Abstract
The cancer-germline gene MAGE-3 codes for tumor-specific antigens recognized on many tumors by T lymphocytes. A MAGE-3 antigen presented by HLA-A1 has been used in several vaccination trials on metastatic melanoma patients. Only a small minority of patients have shown evidence of tumor regression. Attempts to correlate the tumor rejections with the cytotoxic T lymphocyte (CTL) response against the vaccine have been hampered by the low level of these responses. In noncancerous individuals, the frequency of the T cell precursors against antigen MAGE-3.A1 is ≈4 × 10-7 CD8 T cells. The diversity of the T cell receptor repertoire of these anti-MAGE-3.A1 precursors was analyzed in one individual. The results indicate that it is very likely that the repertoire comprises >100 clonotypes. On this basis, it is possible to use not only the frequency of CTL precursors in the blood but also the presence of dominant clonotypes to ascertain in patients the existence of anti-MAGE-3.A1 responses as low as 10-6 of CD8. With this approach, we observed a correlation between tumor regression and anti-MAGE-3.A1 CTL responses in patients vaccinated with a recombinant virus encoding the antigen and also in patients vaccinated with peptide-pulsed dendritic cells. In contrast, for patients showing tumor regression after vaccination with peptide alone, CTL responses were almost never observed. It is possible that even those CTL responses that are below our present detection level can trigger a sequence of events that leads to tumor regression.
Shared tumor-specific antigens encoded by cancer-germline genes such as those of the MAGE family have been used for therapeutic vaccination of cancer patients. A number of small clinical trials on metastatic melanoma patients have been performed with the MAGE-3168-176 antigen EVDPIGHLY, which is presented by HLA-A1 (1, 2). Evidence of tumor regression was observed in ≈20% of the patients, but clinical benefit was limited to ≈10% of the patients.
Our initial work suggested that in most vaccinated patients, even in those who displayed tumor regression, it was difficult to ascertain the existence of an antivaccine T cell response. We nevertheless felt that it was crucial to know whether low-level responses had occurred and whether such cytotoxic T lymphocyte (CTL) responses showed a correlation with tumor regression to understand why most patients failed to show any evidence of regression. We therefore developed a sensitive approach based on in vitro restimulation of blood lymphocytes with the antigenic peptide over 2 weeks, followed by labeling with tetramers. To evaluate precursor frequencies, these mixed lymphocyte–peptide cultures were conducted under limiting dilution conditions. Cells that were labeled with the tetramer were cloned, the lytic specificity of the clones was verified, and their diversity was analyzed by T cell receptor (TCR) sequencing (3). As the interpretation of these analyses was based on both the frequency and the diversity of the anti-MAGE-3.A1 CTL clones, it was necessary to evaluate these parameters in noncancerous individuals.
We describe here the frequency of anti-MAGE-3.A1 CTL precursors and the diversity of their TCR repertoire in such an individual. We also present an overview of the anti-MAGE-3.A1 CTL response of metastatic melanoma patients who were vaccinated with peptides, with a recombinant canarypox ALVAC virus carrying sequences encoding the MAGE-3.A1 antigenic peptide, or with dendritic cells pulsed with the peptide.
Materials and Methods
Cell Lines, Media, and Reagents. The Epstein–Barr-virus-transformed B lymphocyte (EBV-B) cell lines and the tumor cell lines were cultured in Iscove's modified Dulbecco's medium (Life Technologies, Rockville, MD) supplemented with 10% FCS (Life Technologies). All of the media were supplemented with 0.24 mM l-asparagine, 0.55 mM l-arginine, 1.5 mM l-glutamine [alkyladenine DNA glycosylase (AAG)], 100 units/ml penicillin, and 100 μg/ml streptomycin. Human recombinant IL-2 was purchased from Eurocetus (Amsterdam), IL-7 was purchased from PeproTech (Rocky Hill, NJ), and phytohemagglutinin (HA16) was purchased from Murex Diagnostics (Dartford, U.K.). IL-4 was produced in our laboratory.
Isolation of T Cells. Peripheral blood was obtained from hemochromatosis (C282Y mutation) patient LB1965 as standard buffy coat preparations, which were laid down on a 15-ml Lymphoprep layer (Nycomed, Oslo) in 50-ml tubes. To minimize contamination of the peripheral blood mononuclear cells (PBMC) by platelets, the tubes were first centrifuged at 190 × g for 20 min at room temperature. After removal of the top 20–25 ml, which contained most of the platelets, the tubes were centrifuged at 430 × g for 20 min at room temperature. The interphase containing the PBMC was harvested and washed three times (or more) in cold phosphate buffer solution with 2 mM EDTA to eliminate the remaining platelets.
Experiments I and II (Fig. 1 and Table 1) were performed with 1.3 × 108 and 1 × 108 rosetted T cells, respectively. Experiment III was performed with 107 PBMC that were partially depleted from adherent cells, CD4, and B cells, and experiment IV was performed with 107 purified CD8 T cells.
Fig. 1.
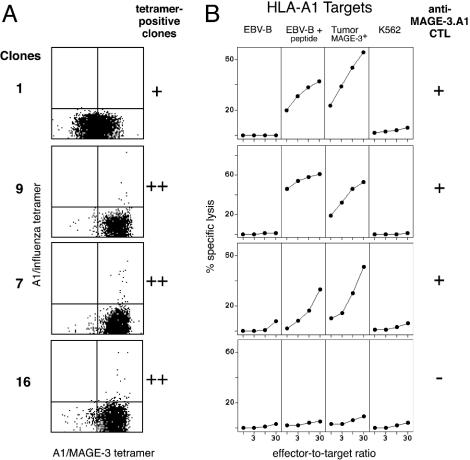
Overview of the procedure to obtain anti-MAGE-3.A1 CTL clones in experiment I. The cells represented in the FACS panel are CD8 T cells. The control tetramer is an HLA-A1 tetramer containing an influenza peptide.
Table 1. Frequency of anti-MAGE-3.A1 CTL precursors in the blood of an individual without cancer.
| Experiment | Total no. of CD8 cells recovered from MACS (A)* | No. of CD8 cells selected by MACS (B)† | Fraction of CD8 cells selected by FACS (C)† | No. of selected cells recovered after FACS (D) | No. of tetramer-positive clones | No. of lytic anti-MAGE-3.A1 CTL clones (E) | Frequency of anti-MAGE-3.A1 CTL among the CD8 T cells‡ |
|---|---|---|---|---|---|---|---|
| I | 2.5 × 107 | 1.5 × 106 | 1 × 10-3 | 310 | 7 | 1 | 2.4 × 10-7 |
| II | 1.9 × 107 | 1.9 × 106 | 6 × 10-4 | 288 | 6 | 6 | 9.7 × 10-7 |
| III | No MACS for the 5.6 × 106 CD8 cells | 1 × 10-4 | 424 | 5 | 1 | 2.0 × 10-7 | |
| IV | No MACS for the 9.8 × 106 CD8 cells | 4 × 10-4 | 382 | 1 | 1 | 1.0 × 10-6 | |
Total recovered = CD8 cells in the positive fraction selected by tetramer-positive CD8 cells in the negative fraction
Selection: with tetramers
For experiments I and II, the frequency of anti-MAGE-3.A1 CTL precursors among CD8 T cells is calculated as follows: (E/D) × (C) × (B/A). For experiment I described in Fig. 1, frequency = (1/310) × (1 × 10-3) × (1.5 × 106/2.5 × 107) = 2.4 × 10-7. For experiments III and IV, which did not include MACS, the frequency of anti-MAGE-3.A1 CTL precursors among CD8 T cells is calculated as follows: (E/D) × (C). Weighed average = 6.0 × 10-7. The average of the frequencies was weighed according to the number of CD8 T cells engaged in each of the four experiments
T lymphocytes were obtained by rosetting with sheep erythrocytes (BioMerieux, Charbonnier les Bains, France) treated with 2-aminoethylisothiouronium (Sigma–Aldrich). Rosetted T cells were treated with NH4Cl (160 mM) to lyse the sheep erythrocytes and then were washed. CD8+ T lymphocytes were isolated from rosetted T cells by positive selection by using an anti-CD8 monoclonal antibody coupled to magnetic microbeads (Miltenyi Biotec, Auburn, CA). The lymphocytes were then sorted through a separation column inserted into a magnet in an AUTOMACS (Miltenyi Biotec) at 1 ml/min.
Tetramer Production and Labeling with Tetramers. Recombinant HLA-A1 molecules were folded in vitro with β2-microglobulin and peptide EVDPIGHLY from MAGE-3 or peptide VSDGGPNLY from the influenza basic polymerase 1. The HLA-A1 molecules were purified by gel filtration, biotinylated, and mixed as described in ref. 4 with Extravidin-PE (Sigma) for the HLA-A1/MAGE-3 tetramer or with streptavidin–allophycocyanin (Molecular Probes) for the influenza control tetramer. For staining, cells were washed, resuspended in PBS with 1% human serum, and incubated for 15 min at room temperature with both HLA-A1 tetramers (20 nM). Anti-CD8 antibodies coupled to FITC (SK1 at 1/50, BD Biosciences) were added, and after a further incubation for 15 min, cells were washed.
Magnetic Cell Sorting (MACS) and Fluorescence-Activated Cell Sorting (FACS). Tetramer-labeled cells (107 cells per 80 μl) were incubated with anti-phycoerythrin microbeads (20 μl) according to the instructions of the manufacturer (Miltenyi Biotec), washed, and sorted through a separation column inserted into a magnet in an AUTOMACS at 0.5 ml/min (Miltenyi Biotec). A FACS-Vantage cell sorter (BD Biosciences) was used for sorting at one cell per microwell, and a FACSCalibur (BD Biosciences) was used for analysis of the T cell clones with cellquest software (BD Biosciences).
Culture Conditions of Cells Sorted by Flow Cytometry. Sorted cells were cultured in microwells in 200 μl of Iscove's modified Dulbecco's medium supplemented with 10% human serum, IL-2 (50 units/ml), IL-4 (5 units/ml), and IL-7 (5 ng/ml). The cells were stimulated weekly by irradiated (100 Gray) allogenic MZ2-MEL.43 tumor cells (1 × 104 cells per, well), previously incubated for 1 h at 37°C with 1 μg/ml peptide and then washed, in the presence of phytohemagglutinin (125 ng/ml) and irradiated allogenic EBV-B cells (LG2-EBV) as feeder cells (3 × 104 cells per well). For experiment II, the stimulating cells were peptide-pulsed autologous dendritic cells instead of peptide-pulsed tumor cells. After 3–5 weeks of proliferation, the CTL clones were restimulated every 1–2 weeks in 2 ml of medium (5 × 105 CTLs per well) with 105 stimulating cells, 1.5 × 106 feeder cells, IL-2, and IL-4.
Cytotoxicity Assay and Production of Cytokines. To analyze the lytic activity of the tetramer-positive clones, target cells were labeled for 1 h with 100 μCi (1 Ci = 37 GBq) of Na(51Cr)O4, washed, and incubated with the effector cells at various effector-to-target ratios. If indicated, the targets were incubated for 5 min with peptide EVDPIGHLY before adding the CTL. Chromium release was measured after 4 h at 37°C. To analyze the production of cytokines on contact with the MAGE-3.A1 antigen, 5,000 cells of each clone were cocultured overnight with 20,000 HLA-A1 EBV-B cells loaded or not loaded with 1 μg/ml peptide and washed. The presence of cytokines in the supernatant was estimated with the Th1/Th2 Cytokine Cytometric Bead Array Kit (BD Biosciences).
TCR Analysis. For TCR analysis, RNA was extracted from each anti-MAGE-3.A1 CTL clone with the Tripure reagent (Roche Molecular Biochemicals) and converted to cDNA at 42°C for 90 min with 200 units of M-murine leukemia virus reverse transcriptase (Life Technologies). TCR Vα and Vβ usage was assessed by PCR amplification with a complete panel of Vα- or Vβ-specific sense primers and Cα and Cβ antisense primer, respectively (5). Primers were chosen on the basis of described panels of TCR V region oligonucleotides and with alignments of TCR sequences available at the International Immunogenetics Database (http://imgt.cines.fr). Each PCR product was purified and sequenced by using a Cα or Cβ antisense primer to obtain a complete identification of complementarity-determining region 3. Because we used EBV-B cells as feeder cells for the amplification of the CTL clones, TCR sequences were not contaminated by TCR expressed by the peripheral blood lymphocytes that are often used as feeder cells.
Probability Calculations. The probability of obtaining a series of n elements without repeat by drawing (with replacement) from a set of d different elements is
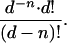 |
The probability of obtaining a series of n elements with one repeat by drawing (with replacement) from a set of d different elements is
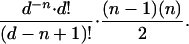 |
The upper estimate of the probability that a series of n elements drawn (with replacement) from a set of d elements (diversity) contains at least one element repeated at least k times is
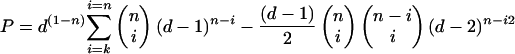 |
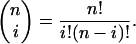 |
This formula was checked by computer simulation for n = 7 and d = 20.
Results
Frequency and Repertoire Diversity of the Naïve Anti-MAGE-3.A1 CTL Precursors. We reported previously an estimate of the frequency of anti-MAGE-3.A1 CTL precursors (CTLp) in the blood of individuals without cancer (6). The approach involved three weekly stimulations under limiting dilution condition of CD8 T cells with autologous phytohemagglutinin-activated T cells incubated with the MAGE-3.A1 peptide. The microcultures were then evaluated for their lytic activity against autologous EBV-B cells pulsed with the peptide. Three individuals were analyzed, leading to an average frequency estimate for anti-MAGE-3.A1 naïve CTLp of 2 × 10-7 of the CD8 T cells. This estimate rested on the small number of 12 positive microcultures.
We wished to estimate not only the frequency but also the T cell receptor diversity of the naïve anti-MAGE-3.A1 CTLp. We developed an approach aimed at obtaining a large number of CTL clones from very low-frequency precursors (Fig. 1). More than 3 billion PBMC were obtained from several blood samples collected from hemochromatosis patient LB1965. An HLA-A1/MAGE-3 tetramer was used to label large numbers of T cells, and the positive cells were enriched by MACS. The selected fraction was then passed through a flow cytometer to further enrich the A1/MAGE-3 tetramer-positive cells among the CD8 cells. A number of positive cells were distributed at one cell per well and were stimulated with irradiated HLA-A1 cells loaded with the MAGE-3 peptide. About 20% of the cloned T cells proliferated, but only a fraction of these cells proved to be tetramer-positive. Only a fraction of these tetramer-positive clones showed lytic activity on cells pulsed with the peptide. These CTL clones invariably lysed the melanoma cells also, indicating that their avidity was high enough to recognize the level of antigen expressed by tumor cells.
Representative tetramer-positive lytic and nonlytic clones are shown in Fig. 2. The tetramer-positive clones that were devoid of anti-MAGE-3.A1 lytic activity were tested for their specific production of IFN-γ, tumor necrosis factor α, IL-2, IL-4, IL-5, or IL-10 upon contact with the MAGE-3.A1 antigen, and all tests were negative. Similar lymphocytes, namely labeled by a given HLA/peptide tetramer but failing to recognize peptide-pulsed cells, are described in ref. 7. Presumably, the multimeric nature of the tetramers permits stable binding on a number of T lymphocytes that have too low an affinity for effective function. For the evaluation of the frequency of anti-MAGE-3.A1 CTLp, we took into account only the tetramer-positive lytic clones.
Fig. 2.
Tetramer analysis and anti-MAGE-3.A1 lytic activity of some tetramer-positive clones. (A) The clones were analyzed 7 days after the last stimulation after labeling with the two tetramers and anti-CD8 antibodies. (B) Lytic activity was tested on day 14 after the last stimulation. Targets were HLA-A1 EBV-B cells, MZ2-MEL.43 melanoma cells expressing HLA-A1 and MAGE-3, and NK target K562. When indicated, MAGE-3 peptide EVDPIGHLY was added at 0.5 μg/ml. CTLs were added at various effector-to-target ratios, and chromium release was measured after 4 h.
We obtained in four experiments a total of nine tetramer-positive clones that had an anti-MAGE-3.A1 lytic activity (Table 1). This led to an estimate of 6 × 10-7 of the CD8 T cells for the anti-MAGE-3.A1 CTLp in this individual, a result that is in good agreement with the frequency of 2 × 10-7 obtained previously by the limiting dilution stimulation approach. On the basis of all our results, we presently consider 4 × 10-7 to be the naïve frequency baseline for our vaccination studies.
To evaluate the diversity of the naïve anti-MAGE-3.A1 repertoire, we examined the TCR sequences of the lytic anti-MAGE-3.A1 CTL clones obtained in this individual. In addition to the 9 anti-MAGE-3.A1 CTL clones obtained in the four experiments mentioned above, we obtained 14 CTL clones in the course of experiments that were not designed for the evaluation of CTL precursor frequency. Among the total of 23 clones, only 2 had the same TCRα and β sequences (Table 2). Three other clones had identical TCRβ nucleotide sequences but different TCRα sequences. The identical TCRβ sequences contain a DRG motif in their complementarity-determining region 3, which is linked to a Jβ 2-7*01 segment. This combination has been observed on 17 anti-MAGE-3.A1 CTLs belonging to 10 different individuals (data not shown). Two other clones had the same TCRβ protein sequences but differed for their nucleotide sequences.
Table 2. Repertoire of anti MAGE-3.A1 CTL precursors in an individual without cancer.
| Clones | Vβ† | Complementarity-determining region 3 | Jβ† | Vα | ||
|---|---|---|---|---|---|---|
| 1 | V4-2*01 | CASSQ | KDRGG | YEQYFG | J2-7*01 | V12-1§ |
| 2 | V9*01 | CASSV | GDTS | EKLFFG | J1-4*01 | V21‡ |
| 3 | V3-1*01 | CASSQ | WGPEA | NTEAFFG | J1-1*01 | |
| 4 | V20-1*01 | CSAR | GTD | SNQPQHFG | J1-5*01 | |
| 5 | V9*01 | CASSV | AGAG | YEQYFG | J2-7*01 | ∥ |
| 6 | V4-2*01 | CASSQ | KDRGG | YEQYFG | J2-7*01 | V21§ |
| 7 | V28*01 | CAS | RGTGGYD | EQYFG | J2-7*01 | |
| 8 | V4-2*01 | CASSQ | KDRGG | YEQYFG | J2-7*01 | V8-6§¶ |
| V20-1*07 | CSAR | DLLS | GELFFG | J2-2*01 | ||
| 9 | V9*01 | CASSV | APSGGG | YEQYFG | J2-7*01 | |
| 10 | V9*01 | CASSV | AGAG | YEQYFG | J2-7*01∥ | |
| 11 | V4-3*04 | CAS | RDSG | SYNEQFFG | J2-1*01 | |
| 12 | V20-1*07 | CSA | AGTGCV | EQYFG | J2-7*01 | |
| 13 | V5-4*01 | CASS | FTTGD | GYTFG | J1-2*01 | |
| 14 | V9*01 | CASSV | GDTS | EKLFFG | J1-4*01 | V21‡ |
| 15 | V7-7*02 | CASS | HDRG | YEQYFG | J2-7*01 | |
| 16 | V29-1*01 | CSVE | V | TGELFFG | J2-2*01 | |
| 17 | V29-1 | CS | FLAGGV | EQGG | J2-1*01 | |
| 18 | V19*03 | CAS | RRDP | TDTQYFG | J2-3*01 | |
| 19 | V20-1 | CSA | PAGNP | SYEQYFG | J2-7*01 | |
| 20 | V4-3*01 | CASSQ | APSRAT | YEQYFG | J2-7*01 | |
| 21 | V9*01 or 02 | CASSV | AQGAED | IQYFG | J2-4*01 | |
| 22 | V28*01 | CAS | KGPGTPEL | GYTFG | J1-2*01 | |
| 23 | V9*01 or 02 | CASSV | GVG | YEQYFG | J2-7*01 | |
Vβ and Jβ were attributed according to the nomenclature available at http://imgt.cines.fr
Clones 2 and 14 express the same TCRαβ nucleotide sequence
Clones 1, 6, and 8 express the same TCRβ nucleotide sequence but express different TCRα
Clone 8 expresses two TCRβ, but V4-2*01/J2-7*01 is most likely the chain that recognizes MAGE-3.A1, because the CDR3 region of this chain is identical to the CDR3 of clone 1
Clones 5 and 10 express the same TCRβ protein sequence but differ for their nucleotide sequences
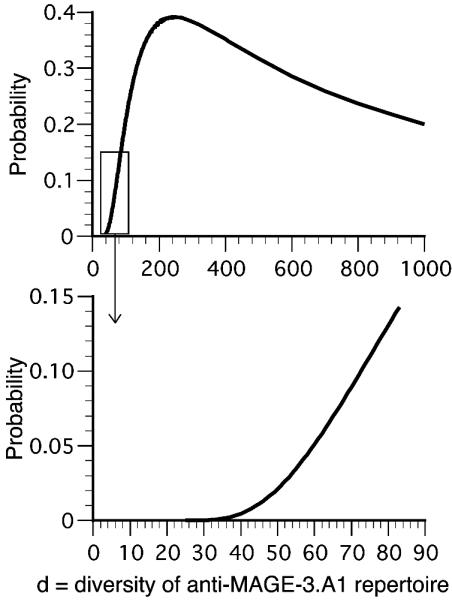
On the basis of this analysis, the diversity of the anti-MAGE-3.A1 TCR repertoire of this individual is at least 22. This minimum estimate is not atypical; we have observed 15, 15, 12, and 12 different anti-MAGE-3.A1 CTL clones in four other patients bearing a melanoma expressing MAGE-3 (data not shown). The probability of obtaining a series of 23 with one repeat by sampling from a repertoire of diversity d is shown in Fig. 3. The diversity that is most likely to yield such a series is around 250, with a probability of 0.4. The probability of this outcome decreases steeply for values <200. It is >15 times lower if the repertoire is <50. In contrast, it decreases only slowly for diversities >250.
Fig. 3.
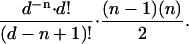 |
On the basis of this evaluation of the diversity of the anti-MAGE-3.A1 naïve TCR repertoire, it is possible to ascertain low-level CTL responses, provided TCR sequence analysis indicates that the same clonotypes are obtained in several independent microcultures. Assuming that the diversity of the naïve TCR repertoire exceeds 50, there is a <5% probability of finding the same clonotype twice in a series of not >8 clones, three times in a series of not >10 clones, or four times in a series of not >20 clones. If such repeats are found, we conclude that the repeated clonotype represents a response to the vaccine (8, 9).
Analysis of Anti-MAGE-3.A1 CTL Responses of Vaccinated Patients. Our present method to evaluate CTL responses of vaccinated patients is outlined in Fig. 4. It involves two weekly stimulations of limiting dilution PBMC microcultures with the MAGE-3.A1 peptide in the presence of IL-2, IL-4, and IL-7. After 2 weeks, the microcultures are tested for the presence of cells stained with the A1/MAGE-3 tetramer. For positive microcultures, the stained cells are cloned and tested for their ability to lyse selectively HLA-A1 tumor cells expressing MAGE-3. Only those microcultures that contain clones that show the adequate lytic ability are considered positive. Patients are considered to have mounted a CTL response only if the postimmunization frequency is higher than 4 × 10-6, i.e., 10 times above the putative naïve level, and if the prevaccination frequency does not exceed 8 × 10-7, i.e., twice the naïve level. If the postimmunization frequency is <4 × 10-6, we still consider a response to have occurred, provided the same clonotype is found at least three times in a set of <10 CTL clones or four times in a set of <20 CTL clones. Patients are considered to be negative for a CTL response if their CTLp frequency is <8 × 10-7 before vaccination and if their postvaccination CTLs do not satisfy either of the criteria mentioned above. Patients with the preimmunization frequency >8 × 10-7 are put in a third category. They are considered to have mounted a spontaneous CTL response before vaccination. It cannot be excluded, however, that they mounted an additional response against the vaccine.
Fig. 4.
Overview of the procedure to evaluate CTL responses of vaccinated patients. Typically, 250,000 PBMC were distributed in each microwell.
Patterns of antivaccine CTL responses observed in patients vaccinated with ALVAC-MAGE are seen in Table 3. The responses to the vaccine were all monoclonal. So was a response observed after peptide vaccination (3, 10). In contrast, we have observed that most patients vaccinated with dendritic cells pulsed with the peptide mount a polyclonal response (9).
Table 3. Anti-MAGE-3.A1 CTL response in 17 HLA-A1 patients vaccinated with ALVAC-MAGE.
| Preimmune anti-MAGE-3.A1 CTL*
|
Postimmune anti-MAGE-3.A1 CTL§
|
||||
|---|---|---|---|---|---|
| Patients | CTLp frequency† | Presence of repeated clonotypes‡ | CTLp frequency | Presence of repeated clonotypes | CTL response¶ |
| Patients with tumor regression (n = 4) | |||||
| EB81 | <1 × 10-7 | 3 × 10-6 | 50 tested: 1 repeated 48 times | + | |
| CP67 | nt | 4 × 10-7 | – | ||
| LAU147 | <4 × 10-7 | 3 × 10-3 | 13 tested: 1 repeated 13 times | + | |
| NAP33 | 3 × 10-7 | 7 × 10-7 | 18 tested: 1 repeated 6 times | + | |
| Total: 3/4 | |||||
| Patients without tumor regression (n = 13) | |||||
| NAP35 | nt | <5 × 10-7 | – | ||
| KUL73 | nt | <5 × 10-7 | – | ||
| LB2196 | <3 × 10-7 | <3 × 10-7 | – | ||
| NAP36 | <1 × 10-7 | 2 × 10-6 | 7 tested: 1 repeated 6 times | + | |
| CP69 | nt | <5 × 10-7 | – | ||
| IGR37 | nt | <1 × 10-6 | – | ||
| BB132 | nt | 1 × 10-7 | – | ||
| VUB39 | 1 × 10-6 | 7 tested: 1 repeated 6 times | 9 × 10-6 | 22 tested: 1 repeated 7 times and 1 12 times | P |
| LAU407 | nt | <3 × 10-7 | – | ||
| NAP37 | 3 × 10-5 | 2 × 10-5 | P | ||
| LAU622 | nt | <3 × 10-7 | – | ||
| UZG10 | nt | <9 × 10-7 | – | ||
| LE1 | nt | <5 × 10-7 | – | ||
| Total: 1/11 | |||||
| Patient without cancer, not vaccinated | |||||
| LB1965 | 6 × 10-7 | 23 tested: 1 repeated 2 times | |||
The preimmune CTL were tested only if either an increased CTLp frequency or repeated CTL clones were observed in the postimmune blood
Frequency of CTL precursors, as measured with the approach shown in Fig. 4. nt, not tested
When possible, tetramer-stained CTL were sorted and cloned. The TCR gene rearrangements of independent anti-MAGE-3.A1 CTL clones were sequenced to analyze the CTL diversity. The table gives the number of specific CTL clones that were tested and the occurrence of repeated clonotypes
Postimmune CTL were isolated from blood collected after four ALVAC vaccinations, except in four cases. The blood from patients NAP37 and NAP36 was collected after two ALVAC vaccinations and after four ALVAC + one peptide vaccinations, respectively. The blood from patients LE1 and LAU407 was collected after four ALVAC + three peptide vaccinations
A CTL response is defined as a significant increase in CTLp frequency, or the presence of a repeated clonotype according to the criteria mentioned in the text. P, spontaneous CTL response prior to vaccination
Table 4 presents a summary of the anti-MAGE-3.A1 CTL responses that we observed in patients who showed evidence of tumor regression and in patients who did not. Among patients vaccinated with ALVAC-MAGE, a CTL response was found in 3 of 4 patients who showed regression and in 1 of 11 patients who did not. Among patients vaccinated with dendritic cells, a CTL response was found in three of three patients who showed regression and in zero of three patients who did not. These are statistically significant correlations. Only one of seven patients vaccinated with the MAGE-3.A1 peptide showed a CTL response.
Table 4. Summary of anti-MAGE-3.A1 CTL responses in vaccinated melanoma patients.
We considered the possibility that our inability to detect an anti-MAGE-3.A1 T cell response in several patients who displayed tumor regression could be due to the fact that these patients made a CTL response against the MAGE-3168-176 peptide presented by an HLA molecule other than A1. Such responder CTLs would not be detected by our A1/MAGE-3 tetramer. We have observed that this can occur with HLA-B35. The MAGE-3 peptide can be presented to CTLs by HLA-B*3501 molecules (11), and we found that one patient of the ALVAC study, LB2196, who happened to be HLA-A1 and B35, had no HLA-A1-restricted CTLs against the peptide but had HLA-B35-restricted CTLs recognizing the peptide. We followed two approaches to examine the possibility that this could occur with other HLA class I molecules. First, in addition to testing the mixed lymphocyte–peptide culture microcultures with tetramers, we screened them with an IFN-γ enzyme-linked immunospot (ELISPOT) assay after restimulation with autologous EBV-B cells pulsed or not pulsed with the MAGE-3168-176 peptide. This screening was applied to three patients showing tumor regression without a detectable CTL response, and no new positive microculture was detected. But the sensitivity of the ELISPOT assay was 20 times lower than that of the tetramer assay, which identified positive microcultures containing as little as 0.05% positive cells. In a second approach, we tested directly whether peptide MAGE-3168-176 could bind to other HLA molecules by examining its ability to be correctly refolded with recombinant HLA and β2 microglobulin chains. We focused on HLA molecules carried by some of the regressor patients, namely A*3201, B*1501, B*4001, C*0303, and C*0701. We did not obtain correctly folded HLA–peptide complexes for any of these HLA molecules, even though each complex was obtained with an appropriate control peptide. Octamers included in the MAGE-3 peptide also failed to bind to HLA-Cw3 and Cw7. These results indicate that the failure to detect anti-MAGE-3.A1 CTLs with the tetramer in many regressing patients is rarely due to the fact that a response is directed against the peptide presented by another HLA molecule.
Of 27 patients who were analyzed before vaccination, 2 patients vaccinated with ALVAC and 2 patients vaccinated with dendritic cells showed a preexisting anti-MAGE-3.A1 CTL response, as determined either by high frequency or by predominant clonotypes. In patient VUB39 (Table 3), the vaccine may nevertheless have initiated the amplification of new naïve precursors, because new clonotypes were observed after vaccination (8). Interestingly, in two of these patients, melanoma diagnosis was established on a lymph node metastasis in the absence of concomitant or prior observation of a primary tumor. Both patients reported the spontaneous disappearance of a pigmented lesion in the drainage territory of the metastatic node several years before the diagnosis. These observations confirm that spontaneous CTL responses can occur against antigens encoded by cancer-germline genes (12) and that they may play a role in the spontaneous regressions of melanomas that were observed occasionally.
Discussion
Our results indicate that the frequency of the naïve anti-MAGE-3.A1 CTL precursors is ≈4 × 10-7. To our knowledge, the only other reported precursor frequency of human naïve T cells against a defined peptide–HLA combination concerned the Melan-AMART-126,35 antigen, which is presented by HLA-A2. It was estimated to be 10-3 of the CD8 T cells (13). This very high frequency is all the more remarkable because the target is a normal melanocytic differentiation antigen, for which a situation of natural tolerance would be expected.
We estimate that the size of the anti-MAGE-3.A1 repertoire is very likely to be >50, with a likely maximum of 250. It is important to realize that the range of plausible values extends as high as 900. As a working hypothesis, we will select the value of 100 for the diversity of the anti-MAGE-3.A1 repertoire, because we feel that the value is unlikely to be lower. Assuming that the diversity of the anti-MAGE-3.A1 TCR repertoire is representative of the whole repertoire, this leads to an estimate of 2.5 × 108 (100/4 × 10-7) for the diversity of the whole human CD8 T cell repertoire. Kourilsky and colleagues (14) estimated on the basis of random TCR sequencing that the Vα 12 T cell population bears ≈6 × 105 different β chains. Considering that Vα 12 T cells represent ≈2.5% of total human T cells, this led to a total TCRα–β diversity of 2.5 × 107 (14). These authors proposed another estimate based on the following reasoning. An estimate of 106 for the total number of different β chains sequences was obtained by gene amplification and sequencing (14). After its β sequence rearrangement, each thymic T lymphocyte engages in a 5-day proliferation of ≈1,000-fold (15). At that point, the α rearrangement occurs, and only 10% of the produced α–β combinations are functional and survive selection, leading to a TCRα–β diversity of 108. This figure is plausible provided that, on the average, the thymus produces a given β chain rearrangement only once, the progeny of which later multiplies outside the thymus for ≈10 cycles to produce a total of ≈1,000 cells in the periphery (16). Because CD8 T cells constitute about one-third of total T cells, their TCR diversity would be 8 × 106 according to the first estimate of Kourilsky and colleagues (14) and 3 × 107 according to their second estimate. Our estimate is 10-fold higher than their higher estimate, which would have predicted an anti-MAGE-3.A1 repertoire of only ≈12 different TCRs.
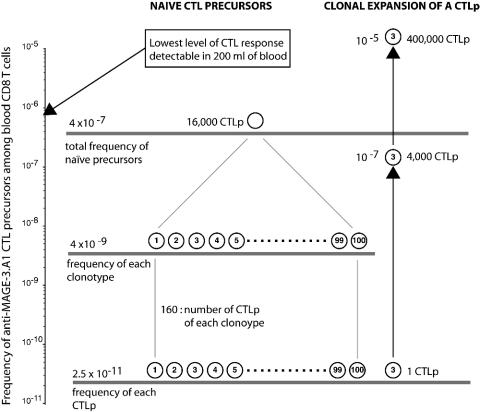
Fig. 5 summarizes the view of the anti-MAGE-3.A1 naïve repertoire that emerges from our analysis: ≈100 different clonotypes comprising ≈160 CTLp each contribute to the naïve CTLp compartment of 16,000 cells, i.e., 4 × 10-7 of the 4 × 1010 CD8 T cells, which is estimated to be the total number of CD8 T cells in the human body. The monoclonal character of many responses that we observe suggests that they are the product of the activation of a single naïve CTLp after vaccination. Even if this cell expands 10,000 times, the total frequency of anti-MAGE-3.A1 CTLp is not increased significantly.
Fig. 5.
Frequencies and diversity of anti-MAGE-3.A1 CTLs. Different clonotypes are indicated by circled numbers. We assume here that humans have a total of 4 × 1010 CD8 T cells (106 PBMC per ml of blood containing 15% CD8 T cells, 5 liters of blood, and 2% of the total lymphocytes located in the blood) (17).
Only a small minority (10–20%) of melanoma patients vaccinated with tumor antigens such as MAGE-3.A1 have shown evidence of tumor regression. Because these vaccination trials have not included randomization with an untreated arm, one cannot rigorously exclude the possibility that the regressions observed are not due to the vaccine. Occasional spontaneous regressions of melanoma have been observed, but the frequency reported is at least 20 times lower than the frequency observed in the vaccination trials. The correlation between CTL responses and tumor regression observed in the ALVAC and dendritic cell trials supports the notion that the rejection is caused by the vaccine. But it does not rigorously prove it, because spontaneous regressions might result in the activation of new CTLs against the tumor.
Our working hypothesis is that low anti-MAGE-3.A1 CTL responses, including some that are below our detection level of 8 × 10-7, i.e., comprising <16,000 CTLs, occasionally trigger a sequence of events that cause tumor rejection. In mice, tumors of 0.5 cm3, i.e., of the same order of magnitude as many cutaneous metastases seen on melanoma patients, can disappear within a few days upon the occurrence of a CTL response with a frequency of ≈10-3, which is directed against a tumor antigen. Because mice contain ≈2 × 107 CD8 T lymphocytes, their tumor rejection appears to be initiated by a group of ≈20,000 specific CTLs. It is therefore possible that, in human patients, such a small number of antivaccine T cells could initiate the rejection response. Their interaction with the tumor cells may create conditions that favor the activation and proliferation of a much larger number of T cells directed against other antigens of the tumor. These T cells could be the effector cells that destroy the majority of the tumor cells.
Acknowledgments
We thank Mrs. S. Ottaviani and C. Wildmann for their precious assistance, Dr. M. Bourgois for helpful discussions about probability calculations, and Mrs. V. Winand and N. Krack for editorial assistance. This work was supported by grants from TELEVIE (to C.L.), Fédération Belge Contre le Cancer, Fondation Salus Sanguinis, and FB Assurances. S.L. was supported by Fonds National de la Recherche Scientifique as a Postdoctoral Researcher.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Therapeutic Vaccines: Realities of Today and Hopes for Tomorrow,” held April 1–3, 2004, at the National Academy of Sciences in Washington, DC.
Abbreviations: CTL, cytotoxic T lymphocyte; CTLp, CTL precursors; EBV-B, Epstein–Barr-virus-transformed B lymphocytes; FACS, fluorescence-activated cell sorting; MACS, magnetic cell sorting; PBMC, peripheral blood mononuclear cells; TCR, T cell receptor.
References
- 1.Marchand, M., van Baren, N., Weynants, P., Brichard, V., Dréno, B., Tessier, M.-H., Rankin, E., Parmiani, G., Arienti, F., Humblet, Y., et al. (1999) Int. J. Cancer 80, 219-230. [DOI] [PubMed] [Google Scholar]
- 2.Thurner, B., Haendle, I., Roder, C., Dieckmann, D., Keikavoussi, P., Jonuleit, H., Bender, A., Maczek, C., Schreiner, D., von den Driesch, P., et al. (1999) J. Exp. Med. 190, 1669-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coulie, P. G., Karanikas, V., Colau, D., Lurquin, C., Landry, C., Marchand, M., Dorval, T., Brichard, V. & Boon, T. (2001) Proc. Natl. Acad. Sci. USA 98, 10290-10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altman, J. D., Moss, P. A. H., Goulder, P. J. R., Barouch, D. H., McHeyzer-Williams, M. G., Bell, J. I., McMichael, A. J. & Davis, M. M. (1996) Science 274, 94-96. [DOI] [PubMed] [Google Scholar]
- 5.Genevée, C., Diu, A., Nierat, J., Caignard, A., Dietrich, P. Y., Ferradini, L., Roman-Roman, S., Triebel, F. & Hercend, T. (1992) Eur. J. Immunol. 22, 1261-1269. [DOI] [PubMed] [Google Scholar]
- 6.Chaux, P., Vantomme, V., Coulie, P., Boon, T. & van der Bruggen, P. (1998) Int. J. Cancer 77, 538-542. [DOI] [PubMed] [Google Scholar]
- 7.Dutoit, V., Rubio-Godoy, V., Pittet, M. J., Zippelius, A., Dietrich, P.-Y., Legal, F. A., Guillaume, P., Romero, P., Cerottini, J.-C., Houghten, R. A., et al. (2002) J. Exp. Med. 196, 207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulie, P. G., Karanikas, V., Lurquin, C., Colau, D., Connerotte, T., Hanagiri, T., Van Pel, A., Lucas, S., Godelaine, D., Lonchay, C., et al. (2002) Immunol. Rev. 188, 33-42. [DOI] [PubMed] [Google Scholar]
- 9.Godelaine, D., Carrasco, J., Lucas, S., Karanikas, V., Schuler-Thurner, B., Coulie, P. G., Schuler, G., Boon, T. & Van Pel, A. (2003) J. Immunol. 171, 4893-4897. [DOI] [PubMed] [Google Scholar]
- 10.Karanikas, V., Lurquin, C., Colau, D., van Baren, N., De Smet, C., Lethé, B., Connerotte, T., Corbière, V., Demoitié, M.-A., Liénard, D., et al. (2003) J. Immunol. 171, 4898-4904. [DOI] [PubMed] [Google Scholar]
- 11.Luiten, R. M., Demotte, N., Tine, J. & van der Bruggen, P. (2000) Tissue Antigens 56, 77-81. [DOI] [PubMed] [Google Scholar]
- 12.Valmori, D., Dutoit, V., Liénard, D., Rimoldi, D., Pittet, M. J., Champagne, P., Ellefsen, K., Sahin, U., Speiser, D., Lejeune, F., et al. (2000) Cancer Res. 60, 4499-4506. [PubMed] [Google Scholar]
- 13.Zippelius, A., Pittet, M. J., Batard, P., Rufer, N., de Smedt, M., Guillaume, P., Ellefsen, K., Valmori, D., Liénard, D., Plum, J., et al. (2002) J. Exp. Med. 195, 485-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arstila, T. P., Casrouge, A., Baron, Y., Even, J., Kanellopoulos, J. & Kourilsky, P. (1999) Science 286, 958-961. [DOI] [PubMed] [Google Scholar]
- 15.Trigueros, C., Ramiro, A. R., Carrasco, Y. R., de Yebenes, V. G., Albar, J. P. & Toribio, M. L. (1998) J. Exp. Med. 188, 1401-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kesmir, C., Borghans, J. A. M., de Boer, R. J., Arstila, T. P., Casrouge, A., Baron, V., Even, J., Kanellopoulos, J. & Kourilsky, P. (2000) Science 288, 1135a. [DOI] [PubMed] [Google Scholar]
- 17.Westermann, J. & Pabst, R. (1990) Immunol. Today 11, 406-410. [DOI] [PubMed] [Google Scholar]