations in DAT-1 that disrupt 6-OHDA accumulation, or that induce Swip, resulting in a significant reduction in DA transport activity (Hardaway et al., 2012; Nass et al., 2005). When DAT-1 (5A and DAT-1 KW-AA constructs were expressed in HEK293T cells, we observed a significant reduction in [3H]DA uptake compared to that achieved with transfection of wildtype HA::DAT-1 (Figure 3F). These results provide direct evidence that a lack of restoration of DA clearance in dat-1(ok157) animals by DAT-1 5A and DAT-1 KW-AA constructs underlies failures to reduce Swip.
C. elegans SEC24 ortholog SEC24.2 is expressed in DA neurons
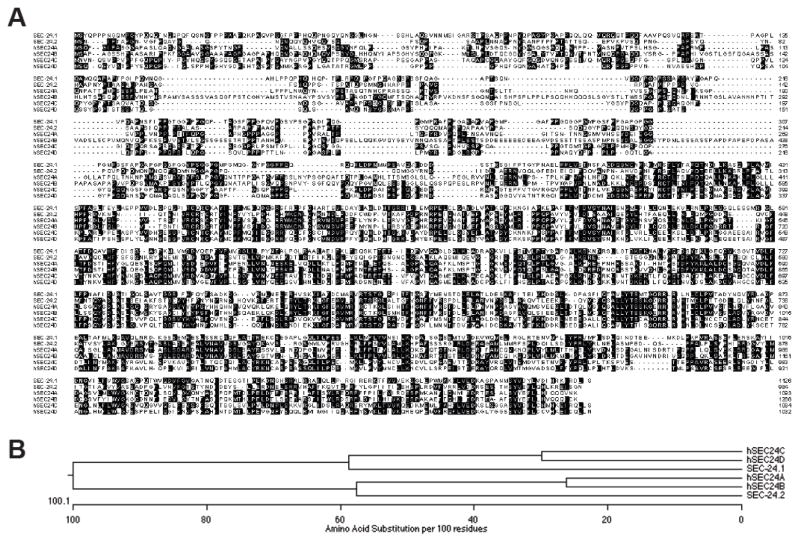
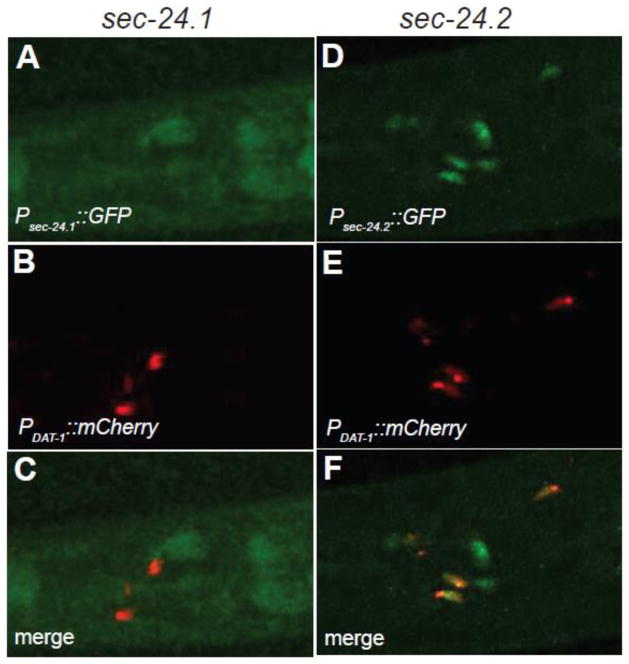
The C. elegans genome expresses two SEC24 orthologs (Figure 4A,B), encoded by sec-24.1 and sec-24.2 genes. SEC-24.1 and SEC-24.2 proteins share only 27% amino acid identity (Figure 4A, B). SEC-24.1 is most similar to human SEC24C (40% identical) and SEC24D (39%), and exhibits lower identity with SEC24A (28%) and SEC24B (30%). On the other hand, SEC-24.2 is most similar to human SEC24A and SEC24B (43% identical), followed by SEC24C (28%) and SEC24D (26%). Tiling array experiments (Spencer et al., 2011) indicate that sec-24.1 has modest levels of expression, the highest being in the hypodermis and coelomocytes, whereas sec-24.2 shows very little enrichment anywhere in the worm and a slightly higher level of expression in larval stage 3 (L3) worms and later. Both genes expression levels in L3-L4 DA neurons was below the L3-L4 reference, with sec-24.2 being the closest. To gain more direct insight into which sec-24 gene product acted in DA neurons to support the export of DAT-1, we fused 1 kB of sequence upstream of the start codons of sec-24.1 and sec-24.2 to GFP and produced transgenic lines transmitting these reporters. Confocal imaging revealed markedly different expression patterns for these genes, with only sec-24.2 expression seen in DA neurons, as defined by a DAT-1 promotor fused to mCherry (Figure 5F). We found that all eight DA neurons were co-labeled by psec24.2:GFP. Remarkably, sec24.2:GFP signal was detected in only a few non-DA cells, suggesting a heretofore unappreciated selectivity of function for SEC24.2, insofar as sec-24.2 expression patterns are mirrored by the upstream 1kB element used in our reporter constructs. These results support a role for SEC-24.2 in somatic export of DAT-1.
Figure 4. Sequence relationships between SEC24 proteins.
A. Amino acid alignment of C. elegans SEC-24.1 and SEC-24.2 proteins with human SEC24 A, B, C, and D proteins. Shading = identical sequences.
B. Dendogram of C. elegans SEC24.1 and SEC24.2 proteins and human SEC24A,B,C, and D proteins, showing the number of amino acid substitutions per 100 residues. Analysis reveals SEC24A and B to be more closely related to SEC24.2 whereas SEC24C and D are more closely related to SEC24.1.
Figure 5. SEC-24.2 is expressed in C. elegans DA neurons.
A–F. Confocal Z stack images of transgenic animals expressing GFP promotor fusions for sec-24.1 (A–C) or sec-24.2 (D–F). Lines were produced to provide for co-expression of mCherry fused to the dat-1 promotor. Only sec-24.2 expression overlaps with dat-1 expression (arrows in F). Data are representative of three independent lines for each sec-24 reporter.
The 591PYRKR595 motif is conserved in human DAT and supports surface trafficking and DA transport function
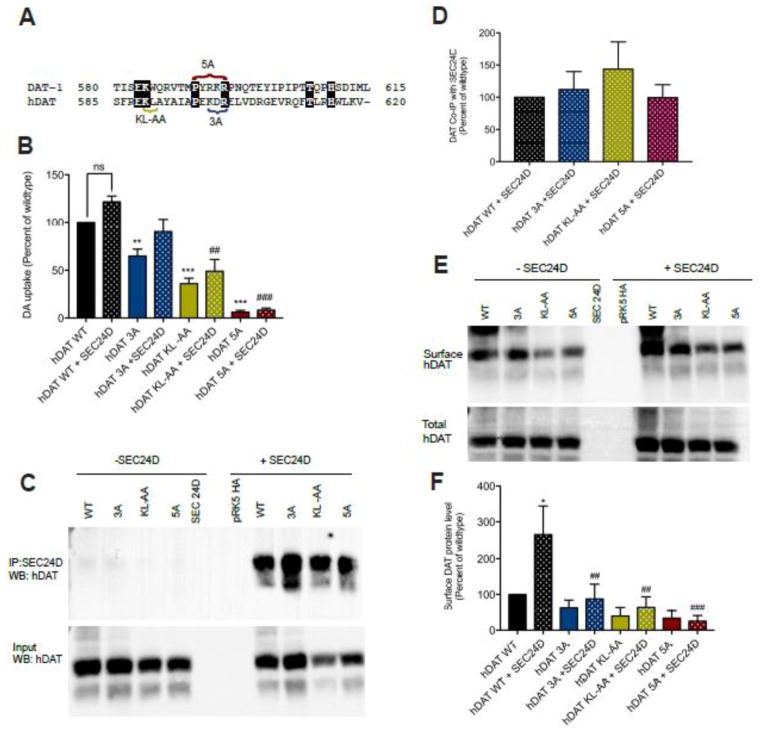
Comparison between C. elegans and human DAT reveals the novel motif is a charged cluster bounded by conserved proline and arginine residues (Figure 6A). The lysine of the SEC24 binding motif is also conserved between C. elegans and human. To assess whether the novel motif is functionally conserved in humans, we mutated the corresponding residues (PEKDR) in an hDAT cDNA construct to alanine (hDAT 5A) and transfected this DNA, as well as constructs bearing alanine substitutions of the 3R motif (KDR, hDAT 3A), or the previously identified SEC-24 binding motif (hDAT KL-AA), in parallel with wildtype sequences into HEK293T cells. The hDAT 5A construct resulted in drastically reduced [3H]DA transport (Figure 6B), without an effect on transporter protein expression (Figure 6C). Mutation of the 3R motif as well as of the KL sequenced resulted in less severe, but nonetheless significant reductions in [3H]DA uptake, also without reductions in transporter protein (Figure 6B, C).
Figure 6. The region covered by the 5A mutation and the SEC24 binding motif are required for SEC24D to enhance surface trafficking of human DAT.
A. Amino acid alignment of the C-terminus of hDAT and C. elegans DAT-1. Shading indicates identical sequences. The sequences in brackets define the sites of mutations made in hDAT KL-AA, hDAT 5A=PEDKR-AAAAA, and hDAT 3A=KDR-AAA).
B. Uptake of [3H]DA in HEK-293T cells transfected with HA::hDAT wildtype and mutants, either alone (no pattern) or with co-transfection of GFP::SEC24D (dot pattern). The novel motif is conserved in hDAT and mutation reduces hDAT activity. Co-expression of SEC24D did not significantly enhance hDAT function. Values represent a mean of transport activities collected in six separate experiments with error bars representing SEM. Data were analyzed using one-way ANOVA with post-tests comparing levels obtained with hDAT with ***P<0.001; ##P<0.01 and ###P<0.0001 for comparison against hDAT+SEC24D.
C–D. hDAT 5A, 3A and KL-AA mutations do not significantly affect the association of hDAT with SEC24D. Co-immunoprecipitation of HA::hDAT with GFP::SEC24D. C. Representative western blot with anti-HA antibody of input (lower panel) and after co-IP with GFP::SEC24D (upper panel). D. Results in C expressed as the mean of the percent of wildtype band intensity for eight separate experiments with error bars representing SEM. Data were analyzed by one-way ANOVA. No significant main effects were detected.
E–F. hDAT 5A, 3A and KL-AA mutations limit the ability of SEC24D to enhance the surface expression of hDAT. E. Representative western blot of surface and total hDAT protein levels, with and without co-transfection with SEC24D. F. Data expressed as the mean of surface mutant DAT protein levels relative to that obtained with hDAT from five separate experiments, with error bars representing SEM. Data were analyzed by one-way ANOVA, where *P<0.05 between hDAT WT and hDAT WT+SEC24D, and ##P<0.01 and ###P<0.0001 between hDATWT+SEC24D and hDAT mutant +SEC24D.
To assess the influence of SEC24 on DAT activity, we co-transfected a GFP::SEC24D construct along with wildtype hDAT or the hDATs 5A, 3A and KL-AA mutants. Co-expression of SEC24D with wildtype hDAT or mutant transporters produced small but non-significant increases in [3H]DA uptake, suggesting that either endogenous SEC24 proteins are sufficient to export DAT proteins or that these transporters “switch their allegiance” to other SEC24 proteins to achieve ER export in the HEK-293T model. To assess the latter possibility, we performed co-immunoprecipitation of HA::hDAT and GFP::SEC24D (Figure 6C, D). As noted above for DA uptake, levels of wildtype and mutant transporters were unaffected by co-expression of SEC24D. Furthermore, we were able to co-immunoprecipitate mutant hDAT proteins with SEC24D comparable to that found with wildtype hDAT co-immunoprecipitation (Figure 6D).
Possibly, the dependence on SEC24 interaction sequences confers a change in DAT proteins that renders the protein nonfunctional as opposed to a trafficking perturbation. To assess whether reduced DA uptake arises from decreased, steady-state hDAT surface expression, we performed a biotinylation assay using a membrane-impermeant biotinylating reagent (Figures 6E,F). In these experiments, though a trend for reduced surface expression was evident for hDAT KL-AA and hDAT 5A relative to wildtype hDAT, these changes did not reach statistical significance. Importantly, whereas SEC24D significantly elevated wildtype hDAT surface expression, this effect was lost with all three hDAT mutants. Thus, although our paradigm did not reveal a role for SEC24D in physical interactions or resulting DA uptake, it afforded detection of a dependence of hDAT surface expression on C-terminal sequences.
DISCUSSION
The powerful influence that DAT exerts over the spatial and temporal features of extracellular DA availability and recycling, along with the growing appreciation of the ability of disrupted DAT to influence risk for brain disorders, has compelled significant effort aimed at identifying structural determinants of DAT regulation. With respect to this issue, the cytoplasmically localized DAT N- and C-termini have drawn the most attention, owing to their size as well as the large number of proteins reported to regulate DAT by interacting directly or indirectly with these domains (Sager and Torres, 2011; Vaughan and Foster, 2013). With respect to the DAT C-terminus, a limited number of discrete sequence elements have been reported to support DAT regulation. Torres and colleagues (Torres et al., 2001) documented the first such element, a Class II PDZ domain recognition sequence (LKV) located at the distal end of the C-terminus of mammalian DAT that supports direct interactions with the PDZ domain protein PICK1. In vitro biochemical, imaging and functional studies of DAT proteins mutated in these sequences support a role of PICK-1 in DAT plasma membrane localization and recycling, ultimately insuring adequate DA transport capacity (Bjerggaard et al., 2004; Madsen et al., 2012; Torres et al., 2001). In vivo studies with DAT LKV-AAA mice support a role for the LKV sequence in sustaining steady-state DAT levels, in part determined by dynamin-mediated endocytosis (Rickhag et al., 2013). In this regard, the ADHD-associated substitution R615C, positioned immediately upstream of the PDZ sequence elevates constitutive DAT endocytosis but abrogates amphetamine and PKC-stimulated endocytosis, possibly a result of disrupted surface microdomain stabilization (Kovtun et al., 2015; Sakrikar et al., 2012).
In a prior study, we explored the role played in vivo by the Class II PDZ domain recognition sequence (613IML615) of the C. elegans DAT-1 protein (McDonald et al., 2007). The rapid and efficient transgenic methods available with the worm model, the capacity to visualize the trafficking of GFP-tagged DAT-1 proteins to DA synapses, and the ability to monitor the behavioral consequences of loss of DAT-1 through the Swip phenotype allowed for a study of both a cellular and functional impact of DAT-1 mutant proteins in vivo. Moreover, the lack of developmental effects arising from loss of DAT-1, as compared to mammals (Kasahara et al., 2015; Kurian et al., 2009), allowed us to utilize DAT-1 deficient animals (dat-1(ok157) as the host for transgene expression, precluding confounds potentially arising from disrupted interactions between mutant and wild type DAT-1 proteins. In following the synaptic localization and functional consequences of DAT-1 proteins mutated to eliminate or occlude these sequences, we found no evidence for an essential role of the IML motif in these processes. In contrast, a larger deletion (Δ25) of the DAT-1 C-terminus resulted in somatic retention, leading to a failure to localize transporters to DA synapses. In this study, we pursued these observations further, narrowing down the critical residues responsible for both DAT-1 localization and functional deficits to the five amino acids 591PYRKR595. Thus, either a deletion spanning these sequences, or the PYRKR-AAAA substitution, resulted in a loss of efficient DAT-1 somatic export and synaptic localization. These changes also induced the Swip phenotype, a paralytic behavior characteristic of animals with a loss of functional DAT-1 protein (McDonald et al., 2007). Although a Swip phenotype was not induced in lines bearing up to a twenty amino acid deletion of the DAT-1 C-terminus, we cannot conclude that the deleted residues do not play a critical role in transporter regulation, owing to the extrachromosomal, multi-copy nature of our expression paradigm, which could have resulted in expression that exceeded that targeted by negative regulators in limiting abundance. Alternatively, the distal twenty amino acids of DAT-1 may have a role in acute, synaptic regulatory events not well captured by the Swip phenotype.
The 591PYRKR595 sequence of DAT-1 aligns with 596PWRDQ600 in Drosophila melanogaster DAT and with 597PEKDR601 in human DAT, and has not been implicated to date in regulatory control of DAT proteins. The region in which the sequence is located is predicted computationally to be relatively unstructured (JPRED4, http://www.compbio.dundee.ac.uk/jpred/, (Drozdetskiy et al., 2015) a finding validated in the X-ray structures of Drosophila DAT (Penmatsa et al., 2013) and Drosophila SERT (Coleman et al., 2016). Across SLC6 family members, only the P591 residue appears highly conserved, raising the question as to whether the proline is actually a key component of an upstream motif (see below). However, a single alanine substitution at this site failed to impact to produce Swip, indicating a non-essential character of this single residue for the ultimate expression of DAT-1 at the synapse. The adjacent tryptophan (W597) in Drosophila DAT has been reported to stabilize an interaction of the C-terminus with intracellular loop 1 (between transmembrane domains 2 and 3) through cation-π interactions, an interaction possibly supported by a DAT-1 pair constituted by Y592 and R91. This interaction may assist in interactions that confer conformational stability during the transport cycle, given that transmembrane 3 forms part of the binding domain for dopamine in Drosophila DAT (Wang et al., 2015). However, the coordination of the pair does not appear to be essential for somatic export, as revealed by the inability of a Y592A substitution to produce Swip. Combination mutations across the novel motif produced modest effects on synapse localization or Swip, suggesting that the residues of the 591PYRKR595 region collectively participate in a binding interface that is critical for export to the synapse.
Given the export phenotype evident with alterations of the 591PYRKR595 sequence, it seems likely that these mutations perturb critical interactions with machinery involved in export to synapses of membrane proteins from the endoplasmic reticulum (ER) or Golgi. Importantly, the 591PYRKR595 sequence lies in close proximity to a two amino acid sequence (584KW585) that aligns with a motif previously implicated in the SEC24-dependent ER export of multiple SLC6 family members as part of the COPII trafficking complex (El-Kasaby et al., 2010; Sucic et al., 2013). Consistent with a role of these residues in DAT-1 export to DA synapses, the KW-AA mutation produced trafficking and Swip effects comparable to the 5A substitution in the 591PYRKR595 sequence. Furthermore, a triple arginine motif has been shown to mediate the interaction between SEC24 isoforms and the α2B-adrenergic receptor, (Dong et al. 2012). A 3R motif is contained within the 591PYRKR595 sequence (593RKR595) and the PYR-AAA mutation produced moderate, but significant, trafficking and behavioral perturbations. We also measured reduced levels of DA in both the DAT-1 5A the KW-AA mutant animals. DA is believed to be stabilized inside presynaptic terminals in synaptic vesicles after DAT-mediated DA uptake. DAT elimination in both mice and worms results in reduced levels of DA, consistent with this idea. Thus, the previously described SEC24 recognition motif clearly supports comparable neurotransmitter transporter export mechanisms in the nematode, though to our knowledge, our findings are the first to demonstrate their role in vivo.
Sucic and colleagues discovered that the sequences aligned with DAT-1 584KW585 dictate SEC24 isoform-specific interactions. The worm possesses two SEC24 isoforms, SEC-24.1 and SEC-24.2. Mammalian DAT has been shown to preferentially interact with SEC24D (Sucic et al., 2011), and sequence alignments indicate that SEC24.1 is more closely related to SEC24D than SEC24.2. Unfortunately, animals bearing loss of function sec-24.1 and sec-24.2 alleles are not viable so a conditional elimination of each isoform should be explored in the future. Using a publically accessible, tiling array data set of worm mRNA expression (WormViz; http://www.vanderbilt.edu/wormdoc/wormmap/WormViz.html) (Spencer et al., 2011), we found a lack of cellular selectivity for either SEC-24 isoform. We therefore generated transgenic reporter lines, driving GFP expression with presumptive promoter elements upstream of the translation start sites of the two proteins. Contrary to expectations, we found that Psec-24.2:GFP displayed a relatively selective pattern of expression, including all 8 DA neurons in the hermaphrodite, validated through dual labeling with the DA-specific reporter Pdat-1:mCherry. In contrast, the expression pattern of Psec-24.2 was diffuse and non-overlapping with the pattern obtained with Pdat-1:mCherry. Although we lack functional corroboration, the expression patterns noted above suggest that DAT-1 export may be facilitated by SEC-24.2, though it is also possible that the patterns of expression of SEC24 isoforms in the worm are not faithfully captured by reporter constructs, or that levels of mRNA may not reflect the true level of SEC-24.1 protein.
In human DAT, DAT-1 591PYRKR595 aligns with 597PEKDR601. To determine whether the latter sequence element is functionally relevant for the control of human DAT membrane trafficking, we co-transfected HEK-293T cells with wildtype or mutant human DAT cDNAs alone or together with SEC24D cDNA. In studies performed in the absence of SEC24D, we observed that full or partial alanine substitutions in the 597PEKDR601 sequence disrupted DA uptake, mirroring findings in transgenic nematodes in vivo. A similar pattern was found in cells co-transfected with SEC24D. As we observed no enhancement of DAT activity in cells co-transfected with SEC24D as compared to DAT constructs on their own, suggesting that endogenous levels of SEC24D may be sufficient to achieve maximal levels of transporter surface expression or DA uptake function. Interestingly, the loss of DA uptake induced by alanine substitutions in the PEKDR sequence was not mirrored by changes in DAT surface expression, which appeared to be on average modestly reduced, though these changes did not reach statistical significance. However, we note that whereas SEC24D could enhance surface levels of wildtype DAT protein, such elevations were not seen with the PEKDR mutants. These findings indicate that endogenous levels of SEC24D in HEK293T cells are insufficient to achieve the full capacity of surface DAT delivery possible with elevated transporter protein expression. Importantly, the lack of a comparable effect using the DAT PEKDR mutants points to a critical role of the PEKDR sequence to surface trafficking, as predicted from worm studies. Differences between wildtype DAT surface trafficking enhancement with SEC24D cotransfection and resulting DA uptake function may reflect a targeting of DAT in the context of excess SEC24D to membrane compartments lacking critical components needed to support DAT function (e.g. interacting proteins, cholesterol, activating kinases). This observation may also relate to the heterologous nature of transporter expression, as compared to a neuronal environment.
Despite evidence that the PEKDR sequence supports intrinsic DAT activity or SEC24D-dependent elevations in DAT surface expression, alanine substitutions of part or all of this sequence clearly failed to disrupt DAT/SEC24D interactions, as explored in co-immunoprecipitation studies. These findings indicate that other sequences (including the KL motif) are more critical to formation or stability of the DAT/SEC24D complex but that the PEKDR sequence can influence how these interactions are utilized. For example, the PEKDR sequences may allow special conformations of DAT or the DAT/SEC24D complex needed to recruit or exclude other proteins that can impact efficient surface expression. SEC24 has been reported to export cargo through distinct determinants that permit interactions with multiple cargo proteins (Miller et al., 2003; Pagant et al., 2015), which can interact with one another. As one possible example, alpha-synuclein has been reported to associate with DAT and limit transporter surface expression by inhibiting DAT trafficking from the ER to the Golgi (Oaks and Sidhu, 2013; Thayanidhi et al., 2010). SNARE proteins also impact ER/Golgi and Golgi-plasma membrane trafficking (Thayanidhi et al., 2010). One SNARE protein, syntaxin 1A has been found to interact with DAT and regulate transporter surface expression (Cervinski et al., 2010). These or other interactions may be influenced by the conformations achieved through SEC24D:DAT interactions which, although not abrogated by PEKDR mutations, may be sufficiently altered to restrict the efficiency of DAT export, surface expression and ultimately DA uptake capacity. Further progress in this area will likely require higher resolution evaluation of SEC24D:DAT complexes. Further progress in this area will likely require higher resolution evaluation of SEC24D:DAT complexes, and ultimately may provide new opportunities to remedy the disrupted trafficking that is evident with disease-associated DAT mutations (Kasture et al., 2016). As this work proceeds, the opportunities of the C. elegans model for assessment of DAT trafficking to identified, mature DA synapses in vivo may prove highly advantageous.
HIGHLIGHTS.
Presynaptic DAT is a critical determinant of DA signaling across phylogeny.
Two C-terminal dictate synaptic localization of C. elegans DAT-1.
Sequence motifs associated with SEC24 support DAT axonal export
SEC24 proteins support DAT trafficking across phylogeny.
Acknowledgments
The authors gratefully acknowledge the excellent support for general laboratory operations provided by Chris Svitek, Angela Steele and Tracy Moore-Jarrett. NIH award MH095044 to RDB supported the research. SR was supported by NIH award MH65215 to RDB. JAH was supported by NIH award MH093102. DPB was supported by NIH award MH DA035559.
Abbreviations
- DA
dopamine
- DAT
dopamine transporter
- C. elegans
Caenhorhabditis elegans
- SWIP
swimming-induced paralysis
- GFP
green fluorescent protein
- ER
endoplasmic reticulum
- hDAT
human dopamine transporter
- ceDAT
C. elegans dopamine transporter
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Apparsundaram S, Sung U, Price RD, Blakely RD. Trafficking-dependent and -independent pathways of neurotransmitter transporter regulation differentially involving p38 mitogen-activating protein kinase revealed sstudies of insulin modulation of neuroepinepherine transport in SK-N-SH cells. J Pharmacol Exp Ther. 2001;299:666–677. [PubMed] [Google Scholar]
- Bjerggaard C, Fog JU, Hastrup H, Madsen K, Loland CJ, Javitch JA, Gether U. Surface targeting of the dopamine transporter involves discrete epitopes in the distal C terminus but does not require canonical PDZ domain interactions. J Neurosci. 2004;24:7024–7036. doi: 10.1523/JNEUROSCI.1863-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonoldi I, Howes OD. The enduring centrality of dopamine in the pathophysiology of schizophrenia: in vivo evidence from the prodrome to the first psychotic episode. Adv Pharmacol. 2013;68:199–220. doi: 10.1016/B978-0-12-411512-5.00010-5. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson A. Perspectives on the discovery of central monoaminergic neurotransmission. Annu Rev Neurosci. 1987;10:19–40. doi: 10.1146/annurev.ne.10.030187.000315. [DOI] [PubMed] [Google Scholar]
- Carvelli L, Blakely RD, DeFelice LJ. Dopamine transporter/syntaxin 1A interactions regulate transporter channel activity and dopaminergic synaptic transmission. Proc Natl Acad Sci U S A. 2008;105:14192–14197. doi: 10.1073/pnas.0802214105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvelli L, McDonald PW, Blakely RD, Defelice LJ. Dopamine transporters depolarize neurons by a channel mechanism. Proc Natl Acad Sci U S A. 2004;101:16046–16051. doi: 10.1073/pnas.0403299101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervinski MA, Foster JD, Vaughan RA. Syntaxin 1A regulates dopamine transporter activity, phosphorylation and surface expression. Neuroscience. 2010;170:408–416. doi: 10.1016/j.neuroscience.2010.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Furman CA, Gnegy ME. Dopamine transporter trafficking: rapid reponse on demand. Future Neurol. 2010;5:123. doi: 10.2217/fnl.09.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Green EM, Gouaux E. X-ray structures and mechanism of the human serotonin transporter. Nature. 2016;532:334–339. doi: 10.1038/nature17629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cousins DA, Butts K, Young AH. The role of dopamine in bipolar disorder. Bipolar Disord. 2009;11:787–806. doi: 10.1111/j.1399-5618.2009.00760.x. [DOI] [PubMed] [Google Scholar]
- Del Campo N, Chamberlain SR, Sahakian BJ, Robbins TW. The roles of dopamine and noradrenaline in the pathophysiology and treatment of attention-deficit/hyperactivity disorder. Biol Psychiatry. 2011;69:e145–157. doi: 10.1016/j.biopsych.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Dong C, Nichols CD, Guo J, Huang W, Lambert NA, Wu G. A triple arg motif mediates alpha(2B)-adrenergic receptor interaction with Sec24C/D and export. Traffic. 2012;13:857–868. doi: 10.1111/j.1600-0854.2012.01351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A, Cole C, Procter J, Barton GJ. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kasaby A, Just H, Malle E, Stolt-Bergner PC, Sitte HH, Freissmuth M, Kudlacek O. Mutations in the carboxyl-terminal SEC24 binding motif of the serotonin transporter impair folding of the transporter. J Biol Chem. 2010;285:39201–39210. doi: 10.1074/jbc.M110.118000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster JD, Vaughan RA. Palmitoylation controls dopamine transporter kinetics, degradation, and protein kinase C-dependent regulation. J Biol Chem. 2011;286:5175–5186. doi: 10.1074/jbc.M110.187872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen FH, Skjorringe T, Yasmeen S, Arends NV, Sahai MA, Erreger K, Andreassen TF, Holy M, Hamilton PJ, Neergheen V, Karlsborg M, Newman AH, Pope S, Heales SJ, Friberg L, Law I, Pinborg LH, Sitte HH, Loland C, Shi L, Weinstein H, Galli A, Hjermind LE, Moller LB, Gether U. Missense dopamine transporter mutations associate with adult parkinsonism and ADHD. J Clin Invest. 2014;124:3107–3120. doi: 10.1172/JCI73778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardaway JA, Hardie SL, Whitaker SM, Baas SR, Zhang B, Bermingham DP, Lichtenstein AJ, Blakely RD. Forward genetic analysis to identify determinants of dopamine signaling in Caenorhabditis elegans using swimming-induced paralysis. G3 (Bethesda) 2012;2:961–975. doi: 10.1534/g3.112.003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardaway JA, Sturgeon SM, Snarrenberg CL, Li Z, Xu XZ, Bermingham DP, Odiase P, Spencer WC, Miller DM, 3rd, Carvelli L, Hardie SL, Blakely RD. Glial Expression of the Caenorhabditis elegans Gene swip-10 Supports Glutamate Dependent Control of Extrasynaptic Dopamine Signaling. J Neurosci. 2015;35:9409–9423. doi: 10.1523/JNEUROSCI.0800-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heshmati M, Russo SJ. Anhedonia and the brain reward circuitry in depression. Curr Behav Neurosci Rep. 2015;2:146–153. doi: 10.1007/s40473-015-0044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Hornykiewicz O. The discovery of dopamine deficiency in the parkinsonian brain. J Neural Transm Suppl. 2006:9–15. doi: 10.1007/978-3-211-45295-0_3. [DOI] [PubMed] [Google Scholar]
- Horschitz S, Hummerich R, Lau T, Rietschel M, Schloss P. A dopamine transporter mutation asssociated with bipolar affective disorder causes inhibition of transporter surface expression. Mol Psychiatry. 2005;10:1104–1109. doi: 10.1038/sj.mp.4001730. [DOI] [PubMed] [Google Scholar]
- Kasahara Y, Arime Y, Hall FS, Uhl GR, Sora I. Region-specific dendritic spine loss of pyramidal neurons in dopamine transporter knockout mice. Curr Mol Med. 2015;15:237–244. doi: 10.2174/1566524015666150330143613. [DOI] [PubMed] [Google Scholar]
- Kasture A, El-Kasaby A, Szollosi D, Asjad HM, Grimm A, Stockner T, Hummel T, Freissmuth M, Sucic S. Functional rescue of a misfolded Drosophila melanogaster dopamine transporter mutant associated with a sleepless phenotype by pharmacological chaperones. J Biol Chem. 2016 doi: 10.1074/jbc.M116.737551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiflin R, Janak PH. Dopamine Prediction Errors in Reward Learning and Addiction: From Theory to Neural Circuitry. Neuron. 2015;88:247–263. doi: 10.1016/j.neuron.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun O, Sakrikar D, Tomlinson ID, Chang JC, Arzeta-Ferrer X, Blakely RD, Rosenthal SJ. Single-quantum-dot tracking reveals altered membrane dynamics of an attention-deficit/hyperactivity-disorder-derived dopamine transporter coding variant. ACS Chem Neurosci. 2015;6:526–534. doi: 10.1021/cn500202c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian MA, Zhen J, Cheng SY, Li Y, Mordekar SR, Jardine P, Morgan NV, Meyer E, Tee L, Pasha S, Wassmer E, Heales SJ, Gissen P, Reith ME, Maher ER. Homozygous loss-of-function mutations in the gene encoding the dopamine transporter are associated with infantile parkinsonism-dystonia. J Clin Invest. 2009;119:1595–1603. doi: 10.1172/JCI39060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Ferro-Novick S, Miller EA. The highly conserved COPII coat complex sorts cargo from the endoplasmic reticulum and targets it to the golgi. Cold Spring Harb Perspect Biol. 2013:5. doi: 10.1101/cshperspect.a013367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen KL, Thorsen TS, Rahbek-Clemmensen T, Eriksen J, Gether U. Protein interacting with C kinase 1 (PICK1) reduces reinsertion rates of interaction partners sorted to Rab11-dependent slow recycling pathway. J Biol Chem. 2012;287:12293–12308. doi: 10.1074/jbc.M111.294702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazei-Robison MS, Couch RS, Shelton RC, Stein MA, Blakely RD. Sequence variation in the human dopamine transporter gene in children with attention deficit hyperactivity disorder. Neuropharmacology. 2005;49:724–736. doi: 10.1016/j.neuropharm.2005.08.003. [DOI] [PubMed] [Google Scholar]
- McDonald PW, Hardie SL, Jessen TN, Carvelli L, Matthies DS, Blakely RD. Vigorous motor activity in Caenorhabditis elegans requires efficient clearance of dopamine mediated by synaptic localization of the dopamine transporter DAT-1. J Neurosci. 2007;27:14216–14227. doi: 10.1523/JNEUROSCI.2992-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald PW, Jessen T, Field JR, Blakely RD. Dopamine signaling architecture in Caenorhabditis elegans. Cell Mol Neurobiol. 2006;26:593–618. doi: 10.1007/s10571-006-9003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melikian HE. Neurotransmitter transporter trafficking: endocytosis, recycling, and regulation. Pharmacol Ther. 2004;104:17–27. doi: 10.1016/j.pharmthera.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R. Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- Nass R, Hahn MK, Jessen T, McDonald PW, Carvelli L, Blakely RD. A genetic screen in Caenorhabditis elegans for dopamine neuron insensitivity to 6-hydroxydopamine identifies dopamine transporter mutants impacting transporter biosynthesis and trafficking. J Neurochem. 2005;94:774–785. doi: 10.1111/j.1471-4159.2005.03205.x. [DOI] [PubMed] [Google Scholar]
- Nass R, Hall DH, Miller DM, 3rd, Blakely RD. Neurotoxin-induced degeneration of dopamine neurons in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2002;99:3264–3269. doi: 10.1073/pnas.042497999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ. Is there a common molecular pathway for addiction? Nat Neurosci. 2005;8:1445–1449. doi: 10.1038/nn1578. [DOI] [PubMed] [Google Scholar]
- Oaks AW, Sidhu A. Parallel mechanisms for direct and indirect membrane protein trafficking by synucleins. Commun Integr Biol. 2013;6:e26794. doi: 10.4161/cib.26794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano A, Letourneur F, Garcia-Estefania D, Carpentier J, Orci L, Paccaud J. Sec24 proteins and sorting at the endoplasmic reticulum. J Biol Chem. 1999;274:7833–7840. doi: 10.1074/jbc.274.12.7833. [DOI] [PubMed] [Google Scholar]
- Pagant S, Wu A, Edwards S, Diehl F, Miller EA. Sec24 is a coincidence detector that simultaneously binds two signals to drive ER export. Curr Biol. 2015;25:403–412. doi: 10.1016/j.cub.2014.11.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penmatsa A, Wang KH, Gouaux E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013;503:85–90. doi: 10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickhag M, Hansen FH, Sorensen G, Strandfelt KN, Andresen B, Gotfryd K, Madsen KL, Vestergaard-Klewe I, Ammendrup-Johnsen I, Eriksen J, Newman AH, Fuchtbauer EM, Gomeza J, Woldbye DP, Wortwein G, Gether U. A C-terminal PDZ domain-binding sequence is required for striatal distribution of the dopamine transporter. Nat Commun. 2013;4:1580. doi: 10.1038/ncomms2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz MC, Lamb RJ, Goldberg SR, Kuhar MJ. Cocaine receptors on dopamine transporters are related to self-administration of cocaine. Science. 1987;237:1219–1223. doi: 10.1126/science.2820058. [DOI] [PubMed] [Google Scholar]
- Sager JJ, Torres GE. Proteins interacting with monoamine transporters: current state and future challenges. Biochemistry. 2011;50:7295–7310. doi: 10.1021/bi200405c. [DOI] [PubMed] [Google Scholar]
- Sakrikar D, Mazei-Robison MS, Mergy MA, Richtand NW, Han Q, Hamilton PJ, Bowton E, Galli A, Veenstra-Vanderweele J, Gill M, Blakely RD. Attention deficit/hyperactivity disorder-derived coding variation in the dopamine transporter disrupts microdomain targeting and trafficking regulation. J Neurosci. 2012;32:5385–5397. doi: 10.1523/JNEUROSCI.6033-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman P. Dopamine D2 receptors as treatment targets in schizophrenia. Clin Schizophr Relat Phychoses. 2010:4. [PubMed] [Google Scholar]
- Smith CJ, Watson JD, Spencer WC, O’Brien T, Cha B, Albeg A, Treinin M, Miller DM., 3rd Time-lapse imaging and cell-specific expression profiling reveal dynamic branching and molecular determinants of a multi-dendritic nociceptor in C. elegans. Dev Biol. 2010;345:18–33. doi: 10.1016/j.ydbio.2010.05.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WC, Zeller G, Watson JD, Henz SR, Watkins KL, McWhirter RD, Petersen S, Sreedharan VT, Widmer C, Jo J, Reinke V, Petrella L, Strome S, Von Stetina SE, Katz M, Shaham S, Ratsch G, Miller DM., 3rd A spatial and temporal map of C. elegans gene expression. Genome Res. 2011;21:325–341. doi: 10.1101/gr.114595.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucic S, El-Kasaby A, Kudlacek O, Sarker S, Sitte HH, Marin P, Freissmuth M. The serotonin transporter is an exclusive client of the coat protein complex II (COPII) component SEC24C. J Biol Chem. 2011;286:16482–16490. doi: 10.1074/jbc.M111.230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucic S, Koban F, El-Kasaby A, Kudlacek O, Stockner T, Sitte HH, Freissmuth M. Switching the clientele: a lysine residing in the C terminus of the serotonin transporter specifies its preference for the coat protein complex II component SEC24C. J Biol Chem. 2013;288:5330–5341. doi: 10.1074/jbc.M112.408237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulzer D, Sonders MS, Poulsen NW, Galli A. Mechanisms of neurotransmitter release by amphetamines: a review. Prog Neurobiol. 2005;75:406–433. doi: 10.1016/j.pneurobio.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Thayanidhi N, Helm JR, Nycz DC, Bentley M, Liang Y, Hay JC. Alpha-synuclein delays endoplasmic reticulum (ER)-to-Golgi transport in mammalian cells by antagonizing ER/Golgi SNAREs. Mol Biol Cell. 2010;21:1850–1863. doi: 10.1091/mbc.E09-09-0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson O, Edgley M, Strasbourger P, Flibotte S, Ewing B, Adair R, Au V, Chaudhry I, Fernando L, Hutter H, Kieffer A, Lau J, Lee N, Miller A, Raymant G, Shen B, Shendure J, Taylor J, Turner EH, Hillier LW, Moerman DG, Waterston RH. The million mutation project: a new approach to genetics in Caenorhabiditis elegans. Genome Res. 2013;23:1749–1762. doi: 10.1101/gr.157651.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres GE, Gainetdinov RR, Caron MG. Plasma membrane monoamine transporters: structure, regulation and function. Nat Rev Neurosci. 2003;4:13–25. doi: 10.1038/nrn1008. [DOI] [PubMed] [Google Scholar]
- Torres GE, Yao WD, Mohn AR, Quan H, Kim KM, Levey AI, Staudinger J, Caron MG. Functional interaction between monoamine plasma membrane transporters and the synaptic PDZ domain-containing protein PICK1. Neuron. 2001;30:121–134. doi: 10.1016/s0896-6273(01)00267-7. [DOI] [PubMed] [Google Scholar]
- Vaughan RA, Foster JD. Mechanisms of dopamine transporter regulation in normal and disease states. Trends Pharmacol Sci. 2013;34:489–496. doi: 10.1016/j.tips.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Penmatsa A, Gouaux E. Neurotransmitter and psychostimulant recognition by the dopamine transporter. Nature. 2015;521:322–327. doi: 10.1038/nature14431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Bellve KD, Fogarty KE, Melikian HE. Ack1 is a dopamine transporter endocytic brake that rescues a trafficking-dysregulated ADHD coding variant. Proc Natl Acad Sci U S A. 2015;112:15480–15485. doi: 10.1073/pnas.1512957112. [DOI] [PMC free article] [PubMed] [Google Scholar]