Abstract
Delivery of antigens by injection of the encoding DNA allows access to multiple antigen-presenting pathways. Knowledge of immunological processes can therefore be used to modify construct design to induce selected effector functions. Expression can be directed to specific intracellular sites, and additional genes can be fused or codelivered to amplify responses. Therapeutic vaccination against cancer adds a requirement to overcome tolerance and to activate a weakened immune repertoire. Induction of CD4+ T helper cells is critical for both antibody and T cell effector responses. To activate immunity against tumor antigens, we fused the tumor-derived sequences to genes encoding microbial proteins. This strategy engages T helper cells from the large antimicrobial repertoire for linked help for inducing antibody against cell-surface tumor antigens. The principle of linked T cell help also holds for induction of epitope-specific antitumor CD8+ T cells, but the microbial sequence has to be minimized to avoid competition with tumor antigens. Epitope-specific DNA vaccination leads to powerful antitumor attack and can activate immunity from a profoundly tolerized repertoire. Vaccine designs validated in preclinical models are now in clinical trial with immune responses detected against both tumor antigens and fused microbial antigens. DNA priming is highly efficient, but boosting may benefit from increased antigen expression. Physical methods including electroporation provide increased expression without introducing additional competing antigens. A wide range of cancers can be targeted, and objective assays of response will determine efficacy.
Therapeutic vaccination was tested first against infectious disease in 1885 by Louis Pasteur, who successfully vaccinated individuals infected with rabies. This was a fortunate setting because the virus grows relatively slowly, giving time for induction of immunity, but it established the principle of vaccination as a treatment. For the bulk of infectious diseases with available vaccines, the preferred setting of prophylactic vaccination was used once the relative safety of vaccination was assured. The consequent benefit for public health is clear, and new vaccines against additional organisms are being sought. The previous treatment of infections by passive transfer of antibodies gradually declined but is still used occasionally for treatment of certain infections (1). Furthermore, antibody therapy has certainly been shown to be effective in the treatment of selected cancers. The advent of monoclonal antibodies allows infusion of measured doses targeted to selected antigens, and antibody is now an important “drug” for the treatment of B cell malignancies (2) and solid tumors.
Passive transfer of specific T cells can be used also to attack infections (3) and cancer (4). For tumors, efficacy of transferred T cells is seen most clearly in allogeneic stem cell transfer, in which donor T cells recognize patient-specific polymorphic antigens expressed by leukemic cells (5). In this setting, an obvious balance between attack on tumor cells or normal host cells has to be struck to avoid damaging graft-versus-host disease. A similar balance applies to active vaccination against cancer, because many candidate tumor antigens are self-proteins overexpressed by tumor cells (6). The success of passive immunity has demonstrated the ability of immune effector pathways to suppress cancer, and the challenge for therapeutic vaccines is to prime and maintain those pathways.
The idea of activating immunity against cancer is not new. There have been many attempts to induce immunity, often by using nonspecific or uncharacterized vaccines, with no objective measure of outcome except observation of clinical status. Because this status is often variable even without treatment, it has been difficult to assess efficacy. The arrival of genomics has transformed this situation. It is now possible to identify new target antigens, develop novel vaccine-delivery systems, and assess specific immune responses objectively and quickly. For cancer, it is feasible to take principles developed in preclinical models into pilot clinical trials, generating data that should be of use for vaccination in general. DNA vaccines provide testable vehicles for inducing immunity against candidate antigens from either infectious diseases or cancer and have now advanced to a pivotal point (7).
DNA Vaccines
DNA vaccines are simple vehicles for in vivo transfection and antigen production, and the subsequent steps to the induction of immunity are now emerging. The first is the activation of the innate immune response caused by the presence of hypomethylated CpG dinucleotide sequences with particular surrounding motifs in the bacterial plasmid backbone (8), which may be a natural response to exposure to bacterial DNA and is a significant operational component of DNA vaccines. The outcome is an outpouring of cytokines including IL-6, IL-12, tumor necrosis factor-α, IFN-γ, and IFN-α (9) and a polarization of the CD4+ T cell response toward T helper 1 (Th1) dominance. The most effective sequence motifs have been delineated by using synthetic oligonucleotides and differ among species. Selected oligonucleotides are now being investigated as adjuvants for protein vaccines, with promising results emerging (8). However, the data from oligonucleotides do not completely explain how plasmid DNA is perceived by the innate immune response. Oligonucleotides are known to require Toll-like receptor 9 (TLR-9) for activity, but DNA vaccines operate normally in TLR-9-/- mice, pointing to the involvement of additional receptors (10). Recognition of bacterial DNA by the innate immune response can be harnessed by DNA vaccines, with no apparent harmful side effects.
In terms of induction of immunity, it is difficult to generalize about DNA vaccines. There is an influence of the site and procedure used for injection, with muscle and skin cells clearly able to act as antigen depots but unable to prime the immune response. It is likely that cross-presentation from these sites to antigen-presenting cells (APCs) is the major route to priming (11, 12), but there is also evidence for direct transfection of APCs, especially when delivery is to skin sites through a gene gun (13). The uncertainty on this point makes rational design more difficult, particularly because the process of cross-presentation is incompletely understood. However, a recent investigation of the route of access of exogenous phagosomes to the MHC class I pathway could have relevance. Phagosomes apparently carry elements of the endoplasmic reticulum (ER), creating organelles capable of antigen processing for induction of cytotoxic T cell responses (14, 15). It is conceivable that transfected depot cells undergoing apoptosis can behave similarly. The process that conveys antigens to the APC seems highly efficient in that DNA vaccines that produce only very low levels of antigen can induce all arms of the immune response (16). However, there may be different requirements for priming or boosting immunity, and to activate antitumor immunity, both processes need to be efficient. It is also essential that tumor cells alone can boost the vaccine-induced response so that continuing pressure is maintained against emergent cells.
Candidate Tumor Antigens
The list of candidate tumor antigens grows daily, largely because of expanding genetic technology including human genome sequencing and gene-expression profiling. Target antigens can be tumor-specific, lineage-specific, or overexpressed by tumor cells. Structural modifications, particularly of carbohydrates, can also create new antigens (7). In some cases, the myriad of antigens generates a spontaneous immune response (17), and serum antibodies in patients have been used to identify potential tumor antigens by serological identification of antigens by recombinant expression profiling (SEREX) (18). There is, in fact, intriguing evidence for an effective immune response able to suppress cancer. These data were manifested clinically as an autoimmune disease, paraneoplastic neurological degeneration, but immunity arises initially against cancer cells and cross-reacts with neuronal cells (19). This example illustrates an important dilemma for vaccination against overexpressed normal antigens, which is how to induce effective immunity against the chosen target without leading to damaging autoimmunity. The precision offered by DNA vaccines will induce focused immunity against selected antigens, and, as they become more powerful, targets will have to be selected carefully to avoid autoimmunity. DNA vaccines can be designed to induce an appropriate effector pathway, including antibody against cell-surface antigens, or cytotoxic T lymphocyte (CTL) responses against intracellular antigens expressed only as MHC class I-associated peptides. There also may be an effector role for CD4+ T cells, especially for MHC class II-expressing hematological tumors. The real question is whether DNA delivery can activate and maintain the high level of immunity required to suppress cancer cell growth. Not only are tumor antigens generally weakly immunogenic, but the immune repertoire in patients may have been tolerized. DNA vaccines offer the opportunity to add genes encoding molecules aimed at overcoming these problems.
“Additives” for DNA Vaccines Against Cancer
It is becoming clear that additional molecules are required to activate effective immunity against weak tumor antigens. Immunology textbooks can supply a long list of candidate additives, and many have been tested (20). The growing understanding of the process of activation of immunity is providing more refined approaches to this problem. Adjuvants have always been required for vaccines using purified proteins, and although there is a natural stimulus of innate pathways from the CpG sequences, additional adjuvant activity could be useful. Knowledge of Toll-like receptors involved in innate immunity is facilitating manipulation of this pathway against cancer (21). Heat-shock proteins are also able to activate innate immunity as well as to deliver antigens to APCs (22). For DNA vaccines, addition of the simple aluminum salts typically used with conventional vaccines has been found to improve performance (23), and codelivery of cytokines either as protein or through DNA has been explored (24). One problem is that these short-acting molecules are synthesized at specific time points during the response, and continuous production at the site of injection may be undesirable. It will take time to assess the efficacy of these approaches and translate results into patients, in whom response to adjuvants may differ from those in mice. In addition to codelivery, DNA vaccines allow fusion of genes encoding activating molecules to the antigen-encoding sequence. For selected molecules this is an advantage, and fusion genes can create single vaccines capable of multiple functions.
DNA Fusion Gene Vaccines
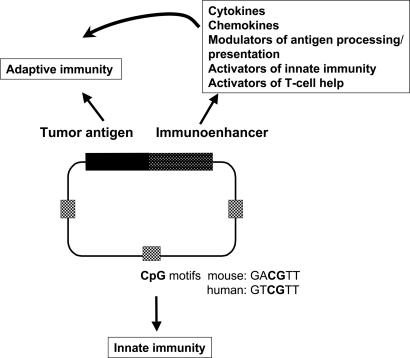
A wide range of genes encoding potentially immunoenhancing proteins involved in different immune pathways is being assessed in the context of fusion genes (Fig. 1). The aim of fusion is to increase the level of adaptive immunity against the tumor antigen and also to direct immune outcome. Fusion gene proteins including cytokines, chemokines, Fc receptors, complement, or antibodies against specific receptors could act by binding to APCs, thereby improving uptake for presentation. However, if direct transfection is important, it is difficult to determine how these proteins will act. It could be more likely then that potentially hazardous autoimmunity against these proteins could be induced (25). Molecules to activate innate immunity may be useful, although the CpG motifs already provide some of this stimulus. Costimulatory molecules such as B7-1/B7-2 to enhance antigen presentation are being assessed, although again it is difficult to know which cells are targets for enhancement. In our present state of ignorance, the best approach is probably empirical, with additional genes tested in models and deductions on the pathways activated made subsequently.
Fig. 1.
DNA fusion gene vaccines to enhance and direct immune responses. DNA vaccines contain CpG sequences within the bacterial DNA backbone able to activate innate immunity in different species. Specific tumor antigen expression is driven by a viral promoter, leading to adaptive immunity. To enhance and direct immune pathways, genes encoding a range of modulating molecules can be fused to the 3′ end of the antigen-encoding sequence to generate activating fusion proteins.
One important aspect that can be manipulated logically is the antigen presentation and processing pathway. If an antibody response is the goal, it is clearly desirable to direct antigen expression to the ER, in which folding and secretion can occur. An appropriate signal sequence can achieve this, but driving any protein to the ER will not always succeed, because some proteins normally located at different sites may be unable to fold and fail to induce antibody (26). To induce CTLs, driving expression to the cytosol for access to the proteasome has logic, but it has been found that addition of a signal sequence does not seem to disadvantage this pathway and in fact can amplify CTL responses (26). The mechanism is likely to involve retrograde transport from the ER to the cytosol (27). For induction of CTLs, addition of genes encoding molecules such as ubiquitin, aimed to enhance degradation and peptide production in the proteasome, can be effective but does not always succeed (28). Similarly, targeting expression to different subcellular pathways such as the endosome or lysosome can amplify CD4+ T cell responses (29). Although there are many ways to present different tumor antigens for induction of immunity, there is one underlying component of the immune response that seems to be crucial for success of all vaccine approaches, and this is the provision of T cell help. DNA vaccines offer the opportunity to activate Th cells and transform weak and ineffective immunity to a powerful antitumor attack.
DNA Fusion Genes to Activate T Cell Help for Antitumor Responses
It has been clear for many years that CD4+ T cells have a major role in helping B cells to produce antibody. More recently, it has become evident that Th cells also control induction and maintenance of CD8+ T cells (30) and can help other CD4+ T cells by a process of linked T cell help (31). It is known also that CD4+ T cells can act as regulatory cells (Treg), suppressing ongoing immunity (32), and in this context could be a disadvantage for immunity against cancer (33). It is necessary therefore to activate Th cells to amplify and maintain antitumor immunity and suppress Tregs that could act against the desired response. In this respect we can learn a great deal from the immune response to infectious organisms.
The pivotal position of CD4+ Th cells has led us to focus on their importance in responses to DNA vaccination and to design fusion genes to induce activating Th responses from the nontolerized immune repertoire (20). Our strategy is to include genes encoding microbial proteins fused to the tumor antigen sequence. Not only is the Th repertoire large, but it is untouched by tolerogenic tumor-derived molecules. By selecting different microbial genes, or subgenic sequences, it is possible to dramatically amplify immunity against tumor cells (7). We have used preclinical models of B cell malignancies to investigate this strategy, first for induction of antibody and CD4+ T cells (34). To focus on induction of CTLs, we moved to a carcinoma model in which known MHC class I-binding peptides offer targets for attack (35). We have been conscious throughout of the need to test the operation of chosen DNA fusion vaccine designs in human subjects.
DNA Fusion Gene Vaccines Against B Cell Malignancies
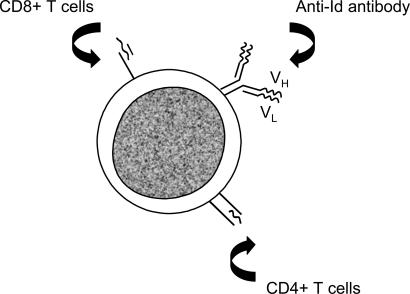
B cell tumors have several advantages for testing vaccine approaches, the main one being the expression of a defined tumor-specific antigen, the idiotypic (Id) Ig (36). In B cell lymphoma, Id Ig is expressed at the cell surface, and the variable region sequences offer clone-specific targets for antibody or T cell recognition (Fig. 2). Although Id Ig is a membrane-bound protein, Id-derived peptides are presented in association with MHC class II and can therefore be targeted by CD4+ T cells (37). Id peptides also may be presented in association with MHC class I molecules (38), but the sequence restriction imposed for binding on peptides that differ between each B cell tumor hampers attempts to induce CD8+ T cells for therapeutic vaccines in human subjects.
Fig. 2.
Target Id Ig antigen expressed at the surface of B cell malignancies. Id Ig is a clonotypic tumor-specific antigen expressed as a cell-surface glycoprotein available for antibody attack. Id peptides can also be expressed in the groove of the MHC class I or II molecules, in which they can be recognized by CD8+ or CD4+ T cells, respectively.
The first attempts to use a defined antibody to treat human B cell malignancy was with anti-Id antibody (39, 40), which remains an effective strategy. The problem is that each Id Ig is different, and it is expensive and technically demanding to make individual anti-Id antibodies. A slightly easier approach is to use the Id Ig as a protein vaccine, which has succeeded in preclinical models (41) and in pilot clinical trials (42). However, even making the Id Ig protein is difficult, and genetic technology appeared more attractive. It is relatively simple to identify and isolate the variable (V) region genes, VH and VL, which encode the Id determinants, on an individual patient basis. Therefore, we chose this antigen as a completely safe, tumor-specific target for testing in a DNA fusion vaccine format.
For convenience, we assembled the variable region genes as a single-chain Fv (scFv) sequence, known to be capable of folding to display the Id determinants of the parental Id Ig (43). Once in a DNA vaccine, it soon became clear that scFv alone was a typical weak antigen, and even injecting human scFv into mice induced only poor antibody responses. We reasoned that addition of a gene to enhance T cell help was required, and we chose a microbial gene for this purpose. Tetanus toxin has been used as an inactivated toxoid vaccine against Clostridium tetani for many years with good results, but there was interest in replacing this with a DNA vaccine encoding a nontoxic region of the toxin, the fragment C (FrC) (44). We therefore obtained the FrC sequence and fused it to the 3′ end of the scFv. We then tested the ability of the fused scFv-FrC gene to induce protection against the parental lymphoma. The results were dramatic: not only was there a striking increase in anti-Id antibody levels, but Id-specific immunity against the lymphoma was clear (34, 45). Similar results have been obtained in other models and with other antigens, including a recent test of a translocation sequence from a leukemia (46).
The principle of fusing a gene encoding a foreign sequence to Id-encoding genes to amplify anti-Id responses and suppress lymphoma growth is clear. Alternative sequences have been investigated by other groups (47–49). One strategy is to use xenogeneic Fcγ sequences fused to autologous variable regions, and for patients, mouse Fcγ has been used. However, early results in patients with lymphoma have shown only weak responses (50). Although we consider FrC to be a strong inducer of linked T cell help, it is not unique. There is also the question of the effect on induction of immunity in the presence of preexisting immunity to tetanus toxoid, which will be common in patients. We modeled this setting by vaccinating mice with toxoid and subsequently vaccinating with DNA scFv-FrC. A slight slowing of the anti-Id antibody response resulted but did not persist after a boosting injection (51). However, we also investigated an immunoenhancing sequence derived from a plant viral coat protein (PVCP) to which there is no detectable preexisting immunity in patients. In the context of DNA scFv fusion vaccines, scFv-PVCP compared well with scFv-FrC in inducing protective immunity. Interestingly, the effector pathways differed, with CD4+ T cell-mediated protection being more evident after scFv-PVCP, whereas antibody was more important after scFv-FrC (52). This result indicates that the nature of the immunoenhancing sequence influences the immunological outcome and provides another strategy for manipulation of immunity.
Clinical Testing
The striking results with DNA scFv-FrC have allowed us to move to a pilot clinical trial of DNA scFv-FrC fusion genes in patients with follicular lymphoma. DNA is injected at an intramuscular site at weeks 0, 1, 2, 4, 8, and 12, with a dose escalation from 500 to 2,500 μg per injection. Individual vaccines are required, and each scFv protein has to be expressed as a recombinant molecule for measuring immune responses, which slows progress. We have made preliminary assessments of responses against both the FrC and Id components of the vaccine in 10 patients. Encouragingly, 8 of 10 have responded to FrC by increasing antibody levels or by an increased T cell response. Although these are memory responses caused by previous exposure to tetanus toxoid, it at least shows that the vaccines are presenting antigen to the immune system. The 2 of 10 who failed to respond had initial splenic involvement with tumor and may have had residual disease. These results confirm the initial observation that vaccination will operate best when tumor burden is very low or absent (42). T cell responses to Id were seen in five of seven of those evaluated among the eight responders to toxoid. All five patients remain in complete remission or with very low volume of local disease at 1–2 years from the start of vaccination.
A similar DNA scFv-FrC fusion gene has also been tested against another B cell malignancy, multiple myeloma (34). This tumor does not express surface Ig but secretes Ig, leading to a serum paraprotein. There is evidence from a mouse plasmacytoma that CD4+ T cells specific for Id determinants can suppress growth of MHC class II-negative myeloma cells by an indirect mechanism (53). In the 5T33 myeloma model, we were able to induce Id-specific protection against tumor by using a DNA scFv5T33-FrC vaccine, with protection mediated by T cells (34). Again, induction of effective immunity entirely depended on fusion of the FrC sequence. Clinically, myeloma is a disease with a poor prognosis. Patients are often treated with high-dose chemotherapy accompanied by a stem cell transplant, which offers the opportunity to vaccinate patients posttransplant. Thus far we have vaccinated one patient and obtained significant immune responses against both FrC and Id Ig. Myeloma is an easier setting for testing responses to individual vaccines than lymphoma, because the serum paraprotein is available for purification and testing of immunity.
Although all the clinical results with DNA scFv-FrC vaccines are preliminary, they provide a platform on which we can build. The required dose and number of injections required remain unknown, and, perhaps not surprisingly with individual vaccines, the dose escalation did not show any trend. No toxicity was evident at any dose apart from a mild fever in some cases, and no significant anti-DNA antibodies have been detected. The kinetics of response are interesting, with proliferative responses often developing at 8–12 weeks after the first injection. Evaluation of significant responses against new vaccines faces some difficulty, especially for cancers with a relatively indolent course. Although many objective tests of immune activation are now available, including measurement of cytokine-producing cells ex vivo, little or no basis of comparison with responses of human subjects to conventional vaccines exists.
DNA Fusion Vaccines to Induce CTL
The majority of candidate tumor antigens are intracellular and expressed only as peptides in the groove of the MHC class I molecule. Only CD8+ T cells can recognize these peptides and, after recognition, develop into effector CTLs capable of killing the target cells. Clearly, a strategy for inducing CTLs is desirable, and peptide vaccines have been used for this purpose with mixed results (54). DNA vaccines are particularly suitable for inducing CTL responses, because the encoded protein enters the MHC class I processing pathway through either direct transfection of APCs or cross-presentation. Minigenes encoding peptide alone have been tested, but, although some CTL activity can be generated, memory responses are poor. The importance of T cell help in priming, but especially in establishing memory, has been described in ref. 30.
It might be concluded that the fusion of FrC to a sequence containing candidate peptides derived from tumor antigens would be ideal for inducing CTLs. However, there are other immunological principles that need to be taken into account. The first is the phenomenon of immunodominance, which is a natural focusing of CTL responses onto a limited number of peptides in a large antigen. Staining CTLs with tetramers containing specific MHC-associated peptides is providing insight into the repertoire of T cells induced in normal human subjects during viral infections (55). It has become clear that, after infection with cytomegalovirus (CMV) or Epstein–Barr virus, a large proportion of CTLs is directed against a very few peptide epitopes (56). Such focusing might seem dangerous, because viruses can mutate sequences and thereby escape CTL attack. However, if they do, it is likely that another set of CTLs will be activated. Perhaps the focusing of the powerful CTL response, which is prone to cross-reactivity, is essential to avoid attack on normal tissue. With this in mind, we needed to be careful that a relatively large (50 kDa in the case of FrC) immunoenhancing sequence did not contribute epitopes, which can compete with the relatively weak tumor-derived epitopes. The goal therefore was to focus the CTL response only onto the tumor-derived sequences but at the same time induce and maintain high levels of Th cells able to amplify this response. To achieve this, we made two changes in the design: first, we minimized the FrC sequence to avoid the generation of competitive epitopes; and second, we modified the tumor-derived sequence such that a precise single immunodominant epitope would be presented optimally.
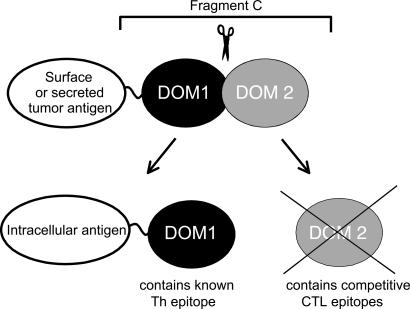
Engineering DNA Fusion Vaccine Design for Maximizing Epitope-Specific Immunity. To engineer the FrC immunoenhancer, we examined the available crystal structure (57). FrC consists of two domains separated by a linking peptide, and the N-terminal domain (DOM1) contains the known “promiscuous” MHC class II-binding peptide, p30, likely to be important in inducing the activating Th cells. The second domain (DOM2) contains several peptide sequences predicted to bind to murine MHC class I from two strains and also to the major human MHC class I haplotype HLA-A2 (26). We have evidence that three of the detected epitopes induce CTLs and that they can compete with fused tumor epitopes (35, 58). We therefore removed DOM2 and retained DOM1 to provide Th support for inducing immunity against a fused tumor antigen (Fig. 3).
Fig. 3.
Minimized DNA fusion vaccines to induce tumor-specific CTLs. Incorporating full-length FrC (Upper) as an immunoenhancing sequence can induce antibody and CD4+ T cell responses against fused tumor antigens. To induce CTLs, competitive peptides located in DOM2 and able to bind to human and mouse MHC class I molecules have been removed (Lower), leaving the Th-inducing sequences in DOM1. This minimized DOM1 sequence can enhance CD8+ responses against intracellular tumor-derived peptides.
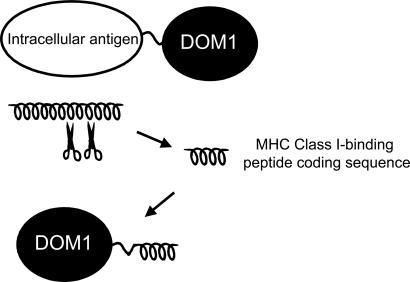
Knowledge of antigen processing was then mined for additional information with potential value for vaccine design. The ability of the cellular machinery to dissect out candidate tumor epitopes from the full backbone sequence of the encoded protein presents an obvious limiting factor for presentation and induction of response. To assist this process we opted to take the peptide sequence and place it in a position in which processing was facilitated. This general strategy would then be applicable to the increasing number of candidate peptides expressed by tumor cells being identified by predictive MHC class I-binding algorithms and binding assays (59). We built on the observation that fixing the C terminus of a peptide directed to the ER facilitates subsequent binding to the MHC class I. The reason for this facilitation seems to be that trimming of the N-terminal amino acids is efficient in that site, liberating C-terminal peptides for efficient binding (60). We therefore fused the sequence encoding the candidate epitope directly to the single domain (DOM) of FrC to encode a DOM-epitope fusion protein with a free C terminus (Fig. 4).
Fig. 4.
Epitope-specific DNA fusion vaccines. To enhance antigen presentation, candidate peptide-encoding sequences have been dissected out from the full sequence encoding an intracellular tumor antigen. Repositioning of the short epitope sequence to the 3′ end of the immunoenhancing DOM1 of FrC facilitates processing and presentation in the ER and increases induction of CD8+ T cells.
Performance of this engineered epitope-specific DNA fusion vaccine has been tested in the carcinoma model, CT26, which expresses a known immunodominant epitope, AH1 (61). Not only were high levels of AH1-specific CTLs induced, but these cells, specific only for a single epitope, were able to kill target tumor cells and completely protect against tumor growth (35). Importantly, injection of tumor cells expanded the CTLs, indicating no requirement for a continuing Th stimulus through FrC for the established response. Subsequently, we found that this design can induce protection against a range of tumors in preclinical models and can induce immunity in a therapeutic setting (unpublished data). An additional hurdle for tumor vaccines is to activate an immune repertoire that may have been subject to tolerogenic pressure, leading to anergy or induction of Treg cells. To assess whether CD8+ T cell responses can be induced from a profoundly tolerized repertoire, we investigated the male-specific transplantation antigens Uty and Smcy (62). The DNA DOM-Uty or DOM-Smcy vaccines could induce high levels of CTLs in females, as expected, but they were capable also of inducing CTLs in tolerant male mice (unpublished data). It therefore seems that the fusion gene design can activate a repertoire under intense tolerogenic pressure.
It could be argued that it is preferable to induce immunity against several tumor-derived epitopes at the same time, particularly to prevent emergence of epitope-negative tumor variants. The “string-of-beads” approach of injecting multiple epitopes together was used with this in mind. However, unless the epitopes are of comparable efficiency in inducing CTLs, it is difficult to determine how this approach will avoid the problem of immunodominance. Evidence from previous investigations of the polyepitope approach is now available showing that immunodominance is particularly important at the boosting stage and leads to suppression of initially primed responses (63). One solution to the problem might be to inject multiple separate DNA vaccines encoding only single or limited epitopes, which has shown promise in the expression library vaccines being explored for infectious diseases (64).
To move toward clinical application, we used transgenic mice expressing the human MHC class I HLA-A2 molecule. Initially, we tested DNA DOM-epitope vaccines by using known immunodominant epitopes from two viruses, CMV and influenza virus. In both cases, high levels of HLA-A2-restricted CTLs have been induced. For CMV, we showed that the CTLs can kill CMV-infected target cells, an important feature before clinical application (unpublished data). There is a significant clinical problem with CMV infection in cancer patients who are being treated with high-dose chemotherapy and allogeneic transplantation, especially if the donor is CMV-seronegative and the recipient is CMV-seropositive; this has provided a setting in which it is ethically acceptable to vaccinate the normal donors before transplantation. For cancer, it is imperative not only to demonstrate that the epitope-specific vaccines can induce high levels of CTLs but also that the epitopes are processed and presented by tumor cells.
Reasons for the Importance of Stimulating T Cell Help. The results in the preclinical models have indicated the critical importance of T cell help, and the principles established are being incorporated into vaccines for patients. It is likely to be even more important in the clinic, because antitumor CD4+ T cells will be limited by the poor availability of epitopes and by tolerogenic pressure. Engaging a new repertoire of Th cells will overcome this problem, and the successful induction of immunity against the male H-Y antigens in male mice supports this conclusion. For both lymphoma and myeloma, splitting the genes encoding tumor scFv antigen from that encoding FrC into two separate constructs led to a complete loss of response to Id determinants with retention of the response to FrC (34, 45), which demonstrates that the Th epitopes must be delivered to the same APC as the tumor antigen. The mechanism of priming is likely to involve “licensing” of the APC, presumed to be a dendritic cell, after specific recognition by the FrC-specific Th cells. APCs and T cells become reciprocally activated through the costimulatory molecules B7-1/B7-2, which bind to CD28. A further range of pairs of interacting molecules expressed by APCs and T cells, respectively, are then expressed, including CD40/CD40L and OX-40L/OX40 (31). Maintenance of APC function by these interactions is likely to be important for presentation of the accompanying weak tumor antigens delivered by the DNA fusion vaccine (65). There are known promiscuous epitopes in FrC able to bind to a wide range of murine and human MHC class II molecules (66). However, when we replaced the full-length FrC sequence in the vaccine with a single Th epitope, there was a large reduction in efficacy (58), which points to multiple elements within FrC that are required for inducing the high levels of T cell help.
Induction of CD4+ T cells specific for FrC, or for alternative immunoenhancing sequences, seems necessary for priming antibody, CD4+, and CD8+ responses against tumor antigens. Preliminary evidence indicates that fusion of the DOM sequence to the epitope-encoding sequence induces higher CD8+ responses than separate vectors, suggesting again that delivery to the same APC is desirable. The question of whether the repeated stimulation of immunoenhancing Th cells is required at the stage of boosting is not settled yet. Preliminary results indicate that it is mandatory for antibody responses but not for CD8+ responses. The latter is important, because tumor cells must be capable of stimulating a continuing immune response. Ideally, epitope presentation by tumor cells will lead to both the death of the cell and boosting of a continuing immune response.
Clinical Translation of DNA Fusion Gene Vaccination
The common problem of translating results in preclinical models to human subjects is particularly acute for DNA vaccines. One reason for this is that certain critical limitations such as dose and volume of DNA injected are difficult to meet. For example, it is known that if the optimal volume of 50 μl used for intramuscular injection in mice is reduced to 5 μl, the response is reduced dramatically (67). It is clearly impossible to increase the 50 μl appropriately on a weight basis in human subjects, and it is likely that our volume of 1 ml is not ideal. There are two simple physical strategies that could overcome this limitation: electroporation and injection of DNA attached to microparticles. Both have been described in detail, and it is clear that they amplify response to naked DNA dramatically (67–69). The mechanism is unclear but in both cases likely to include increased transfection rates together with increased inflammatory responses, possibly including attraction of APCs to the injection site. Both electroporation and microparticle injection are in current clinical use for different applications, making their addition to DNA vaccination feasible.
A popular approach, especially in the field of infectious diseases, is the so-called prime/boost vaccination strategy (70). It seems that priming with naked DNA, followed by boosting with the same antigen delivered through a viral vector, is particularly effective in amplifying immune responses. Nonreplicating pox viruses or adenoviruses are preferred vectors, and modified vaccinia Ankara is particularly attractive as a vehicle, because a number of host range and virulence genes have been deleted by extensive passaging in vitro (71). The principle has been tested in preclinical models and in human subjects by using vaccines against HIV and malaria (72, 73). For malaria, assessment of the efficacy of a range of heterologous prime/boost approaches has been made in malaria-naive subjects and in semiimmune individuals in The Gambia. The opportunity to challenge volunteer vaccinees with sporozoites allows rapid evaluation of the many vector combinations, and encouraging data are emerging already (73). Although these exciting results have clear relevance for cancer, there is the concern that it could be difficult to use live viral delivery systems in immunosuppressed patients. There is also the problem that the ensuing immunity against components of the viral vector will suppress the additional boosts likely to be needed to control emergent cancer cells. Similar considerations apply to the use of boosting with bacterial vectors, although the attraction of this approach for tumors in mucosal sites makes investigation highly desirable (74).
Conclusions
Therapeutic vaccination against cancer faces many difficulties, some of which are shared with vaccination against persistent infection. However, the tools of genomics provide ways to circumvent many of the problems. DNA vaccines can deliver tumor antigens to specific processing pathways, and simple manipulation can add activating molecules to ensure the high level of immunity undoubtedly required to suppress cancer growth. There are many candidate target antigens, but most are weakly immunogenic and need repackaging in an immunogenic form.
We focused on a gene fusion strategy aimed at inducing T cell help for antitumor immunity from a nontolerized antimicrobial repertoire. Modified designs are needed to activate antibody, CD4+ T cells, or CD8+ T cells. It is likely that multiple vaccines will be required to focus attack on different antigenic targets, which should ensure that tumor cells cannot escape by deleting target molecules. It is now feasible to use precise DNA vaccines to suppress the most vicious murine tumors, even in a therapeutic setting in which there is little time to activate immunity. Activation of immunity from a profoundly tolerized repertoire is also feasible, but for clinical application it is clearly desirable to reduce tumor load to a minimum. Vaccination should be seen at present as a means to remove residual disease and continuously survey the body for emergent tumor cells.
Strategies to increase antigen expression, particularly at the point of boosting, should be part of future studies. Safety seems to be assured, although antigens that are also expressed by normal cells will have to be investigated carefully before reaching the clinic. Transgenic mice expressing human antigens can be used to address some of these issues. After a hesitant start, active vaccination may soon follow passive immunotherapy into clinical practice.
Acknowledgments
These studies were supported by the Leukaemia Research Fund (United Kingdom), Tenovus, Cancer Research UK, and the Multiple Myeloma Research Foundation (U.S.).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Therapeutic Vaccines: Realities of Today and Hopes for Tomorrow,” held April 1–3, 2004, at the National Academy of Sciences in Washington, DC.
Abbreviations: Th, T helper; APC, antigen-presenting cell; ER, endoplasmic reticulum; CTL, cytotoxic T lymphocyte; Treg, T cell acting as a regulatory cell; Id, idiotypic; scFv, single-chain Fv; FrC, fragment C; CMV, cytomegalovirus; DOM, single domain; DOM1, N-terminal domain; DOM2, second domain.
References
- 1.Krause, R. M. (1999) Vaccine 17, Suppl. 3, S64-S67. [DOI] [PubMed] [Google Scholar]
- 2.Maloney, D. G., Liles, T. M., Czerwinski, D. K., Waldichuk, C., Rosenberg, J., Grillo-Lopez, A. & Levy, R. (1994) Blood 84, 2457-2466. [PubMed] [Google Scholar]
- 3.Riddell, S. R. & Greenberg, P. D. (2000) J. Antimicrob. Chemother. 45, Suppl. T3, 35-43. [DOI] [PubMed] [Google Scholar]
- 4.Rooney, C. M., Aguilar, L. K., Huls, M. H., Brenner, M. K. & Heslop, H. E. (2001) Curr. Top. Microbiol. Immunol. 258, 221-229. [DOI] [PubMed] [Google Scholar]
- 5.Goulmy, E. (1997) Immunol. Rev. 157, 125-140. [DOI] [PubMed] [Google Scholar]
- 6.Nanda, N. K. & Sercarz, E. E. (1995) Cell 82, 13-17. [DOI] [PubMed] [Google Scholar]
- 7.Stevenson, F. K., Rice, J. & Zhu, D. (2004) Adv. Immunol. 82, 49-103. [DOI] [PubMed] [Google Scholar]
- 8.Krieg, A. M. (2002) Annu. Rev. Immunol. 20, 709-760. [DOI] [PubMed] [Google Scholar]
- 9.Wagner, H. (1999) Adv. Immunol. 73, 329-368. [DOI] [PubMed] [Google Scholar]
- 10.Heit, A., Maurer, T., Hochrein, H., Bauer, S., Huster, K. M., Busch, D. H. & Wagner, H. (2003) J. Immunol. 170, 2802-2805. [DOI] [PubMed] [Google Scholar]
- 11.Corr, M., Lee, D. J., Carson, D. A. & Tighe, H. (1996) J. Exp. Med. 184, 1555-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu, T. M., Ulmer, J. B., Caulfield, M. J., Deck, R. R., Friedman, A., Wang, S., Liu, X., Donnelly, J. J. & Liu, M. A. (1997) Mol. Med. 3, 362-371. [PMC free article] [PubMed] [Google Scholar]
- 13.Porgador, A., Irvine, K. R., Iwasaki, A., Barber, B. H., Restifo, N. P. & Germain, R. N. (1998) J. Exp. Med. 188, 1075-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houde, M., Bertholet, S., Gagnon, E., Brunet, S., Goyette, G., Laplante, A., Princiotta, M. F., Thibault, P., Sacks, D. & Desjardins, M. (2003) Nature 425, 402-406. [DOI] [PubMed] [Google Scholar]
- 15.Guermonprez, P., Saveanu, L., Kleijmeer, M., Davoust, J., Van Endert, P. & Amigorena, S. (2003) Nature 425, 397-402. [DOI] [PubMed] [Google Scholar]
- 16.Gurunathan, S., Klinman, D. M. & Seder, R. A. (2000) Annu. Rev. Immunol. 18, 927-974. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg, S. A. (1998) Cancer J. Sci. Am. 4, Suppl. 1, S1-S4. [PubMed] [Google Scholar]
- 18.Preuss, K. D., Zwick, C., Bormann, C., Neumann, F. & Pfreundschuh, M. (2002) Immunol. Rev. 188, 43-50. [DOI] [PubMed] [Google Scholar]
- 19.Darnell, R. B. & Posner, J. B. (2003) N. Engl. J. Med. 349, 1543-1554. [DOI] [PubMed] [Google Scholar]
- 20.Zhu, D. & Stevenson, F. K. (2002) Curr. Opin. Mol. Ther. 4, 41-48. [PubMed] [Google Scholar]
- 21.Seya, T., Akazawa, T., Uehori, J., Matsumoto, M., Azuma, I. & Toyoshima, K. (2003) Anticancer Res. 23, 4369-4376. [PubMed] [Google Scholar]
- 22.Srivastava, P. (2002) Nat. Rev. Immunol. 2, 185-194. [DOI] [PubMed] [Google Scholar]
- 23.Ulmer, J. B., DeWitt, C. M., Chastain, M., Friedman, A., Donnelly, J. J., McClements, W. L., Caulfield, M. J., Bohannon, K. E., Volkin, D. B. & Evans, R. K. (1999) Vaccine 18, 18-28. [DOI] [PubMed] [Google Scholar]
- 24.Kim, J. J., Yang, J. S., Dang, K., Manson, K. H. & Weiner, D. B. (2001) Clin. Cancer Res. 7, Suppl. 3, S882-S889. [PubMed] [Google Scholar]
- 25.Chen, T. T. & Levy, R. (1995) J. Immunol. 154, 3105-3117. [PubMed] [Google Scholar]
- 26.Rice, J., King, C. A., Spellerberg, M. B., Fairweather, N. & Stevenson, F. K. (1999) Vaccine 17, 3030-3038. [DOI] [PubMed] [Google Scholar]
- 27.Bacik, I., Snyder, H. L., Anton, L. C., Russ, G., Chen, W., Bennink, J. R., Urge, L., Otvos, L., Dudkowska, B., Eisenlohr, L. & Yewdell, J. W. (1997) J. Exp. Med. 186, 479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez, F. & Whitton, J. L. (2000) Virology 268, 233-238. [DOI] [PubMed] [Google Scholar]
- 29.Rowell, J. F., Ruff, A. L., Guarnieri, F. G., Staveley-O'Carroll, K., Lin, X., Tang, J., August, J. T. & Siliciano, R. F. (1995) J. Immunol. 155, 1818-1828. [PubMed] [Google Scholar]
- 30.Janssen, E. M., Lemmens, E. E., Wolfe, T., Christen, U., von Herrath, M. G. & Schoenberger, S. P. (2003) Nature 421, 852-856. [DOI] [PubMed] [Google Scholar]
- 31.Gerloni, M., Xiong, S., Mukerjee, S., Schoenberger, S. P., Croft, M. & Zanetti, M. (2000) Proc. Natl. Acad. Sci. USA 97, 13269-13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi, T. & Sakaguchi, S. (2003) Curr. Mol. Med. 3, 693-706. [DOI] [PubMed] [Google Scholar]
- 33.Golgher, D., Jones, E., Powrie, F., Elliott, T. & Gallimore, A. (2002) Eur. J. Immunol. 32, 3267-3275. [DOI] [PubMed] [Google Scholar]
- 34.King, C. A., Spellerberg, M. B., Zhu, D., Rice, J., Sahota, S. S., Thompsett, A. R., Hamblin, T. J., Radl, J. & Stevenson, F. K. (1998) Nat. Med. 4, 1281-1286. [DOI] [PubMed] [Google Scholar]
- 35.Rice, J., Buchan, S. & Stevenson, F. K. (2002) J. Immunol. 169, 3908-3913. [DOI] [PubMed] [Google Scholar]
- 36.George, A. J. & Stevenson, F. K. (1989) Int. Rev. Immunol. 4, 271-310. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, A., George, A. J., King, C. A. & Stevenson, F. K. (1990) J. Immunol. 145, 3937-3943. [PubMed] [Google Scholar]
- 38.Cao, W., Myers-Powell, B. A. & Braciale, T. J. (1994) J. Exp. Med. 179, 195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamblin, T. J., Abdul-Ahad, A. K., Gordon, J., Stevenson, F. K. & Stevenson, G. T. (1980) Br. J. Cancer 42, 495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, R. A., Maloney, D. G., Warnke, R. & Levy, R. (1982) N. Engl. J. Med. 306, 517-522. [DOI] [PubMed] [Google Scholar]
- 41.Stevenson, F. K., George, A. J. & Glennie, M. J. (1990) Chem. Immunol. 48, 126-166. [PubMed] [Google Scholar]
- 42.Hsu, F. J., Caspar, C. B., Czerwinski, D., Kwak, L. W., Liles, T. M., Syrengelas, A., Taidi-Laskowski, B. & Levy, R. (1997) Blood 89, 3129-3135. [PubMed] [Google Scholar]
- 43.Hawkins, R. E., Zhu, D., Ovecka, M., Winter, G., Hamblin, T. J., Long, A. & Stevenson, F. K. (1994) Blood 83, 3279-3288. [PubMed] [Google Scholar]
- 44.Anderson, R., Gao, X. M., Papakonstantinopoulou, A., Fairweather, N., Roberts, M. & Dougan, G. (1997) Vaccine 15, 827-829. [DOI] [PubMed] [Google Scholar]
- 45.Spellerberg, M. B., Zhu, D., Thompsett, A., King, C. A., Hamblin, T. J. & Stevenson, F. K. (1997) J. Immunol. 159, 1885-1892. [PubMed] [Google Scholar]
- 46.Padua, R. A., Larghero, J., Robin, M., le Pogam, C., Schlageter, M. H., Muszlak, S., Fric, J., West, R., Rousselot, P., Phan, T. H., et al. (2003) Nat. Med. 9, 1413-1417. [DOI] [PubMed] [Google Scholar]
- 47.Zhu, D., Rice, J., Savelyeva, N. & Stevenson, F. K. (2001) Trends Mol. Med. 7, 566-572. [DOI] [PubMed] [Google Scholar]
- 48.Ruffini, P. A., Neelapu, S. S., Kwak, L. W. & Biragyn, A. (2002) Haematologica 87, 989-1001. [PubMed] [Google Scholar]
- 49.Syrengelas, A. D., Chen, T. T. & Levy, R. (1996) Nat. Med. 2, 1038-1041. [DOI] [PubMed] [Google Scholar]
- 50.Timmerman, J. M., Singh, G., Hermanson, G., Hobart, P., Czerwinski, D. K., Taidi, B., Rajapaksa, R., Caspar, C. B., Van Beckhoven, A. & Levy, R. (2002) Cancer Res. 62, 5845-5852. [PubMed] [Google Scholar]
- 51.Forconi, F., King, C. A., Sahota, S. S., Kennaway, C. K., Russell, N. H. & Stevenson, F. K. (2002) Immunology 107, 39-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Savelyeva, N., Munday, R., Spellerberg, M. B., Lomonossoff, G. P. & Stevenson, F. K. (2001) Nat. Biotechnol. 19, 760-764. [DOI] [PubMed] [Google Scholar]
- 53.Lauritzsen, G. F. & Bogen, B. (1993) Cell Immunol. 148, 177-188. [DOI] [PubMed] [Google Scholar]
- 54.Parmiani, G., Castelli, C., Dalerba, P., Mortarini, R., Rivoltini, L., Marincola, F. M. & Anichini, A. (2002) J. Natl. Cancer Inst. 94, 805-818. [DOI] [PubMed] [Google Scholar]
- 55.Xu, X. N. & Screaton, G. R. (2002) J. Immunol. Methods 268, 21-28. [DOI] [PubMed] [Google Scholar]
- 56.Rickinson, A. B. & Moss, D. J. (1997) Annu. Rev. Immunol. 15, 405-431. [DOI] [PubMed] [Google Scholar]
- 57.Umland, T. C., Wingert, L. M., Swaminathan, S., Furey, W. F., Schmidt, J. J. & Sax, M. (1997) Nat. Struct. Biol. 4, 788-792. [DOI] [PubMed] [Google Scholar]
- 58.Rice, J., Elliott, T., Buchan, S. & Stevenson, F. K. (2001) J. Immunol. 167, 1558-1565. [DOI] [PubMed] [Google Scholar]
- 59.Rammensee, H. G., Weinschenk, T., Gouttefangeas, C. & Stevanovic, S. (2002) Immunol. Rev. 188, 164-176. [DOI] [PubMed] [Google Scholar]
- 60.Snyder, H. L., Bacik, I., Yewdell, J. W., Behrens, T. W. & Bennink, J. R. (1998) Eur. J. Immunol. 28, 1339-1346. [DOI] [PubMed] [Google Scholar]
- 61.Huang, A. Y., Gulden, P. H., Woods, A. S., Thomas, M. C., Tong, C. D., Wang, W., Engelhard, V. H., Pasternack, G., Cotter, R., Hunt, D., et al. (1996) Proc. Natl. Acad. Sci. USA 93, 9730-9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Simpson, E., Scott, D. & Chandler, P. (1997) Annu. Rev. Immunol. 15, 39-61. [DOI] [PubMed] [Google Scholar]
- 63.Palmowski, M. J., Choi, E. M., Hermans, I. F., Gilbert, S. C., Chen, J. L., Gileadi, U., Salio, M., Van Pel, A., Man, S., Bonin, E., et al. (2002) J. Immunol. 168, 4391-4398. [DOI] [PubMed] [Google Scholar]
- 64.Singh, R. A., Wu, L. & Barry, M. A. (2002) J. Immunol. 168, 379-391. [DOI] [PubMed] [Google Scholar]
- 65.Stevenson, F. K., Rice, J., Ottensmeier, C. H., Thirdborough, S. M. & Zhu, D. (2004) Immunol. Rev. 199, 156-180. [DOI] [PubMed] [Google Scholar]
- 66.Panina-Bordignon, P., Tan, A., Termijtelen, A., Demotz, S., Corradin, G. & Lanzavecchia, A. (1989) Eur. J. Immunol. 19, 2237-2242. [DOI] [PubMed] [Google Scholar]
- 67.Dupuis, M., Denis-Mize, K., Woo, C., Goldbeck, C., Selby, M. J., Chen, M., Otten, G. R., Ulmer, J. B., Donnelly, J. J., Ott, G., et al. (2000) J. Immunol. 165, 2850-2858. [DOI] [PubMed] [Google Scholar]
- 68.O'Hagan, D., Singh, M., Ugozzoli, M., Wild, C., Barnett, S., Chen, M., Schaefer, M., Doe, B., Otten, G. R. & Ulmer, J. B. (2001) J. Virol. 75, 9037-9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mathiesen, I. (1999) Gene Ther. 6, 508-514. [DOI] [PubMed] [Google Scholar]
- 70.Ramshaw, I. A. & Ramsay, A. J. (2000) Immunol. Today 21, 163-165. [DOI] [PubMed] [Google Scholar]
- 71.Blanchard, T. J., Alcami, A., Andrea, P. & Smith, G. L. (1998) J. Gen. Virol. 79, 1159-1167. [DOI] [PubMed] [Google Scholar]
- 72.Hanke, T., McMichael, A. J., Mwau, M., Wee, E. G., Ceberej, I., Patel, S., Sutton, J., Tomlinson, M. & Samuel, R. V. (2002) Vaccine 20, 1995-1998. [DOI] [PubMed] [Google Scholar]
- 73.McConkey, S. J., Reece, W. H., Moorthy, V. S., Webster, D., Dunachie, S., Butcher, G., Vuola, J. M., Blanchard, T. J., Gothard, P., Watkins, K., et al. (2003) Nat. Med. 9, 729-735. [DOI] [PubMed] [Google Scholar]
- 74.Niethammer, A. G., Primus, F. J., Xiang, R., Dolman, C. S., Ruehlmann, J. M., Ba, Y., Gillies, S. D. & Reisfeld, R. A. (2001) Vaccine 20, 421-429. [DOI] [PubMed] [Google Scholar]