Abstract
Bacteroides are predominant human colonic commensals, but the principal pathogenic species, Bacteroides fragilis (BF), lives closely associated with the mucosal surface, whereas a second major species, Bacteroides thetaiotaomicron (BT), concentrates within the colon. We find corresponding differences in their genomes, based on determination of the genome sequence of BF and comparative analysis with BT. Both species have acquired two mechanisms that contribute to their dominance among the colonic microbiota: an exceptional capability to use a wide range of dietary polysaccharides by gene amplification and the capacity to create variable surface antigenicities by multiple DNA inversion systems. However, the gene amplification for polysaccharide assimilation is more developed in BT, in keeping with its internal localization. In contrast, external antigenic structures can be changed more systematically in BF. Thereby, at the mucosal surface, where microbes encounter continuous attack by host defenses, BF evasion of the immune system is favored, and its colonization and infectious potential are increased.
The Bacteroides are saccharolytic Gram-negative obligate anaerobes belonging to the phylum Bacteroidetes, also known as the Cytophaga-Flavobacteria-Bacteroides group. Bacteroides are among the most predominant genera among intestinal microbiota.
The large intestine ecological niche harbors huge numbers of microbes with diverse properties, in a community that exhibits comprehensive host-microbe and microbe-microbe interactions. The environment in the large intestine is not uniform: the environment close to the host intestinal epithelium is very different from that of the central space of the gut. The composition of the bacterial community also differs significantly, suggesting that microbes use distinct and appropriate adaptation strategies to establish a successful niche. In fact, among the Bacteroides, which together account for ≈30% of fecal isolates (1), the viable cell number of Bacteroides fragilis (BF) is 10- to 100-fold smaller than those of other intestinal Bacteroides such as B. thetaiotaomicron (BT), B. distasonis, and B. vulgatus (1), but BF is the most frequent at the mucosal surface (2). It also is the most frequent isolate from clinical specimens, particularly from abdominal cavity infections, and is regarded as the most virulent Bacteroides species.
Here, we report the complete genome sequence of BF strain YCH46. Genome analysis of BF strain YCH46 and comparisons with other sequenced bacteria of the Cytophaga-Flavobacteria-Bacteroides group, especially BT (3), rationalizes the differential adaptation strategies of commensal Bacteroides species to parts of the intestinal structure with more or less infectious potential.
Methods
Genome Sequencing. BF strain YCH46 was isolated from a patient with septicemia at Yamaguchi Central Hospital, Yamaguchi Prefecture, Japan (4, 5). For preparing the genomic DNA, a single colony grown on a Gifu anaerobic medium (GAM) agar plate (Nissui Pharmaceutical, Tokyo) was inoculated into 5 ml of semiliquid GAM broth and cultivated at 37°C for 24 h. A portion (0.3 ml) of the preculture was then inoculated into 15 ml of fresh GAM broth and cultivated until the late-logarithmic phase. The genomic DNA was prepared with an Easy-DNA kit (Invitrogen) to construct two types of pUC18-based genomic shotgun libraries containing 1- to 2-kb or 4- to 5-kb inserts. The whole genome sequence was obtained by assembling 101,722 end sequences (9.6-fold coverage) from both shotgun libraries. Each clone was sequenced from both ends on automated sequencers (Applied Biosystems PRISM 3700 and Megabase1000). Sequence assembly was accomplished with phred/phrap/consed (6-8). Gap closure was made by PCR direct sequencing with oligonucleotide primers designed to anneal to each end of the neighboring contigs. Overall accuracy of the finished sequence was estimated to have an error rate of <1 per 10,000 bases (phrap score ≥40).
Sequence Analyses. Protein coding regions (ORFs) >150 bp were identified by the combination of genome gambler 1.51 (9), critica (10), genehacker (11), and glimmer 2.0 (12) programs. In the initial step of ORF prediction, 422 ORFs, which showed high similarity to the proteins in the Clusters of Orthologous Groups of Proteins (COGs) database (13), were selected and used to create the training data set for glimmer, critica, and genehacker programs. Individual predicted ORFs were reviewed manually for the presence of start codons (ATG, TTG, and GTG) and ribosome-binding sequences (AGAAAGGAGG, GenBank accession no. M61006). Protein functional annotation was based on homology searches against public protein databases by blastp (14). tRNA genes were identified by trna-scan (15). Functional classification of ORFs was made by homology search against COGs by using blastp. The annotated complete genome sequence is available at the GenBank database (accession no. AP006841 for the chromosome and accession no. AP006842 for the plasmid pBFY46).
Comparative Genomics. Genome sequences of BT strain VPI-5482 and Porphyromonas gingivalis (PG) strain W83 were obtained through the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). To compare the gene repertoires and the paralogous gene families among Cytophaga-Flavobacteria-Bacteroides bacteria, all of the proteomes of BF strain YCH46, BT strain VPI-5482, and PG strain W83 were mixed and clustered on the basis of the amino acid sequence homology by using blastclust (threshold: identity ≥30% and alignment length ≥60%). ORFs on mobile genetic elements (415, 410, and 120 ORFs in BF, BT, and PG, respectively) were excluded from this analysis. Orthologous genes were identified by the bidirectional best-hit analysis.
Genome Searches of Invertible Regions. To detect invertible regions on the BF strain YCH46 genome, we first listed the contigs that consisted of at least two shotgun reads but were excluded from the final assembly as chimeric contigs. Of these contigs, we selected those with two portions, one matching to the final sequence assembly and the other (>100 bp in length) inverse to a nearby sequence at >95% identity. The region surrounding the chimeric junction was then examined to identify inverted repeat sequences (IRs), which presumably mediate the DNA inversion. Identified IRs were classified into five classes according to the internal consensus motif sequences. Using the identified five consensus motif sequences, we further screened the genome sequence of BF strain YCH46, and those of BT strain VPI-5482 and PG strain W83, to identify all possibly invertible regions. IRs that flank the four invertible promoters of capsular polysaccharide biosynthesis loci (PS loci) in BT strain VPI-5482 also were included in this screening. In this case, because all four IRs had an identical sequence, we used an internal sequence, GTTAC{N7}GTAAC, which may be important for recombination, as the motif sequence. From a large number of candidates obtained from this initial screening, we selected genomic regions that were <5 kb and flanked by IRs with complete match or one-base mismatch to the consensus motif. Once an IR was identified in a region where genes for SusC/D homologs were clustered, the entire region, especially the upstream regions of each gene for SusC/D homolog, were examined intensively for additional IRs.
To confirm that DNA inversions actually occur at each candidate region, we examined the genomic DNA of BF strain YCH46 by PCR by using a set of orientation-specific primer pairs (Fig. 5 and Tables 1-3, which are published as supporting information on the PNAS web site). PCR amplification was performed by using 200 ng of genomic DNA and AmpliTaq Gold (PerkinElmer) in the following setting: preheating (96°C for 10 min), 35 cycles of DNA denaturation (96°C for 15 sec)/primer annealing (50°C for 30 sec)/DNA extension (72°C for 1 min), and an additional extension step (72°C for 1 min). PCR products were separated on 2.0% agarose gel electrophoresis and visualized by ethidium bromide staining. DNA fragments >5 kb were amplified by using LA Taq (Takara Shuzo, Tokyo) in the following setting: preheating (94°C for 1 min), 35 cycles of DNA denaturation (94°C for 30 sec)/primer annealing and DNA extension (60°C for 25 min), and an additional extension step (60°C for 10 min).
Results and Discussion
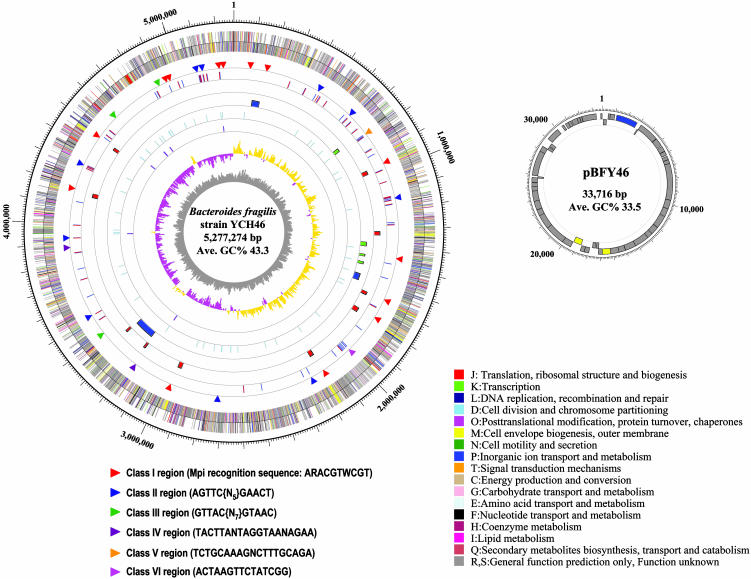
General Genome Features. BF strain YCH46 is a clinical strain isolated from a patient with septicemia. The chromosome, with an average G+C content of 43.3%, is a single circular DNA of 5,277,274 bp (Fig. 1). The size is 1 Mb smaller than that of BT strain VPI-5482 (6.2 Mb) but much larger than the oral pathogenic bacterium PG strain W83 (16), another sequenced member of the Cytophaga-Flavobacteria-Bacteroides group (2.3 Mb). Unlike many other bacterial genomes, the vicinity of the putative replication origin, assigned by GC skewing analysis (17), lacks dnaA, dnaN, recF, gyrB, and gyrA genes. Instead, they are dispersed on the chromosome. Similar relative positions of these genes are observed in the chromosomes of BT and PG. The chromosome contains 4,578 protein-coding ORFs with an average size of 1,032 bp, covering 90% of the whole chromosome sequence. It also contains six ribosomal RNA operons (16S-tRNAIle-tRNAAla-23S-5S), all located on the leading strand and transcribed in the same direction, from the replication origin toward the terminus. Specificity for all amino acids is provided by 74 tRNA genes.
Fig. 1.
Circular maps of the chromosome and the pBFY46 plasmid of BF strain YCH46. Each circle represents (from inside out): G+C content, GC skew, rRNA operon (all consisted of 16S-23S-5S rRNA genes), tRNA genes, conjugative (blue bars) and mobilizable (green bars) transposons, capsular PS loci, SusC (blue bars) and SusD (red bars) families of outer membrane proteins, invertible regions, predicted genes transcribed into the counterclockwise direction, and those into clockwise direction. Classification of the invertible regions based on the motif sequence within IRs is indicated by the color of triangles. The inner and outer circles of the plasmid represent predicted genes transcribed into the counterclockwise direction and those into clockwise direction, respectively. All of the predicted genes are colored according to the Clusters of Orthologous Groups of Proteins functional classification (13).
BF strain YCH46 possesses a variety of mobile genetic elements. They include a 33.7-kb plasmid (pBFY46), three conjugative transposons (CTnYCH46-1, CTnYCH46-2, and CTnYCH46-3), four mobilizable transposons (MTnYCH46-1, MTnYCH46-2, MTnYCH46-3, and MTnYCH46-4), one 36.8-kb prophage remnant, and 14 transposases (13 on the chromosome and one on pBFY46). The plasmid pBFY46 encodes 47 ORFs. A large proportion (43%) of the predicted ORFs showed no significant homology with any proteins in databases, but a set of genes for DNA transfer was identified. Also the G+C content (33.5%) is significantly lower than the chromosome. Thus, this plasmid was likely acquired by conjugal transfer from other species. In some BF strains, Hin invertase-homologue (FinB), which binds to the IRs of invertible promoters for capsular polysaccharide biosynthesis, is encoded on a plasmid (18), but no gene corresponding to finB was found in pBFY46. Among the three CTns, CTnYCH46-1 is a member of the CTnDOT family, one of the major CTn families in Bacteroides (19, 20), and carries a tetracycline-resistant gene of a “ribosomal protection type” (tetQ), accounting for the tetracycline resistance of strain YCH46. The other two CTns diverge from other Bacteroides CTns and carry no apparent drug-resistant genes.
Gene Expansion in Bacteroides Genomes. Excluding 415 ORFs on mobile genetic elements, 1,244 (29.9%) of the predicted 4,163 ORFs in BF comprise 378 groups of paralogous gene families. The remaining 2,919 are singleton genes. In BT, 36.9% of the 4,368 ORFs constitute 457 paralogous gene families, whereas in PG, only 14.3% of the 1,789 ORFs are classified into 102 paralogous families (Fig. 6, which is published as supporting information on the PNAS web site). Nearly 30% of the single genes in BF are conserved in the other two species, and 31.8% are conserved in BT but not in PG. Of the 1,392 ORFs that are present in BF but not in BT and PG, only 291 are functionally assigned on the basis of sequence homology to known proteins; 844 (60.6%) have no significant homology to any protein sequence in databases. To explain the difference in pathogenic potentials between the two Bacteroides species, the only candidates found for virulence-related genes in the 291 functionally assigned ORFs were 60 genes involved in capsular polysaccharide biosynthesis. In addition, two hemolysin-like proteins were encoded on the mobile genetic elements.
Both Bacteroides species have expanded similar paralogous groups (Fig. 7 and Table 4, which are published as supporting information on the PNAS web site). They include those involved in utilization of dietary polysaccharide (glycosylhydrolases, polysaccharide-binding proteins, and transporters), environmental sensing and signal transduction (extracytoplasmic function-type sigma factors and their cognate antisigma factors, and one- or two-component signal transduction systems), and capsular polysaccharide biosynthesis. Genes for polysaccharide utilization and environmental sensing are frequently colocalized in BF, as in BT, probably favoring regulatory coordination of gene expression and nutrient availability.
Gene duplication for polysaccharide utilization is a feature commonly observed in colonic inhabitants (21, 22), reflecting the fact that the lower intestinal tract is an environment poor in monosaccharides and disaccharides, which have already been absorbed by the host and the microbiota in the upper intestinal tract. Compared with other sequenced colonic microorganisms such as Bifidobacterium longum and Clostridium perfringens, both Bacteroides species contain much larger numbers of polysaccharide-degrading enzymes with a wide range of substrate specificities (132 in BF and 259 in BT).
In addition, the SusC family of outer membrane proteins constitutes the largest paralogous family in both Bacteroides (54 members in BF and 79 in BT, see Table 4). As seen in BT, about half the genes for SusC family members (22 members) are paired with genes encoding the SusD family of outer membrane proteins. These SusC- and SusD-family proteins likely comprise a large cohort of polysaccharide receptors that mediate the binding of a wide range of polysaccharides to the bacterial cell surface and their subsequent degradation in the periplasmic space (23-26). The presence of such polysaccharide utilization systems, which can minimize the diffusion of digested products and cross-feeding of competitors, highly favor Bacteroides in competition for growth in the colon. Much greater gene expansion for polysaccharide utilization in BT than BF mirrors the differences in their cell numbers in the colon.
Capsular Polysaccharide Biosynthesis. Bacteroides are unique in the production of multiple capsular polysaccharides, each exhibiting distinct structure and antigenicity. These capsular polysaccharides are known to be profoundly implicated in the virulent formation of abscesses by BF (27-29). BF strain YCH46 and BT strain VPI-5482 possess nine and seven capsular PS loci, respectively (Table 5, which is published as supporting information on the PNAS web site). Among these, the PS-7 locus in BF strain YCH46 is apparently disrupted by the insertion of CTnYCH46-3. PS loci in Bacteroides share a common genetic structure; a gene encoding putative transcriptional regulator (UpxY homolog) (30-32) is always located upstream of a set of genes for polysaccharide biosynthesis and transport. In most cases, a gene encoding a UpxZ homolog is located between them.
Expression of PS loci in BF also is unique in that they are regulated in an on-off manner by a master protein, through reversible inversions of promoters (32). Inversions occur at IRs that flank each promoter sequence. A serine-type site-specific recombinase, Mpi, has been identified as a recombinase responsible for the promoter inversions of seven PS loci (33). Of the nine PS loci on the BF strain YCH46 genome, seven are preceded by invertible promoters that are flanked by 19- to 23-bp IRs containing the ARACGTWCGT consensus sequence that is recognized by Mpi (33) (Table 5). The mpi gene (BF2765) is not colocalized with any PS loci. By reanalyzing the random shotgun clones used for genome sequencing, we identified promoter regions with either orientation in many clones, although the proportions of clones with on-off orientations differed from one locus to another. All clones derived from the PS-1 locus had the off orientation, but PCR analyses demonstrated that this region also underwent DNA inversion (data not shown). PCR and sequencing analyses of PS-7 locus, which lacks invertible promoter, indicated that in some populations of BF strain YCH46 cells, CTnYCH46-3 was precisely excised (data not shown). This may be another mechanism of phase variation of capsular polysaccharide production.
Because the DNA for genome sequencing was prepared from a late-logarithmic phase culture raised from a single colony, promoter inversions must occur very frequently, generating BF strain YCH46 cells with diverse capsular polysaccharide expression patterns during short periods of cultivation. In BT strain VPI-5482, we identified putatively invertible promoters at four PS loci, but IRs associated with them contained a consensus sequence different from that in BF. In addition, each invertible promoter of BT is immediately preceded by a tyrosine-type, site-specific recombinase (BT0375, BT0595, BT1657, and BT1726), suggesting that on-off switching of each PS locus is locally controlled by these recombinases. This finding contrasts with the on-off switching of PS loci in BF, globally controlled by Mpi.
Capsular polysaccharides of BF themselves can induce abscess formation, and the presence of both a positively charged free amino group and at least one negatively charged group in the repeating unit is essential for the activity (34). Of the nine PS loci in BF strain YCH46, six (other than PS-4, PS-5, and PS-7) contain one or two aminotransferase genes that are probably responsible for the creation of free amino groups in repeating units. Moreover, four of them (PS-2, PS-3, PS-6, and PS-9) contain dehydrogenase genes that are known to be involved in the incorporation of negatively charged sugars (30), indicating that BF strain YCH46 produces at least four kinds of abscess-inducible zwitterionic capsular polysaccharides. In contrast, only one locus (PS-3) contains an aminotransferase gene (BT0612) in BT strain VPI-5482. This difference correlates with the higher virulence of BF.
Multiple DNA Inversion in Bacteroides Genomes. It is known that Mpi mediates the DNA inversions not only in the promoter regions of PS loci but also in at least six other promoter regions (33). By a systematic review of chimeric contigs that were excluded from the final sequence assembly and by a genomewide search for IRs with consensus sequences that were identified by the chimeric contig analysis, we identified as many as 31 invertible regions on BF strain YCH46 chromosome (Fig. 1, and Fig. 8 and Table 6, which are published as supporting information on the PNAS web site). PCR analyses of the YCH46 genomic DNA demonstrated that all of the identified regions actually underwent DNA inversions (see Fig. 3, and Figs. 9-11, which are published as supporting information on the PNAS web site). These invertible regions can be grouped into six classes according to internal motif sequences within IRs (Fig. 1). Class I regions, which include seven invertible promoter regions of PS loci and seven additional regions, contain IRs with Mpi recognition sequences, and class III regions contain IRs with the same motif sequences as those found in the invertible PS promoters of BT. The other four classes are invertible elements with different motif sequences.
Many of the identified DNA inversions affect the expression of genes involving in the synthesis of surface architectures, such as capsular polysaccharides, SusC and SusD homologs, and other outer membrane proteins (Table 6 and Table 7, which is published as supporting information on the PNAS web site). They appear to confer a highly advantageous genetic system for evasion of host immune response.
As for SusC homologs, expression of at least 20 homologs, most of which are paired with SusD homologs, are controlled by DNA inversions. In addition, expression of several transporters, signal transduction systems, and carbohydrate degradation systems also is affected by DNA inversion, suggesting that the bacterium also uses DNA inversions to adapt to environmental changes in nutrient levels. Unexpectedly, expression of the chaperone GroES/EL also is under the control of inversion, but the physiological role of this switch is unknown.
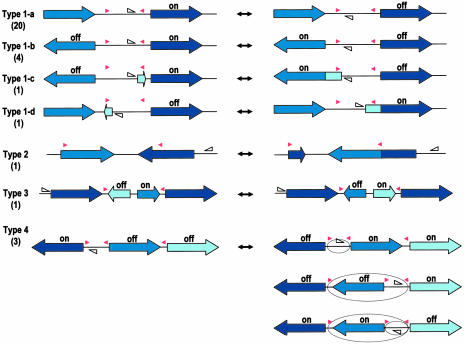
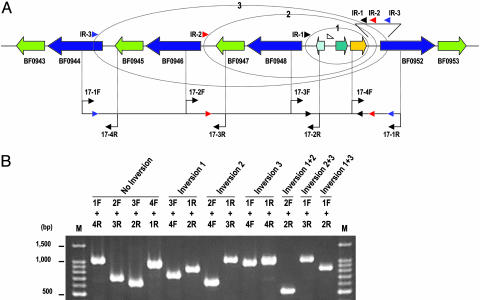
The DNA inversions regulate gene expressions not only by simple on-off switching of promoter orientation. Invertible regions can be classified into four types, according to their mode of regulation (Fig. 2). The most abundant type I inversions regulate the expression of downstream genes monodirectionally (1-a and 1-d) or bidirectionally (1-b and 1-c), by inverting promoter-containing segments. In the cases of types 1-c and 1-d, promoter inversion is accompanied by a fusion to the downstream gene of a small ORF encoded in the flippable segment. Type 2 inversion generates two kinds of hybrid proteins with different C-terminal sequences, by fusing the two genes that are present in a tail-to-tail orientation. Type 3 modulates the operon structure by changing the orientation of genes that are located in the middle of the operon. Type 4 constitutes a shufflon-like multiple inversion system, where several segments flanked by IRs are inverted individually or in groups to deliver the promoter to one of the gene cassettes (35). All three type 4 systems identified on the BF strain YCH46 genome involve the selective expression of SusC and SusD homologs (Figs. 3, 10, and 11). In the locus shown in Fig. 3, a segment containing a promoter and an antisigma factor and extracytoplasmic function-sigma factor genes can be delivered to either of the four SusC/D gene cassettes by various combinations of inversions, via three different IRs each containing class II consensus sequences.
Fig. 2.
Functional classification of DNA inversions identified in Bacteroides genomes. DNA inversions control the expressions of genes downstream or between the IRs (indicated by red arrowheads) in several ways. Locations and directions of each promoter are indicated by open triangles. Type I inversions mediate the on-off switching of the downstream genes monodirectionally (1-a and 1-d) or bidirectionally (1-b and 1-c) by changing the promoter orientation. In the cases of 1-c and 1-d, a small ORF encoded in the invertible segment fuses to the downstream gene by DNA inversion to create an N-terminal extension. In many cases, signal sequences are added to the downstream genes. Type 2 mediates the formation of two types of hybrid proteins with different C-terminal sequences. Type 3 modulates the operon structure. Type 4 is a shufflon-like multiple inversion system. In this case, segments flanked by IRs are inverted independently or in groups to deliver the promoter to either of the gene cassettes. The numbers of each type of invertible region in the chromosome of BF strain YCH46 are shown in parentheses.
Fig. 3.
A shufflon-like DNA inversion system in BF regulates variable expression patterns of SusC/D family proteins. (A) The gene organization of SusC/D cluster 1 and the locations of PCR primers used to detect DNA inversions are shown. Genes for SusC, SusD, antisigma factor, extracytoplasmic function-type sigma factor, and hypothetical proteins are indicated by blue, light green, green, yellow, and light blue arrows, respectively. The position of a Bacteroides consensus promoter sequence (41) is indicated by an open triangle. Circles represent DNA inversions at each IR (IR-1, IR-2, and IR-3). (B) Detection of the possible inversion patterns in SusC/D cluster 1 by PCR. PCR products obtained by various combinations of primers were analyzed by agarose gel electrophoresis. Primer pairs used for each amplification are indicated above each lane. Details, including predicted size for each product, are available in Fig. 5.
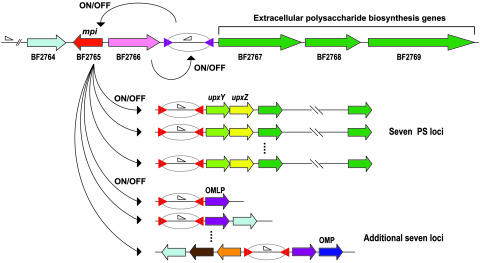
Genome searches of BT strain VPI-5482 and PG strain W83 for IRs containing the six kinds of consensus motif sequences that we identified in BF allowed us to identify 20 possibly invertible regions in BT (including four PS loci), but none in PG (Fig. 8 and Table 7). Remarkably, most of the invertible regions of BT (15 of 20) are colocalized with tyrosine-type site-specific recombinases, again implying that DNA inversions in these regions are locally controlled by cognate recombinases. This finding contrasts sharply with BF strain YCH46, where only five invertible regions are colocalized with site-specific recombinases. Nearly half of the invertible regions on the strain YCH46 genome (14 regions) are flanked with class I IRs regulated by Mpi. Nine invertible regions with class II IRs may also be controlled by some other globally acting recombinase, although the responsible enzyme remains to be identified. Interestingly, the mpi gene is paired with a tyrosine-type site-specific recombinase gene (BF2766) in a head-to-head orientation on the BF strain YCH46 genome (Fig. 4). The BF2766 is followed immediately by a flippable promoter segment flanked by a class IV IR, and thus apparently mediates the on-off switching of the downstream genes for extracellular polysaccharide biosynthesis. Because the intervening region between mpi and BF2766 is very short (15 bp), BF2766-mediated inversion probably controls the expression of Mpi as well, which in turn regulates the expression of 14 loci, including seven PS loci in a hierarchical manner, allowing a coordinate alteration of surface structures.
Fig. 4.
A hypothetical model for the hierarchical control of DNA inversions in the BF genome. Genes for a globally acting site-specific recombinase, Mpi (BF2765), and a tyrosine-type site-specific recombinase (BF2766) are indicated by red and pink arrows, respectively. IRs are indicated by purple or red arrowheads, and promoter sequences are indicated by open triangles. The BF2766 recombinase mediates the inversion of the downstream promoter-containing segment, and thus regulates the selective expression of the accessory PS locus and the mpi gene. The expressed Mpi recombinase in turn regulates the on-off switching of seven PS loci and seven additional loci that are scattered on the BF genomes. Genes for polysaccharide biosynthesis and transport are indicated by green arrows. UpxYgene homologs, UpxZ homologs, outer membrane lipoproteins, an outer membrane protein, an electron transport protein, a polysaccharide deacetylase, and hypothetical proteins are indicated by light green, yellow, purple, blue, orange, brown, and light blue arrows, respectively.
To evade host immune responses and maintain persistent infections, many pathogens have evolved diverse genetic systems for the rapid generation of subpopulations with distinct surface antigenicities (36). On-off switching of gene expression is mediated by variable DNA methylation of regulatory elements, addition and subtraction of base pairs or repeating units (slipped-strands mispairing) in promoters or translational frames, or reversible inversion of genetic elements. Some bacteria such as Mycoplasma and Campylobacter fetus use shufflon-like DNA inversion systems to create variable expression patterns of surface proteins (37, 38). In several pathogens such as Neisseria and trypanosomes, intragenomic recombination or gene conversion generate extensive repertoires of antigenically variant surface molecules (39, 40). In all of these cases, however, the locus responsible for the hypermutability is rather confined to a specific region within the genome (36).
Bacteroides, predominant human and animal colonic commensals, have rather developed numerous DNA inversion systems that are dispersed throughout the genome. Using these systems, Bacteroides can generate a large set of subpopulations selectively expressing a wide range of cell surface structures. This capability, together with the exceptionally high-level expansion of the genes for using dietary polysaccharides, provides a unique strategy for the microbes to establish dominance in the colonic milieu, with its changing nutritional conditions and adaptive host immune system. Notable variations in the two Bacteroides include more prominent gene amplification for polysaccharide utilization in BT and more systematic genetic systems to change surface antigenicity in BF. Correspondingly, BT is more numerous in the normal colon, whereas BF can survive the more intimate association with the intestinal mucosa, evading intensive attack by host immunity and favoring greater infectious potential.
Supplementary Material
Acknowledgments
We thank Drs. H. Yoshikawa, N. Ogasawara, and H. Hayashi for supporting the project and Dr. K. Kurokawa for valuable suggestions in sequence analyses. This work was supported by the Research for the Future Program of the Japan Society for the Promotion of Science (Grants JSPS-RFTF 00L01411 and JSPS-RFTF 00L01412).
Author contributions: T.K. designed research; T.K., A.Y., H.H., H.N., H.T., N.O., S.K., M.H., T.H., and Y.O. performed research; T.K., A.Y., H.H., H.N., H.T., N.O., S.K., M.H., T.H., and Y.O. analyzed data; and T.K. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BF, Bacteroides fragilis; BT, B. thetaiotaomicron; PG, Porphyromonas gingivalis; IR, inverted repeat sequence; CTn, conjugative transposon; PS loci, polysaccharide biosynthesis loci.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AP006841 and AP006842).
References
- 1.Salyers, A. A. (1984) Annu. Rev. Microbiol. 38, 293-313. [DOI] [PubMed] [Google Scholar]
- 2.Namavar, F., Theunissen, E. B. M., Verweij-Van Vught, A. M. J. J., Peerbooms, P. G. M., Bal, M., Hoitsma, H. F. W. & Maclaren, D. M. (1989) J. Med. Microbiol. 29, 171-176. [DOI] [PubMed] [Google Scholar]
- 3.Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., Hooper, L. V. & Gordon, J. I. (2003) Science 299, 2074-2076. [DOI] [PubMed] [Google Scholar]
- 4.Akimoto, S., Ono, T., Tsutsui, H., Kinouchi, T., Kataoka, K. & Ohnishi, Y. (1994) Biochem. Biophys. Res. Commun. 203, 914-921. [DOI] [PubMed] [Google Scholar]
- 5.Kuwahara, T., Sarker, M. R., Ugai, H., Akimoto. S., Shaheduzzaman, S. M., Nakayama, H., Miki, T. & Ohnishi, Y. (2002) FEMS Microbiol. Lett. 207, 193-197. [DOI] [PubMed] [Google Scholar]
- 6.Ewing, B. & Green, P. (1998) Genome Res. 8, 186-194. [PubMed] [Google Scholar]
- 7.Ewing, B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 8.Gordon, D., Abajian, C. & Green, P. (1998) Genome Res. 8, 195-202. [DOI] [PubMed] [Google Scholar]
- 9.Sakiyama, T., Takami, H., Ogasawara, N., Kuhara, S., Kozumi, T., Doga, K., Ohyama, A. & Horikoshi, K. (2000) Biosci. Biotechnol. Biochem. 64, 670-673. [DOI] [PubMed] [Google Scholar]
- 10.Badger, J. H. & Olsen, G. J. (1999) Mol. Biol. Evol. 16, 512-524. [DOI] [PubMed] [Google Scholar]
- 11.Yada, T., Nakao, M., Totoki, Y. & Nakai, K. (1999) Bioinformatics 15, 987-993. [DOI] [PubMed] [Google Scholar]
- 12.Frishman, D., Mironov, A., Mewes, H. W. & Gelfand, M. (1998) Nucleic Acids Res. 26, 2941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001) Nucleic Acids Res. 29, 22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nelson, K. E., Fleischmann, R. D., DeBoy, R. T., Paulsen, I. T., Fouts, D. E., Eisen, J. A., Daugherty, S. C., Dodson, R. J., Durkin, A. S., Gwinn, M., et al. (2003) J. Bacteriol. 185, 5591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lobry, J. R. (1996) Mol. Biol. Evol. 13, 660-665. [DOI] [PubMed] [Google Scholar]
- 18.Patrick, S., Parkhill, J., McCoy, L. J., Lennard, N., Larkin, M. J., Collins, M., Sczaniecka, M. & Blakely, G. (2003) Microbiology 149, 915-924. [DOI] [PubMed] [Google Scholar]
- 19.Bedzyk, L. A., Shoemaker, N. B., Young, K. E. & Salyers, A. A. (1992) J. Bacteriol. 174, 166-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikolich, M. P., Shoemaker, N. B. & Salyers, A. A. (1994) J. Bacteriol. 176, 6606-6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., Zwahlen, M.-C., Desiere, F., Bork, P., Delley, M., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimizu, T., Ohtani, K., Hirakawa, H., Ohshima, K., Yamashita, A., Shiba, T., Ogasawara, N., Hattori, M., Kuhara, S. & Hayashi, H. (2002) Proc. Natl. Acad. Sci. USA 99, 996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson, K. L. & Salyers, A. A. (1989) J. Bacteriol. 171, 3192-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Elia, J. N. & Salyers, A. A. (1996) J. Bacteriol. 178, 7180-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reeves, A. R., D'Elia, J. N., Frias, J. & Salyers, A. A. (1996) J. Bacteriol. 178, 823-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho, K. H. & Salyers, A. A. (2001) J. Bacteriol. 183, 7224-7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindberg, A. A., Weintraub, A., Kasper, D. L. & Lonngren, J. (1982) Scand. J. Infect. Dis. Suppl. 35, 45-52. [PubMed] [Google Scholar]
- 28.Onderdonk, A. B., Kasper, D. L., Cisneros, R. L. & Bartlett, J. (1997) J. Infect. Dis. 136, 82-89. [DOI] [PubMed] [Google Scholar]
- 29.Coyne, M. J., Kalka-Moll, W., Tzianabos, A. O., Kasper, D. L. & Comstock, L. E. (2000) Infect. Immun. 68, 6176-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Comstock, L. E., Pantosti, A. & Kasper, D. L. (2000) Infect. Immun. 68, 6182-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coyne, M. J., Tzianabos, A. O., Mallory, B. C., Carey, V. J., Kasper, D. L. & Comstock, L. E. (2001) Infect. Immun. 69, 4342-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krinos, C. M., Coyne, M. J., Weinacht, K. G., Tzianabos, A. O., Kasper, D. L. & Comstock, L. E. (2001) Nature 414, 555-558. [DOI] [PubMed] [Google Scholar]
- 33.Coyne, M. J., Weinacht, K. G., Krinos, C. M. & Comstock, L. E. (2003) Proc. Natl. Acad. Sci. USA 100, 10446-10451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzianabos, A. O., Onderdonk, A. B., Rosner, B., Cisneros, R. L. & Kasper, D. L. (1993) Science 262, 416-419. [DOI] [PubMed] [Google Scholar]
- 35.Komano, T. (1999) Annu. Rev. Genet. 33, 171-191. [DOI] [PubMed] [Google Scholar]
- 36.Deitsch, K. W., Moxon, E. R. & Wellems, T. E. (1997) Microbiol. Mol. Biol. Rev. 61, 281-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhugra, B., Voelker, L. L., Zou, N., Yu, H. & Dybvig, K. (1995) Mol. Microbiol. 18, 703-714. [DOI] [PubMed] [Google Scholar]
- 38.Dworkin, J. & Blaser, M. J. (1997) Mol. Microbiol. 26, 433-440. [DOI] [PubMed] [Google Scholar]
- 39.Rudel, T., Scheurerpflug, I. & Meyer, T. F. (1992) Mol. Microbiol. 6, 3439-3450. [DOI] [PubMed] [Google Scholar]
- 40.Roth, C., Jacquemot, C., Giroud, C., Bringaud, F., Eisen, H. & Baltz, T. (1991) Res. Microbiol. 142, 725-730. [DOI] [PubMed] [Google Scholar]
- 41.Bayley, D. P., Rocha, E. R. & Smith, C. J. (2000) FEMS Microbiol. Lett. 193, 149-154. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.