Abstract
Ubiquitin family peptide modifications regulate the functions and stabilities of many proteins. We have developed an approach for the visualization of ubiquitinated proteins in living cells designated ubiquitin-mediated fluorescence complementation (UbFC). This approach is based on complementation among fragments of fluorescent proteins when they are brought together by the covalent conjugation of ubiquitin fused to one fragment to a substrate protein fused to a complementary fragment. The UbFC strategy enables simultaneous visualization of proteins modified by different ubiquitin family peptides and comparison of their effects on protein localization. Visualization of ubiquitinated Jun revealed that it was localized predominantly to cytoplasmic structures. In contrast, Jun conjugated to small ubiquitin-related modifier 1 (SUMO1) was localized to subnuclear foci. Comparison of the distribution of ubiquitinated Jun with markers for various cytoplasmic compartments revealed that ubiquitinated Jun was localized to lysosomal vesicles. Fractionation of cell lysates confirmed that the majority of ubiquitinated Jun partitioned to the cytoplasmic fraction, and density gradient centrifugation analysis demonstrated that it cosedimented with lysosomal β-hexosaminidase activity. Mutation of a recognition sequence for the E3 ligase Itch/AIP4 prevented Jun ubiquitination and stabilized it in cells. Inhibition of lysosomal protein degradation by bafilomycin or chloroquine stabilized Jun but had no effect on the stability of mutated Jun that was not ubiquitinated by Itch/AIP4. The visualization of ubiquitinated Jun in living cells has uncovered a lysosomal pathway for Jun degradation that involves ubiquitination by Itch/AIP4.
The small peptide ubiquitin is covalently linked to lysine residues on many proteins (1, 2). Ubiquitination was originally identified as a signal for proteasomal degradation (3, 4). It also regulates other cellular processes, including protein trafficking and transcription activation (5–9). Several peptides related to ubiquitin have been identified (10–12). Modifications by different peptides have distinct effects on the functions of the modified proteins. The mechanisms whereby ubiquitin family peptides affect protein function are being investigated.
Many transcription regulatory proteins can be modified by ubiquitin family peptides (6, 8, 9, 13–22). Ubiquitination is thought to control the rates of transcription factor turnover by targeting them for degradation by proteasomes (13–16). However, ubiquitination can also regulate transcription through mechanisms other than transcription factor degradation (6, 9, 18). Ubiquitin family peptides have been implicated in the control of the subnuclear localization of transcription factors (19–22). The relationship between the control of subcellular localization and the regulation of transcription activation remains to be elucidated.
Jun is a transcription regulatory protein whose expression is induced by many extracellular stimuli (23). The transient response of the cell to these stimuli is controlled in part by the rapid degradation of Jun after synthesis. Jun ubiquitination has been studied in both cell extracts and reconstituted ubiquitination reactions (13–16). Ubiquitinated Jun is thought to be degraded by proteasomes because proteasome inhibitors stabilize Jun (15). However, ubiquitinated Jun has not been directly visualized in living cells, and alternative pathways for Jun degradation have not been investigated.
Materials and Methods
Plasmid Construction. The sequences encoding amino acids residues 1–172 of enhanced yellow fluorescent protein (designated YN) or enhanced cyan fluorescent protein (designated CN) were fused to the 5′ ends of the coding regions for ubiquitin and small ubiquitin-related modifier 1 (SUMO1) by using ANSSIDLISVPVEYPYDVPDYASR linkers. The chimeric coding regions were cloned into the pFLAG-CMV2 (Sigma) to produce plasmids encoding YN-Ub, YN-SUMO1, and CN-SUMO1. The plasmids encoding Jun and Jun257–318 fused to amino acid residues 155–238 of enhanced cyan fluorescent protein (CFP) were as described (24, 25) and are designated Jun-CC and bJun-CC. The plasmid encoding Jun-CFP was constructed by inserting the coding region of Jun into pECFP (Clontech). The Y170F mutation in Jun (JunY170F), the deletion of G100 and G101 in SUMO1 (SUMOΔG), and the K48R, K63R, G75K, and G76L mutations in ubiquitin (UbMut) were generated by using PCR and were confirmed by DNA sequencing.
Cells and Antibodies. COS-1, NIH 3T3, and HEK293T cells were cultured as recommended by the American Type Culture Collection. Polyclonal anti-c-Jun, anti-Rab6, anti-TSG101, anti-Myc, and antihemagglutinin antibodies were from Santa Cruz Biotechnology. Monoclonal LAMP1 (Clone H4A3), LAMP2 (clone ABL-93) (Developmental Studies Hybridoma Bank at the University of Iowa, Iowa City), anti-Xpress (Invitrogen), and anti-Flag (Sigma) antibodies were used.
Analysis of Fluorescence Complementation. Cells transfected with plasmids encoding the indicated combinations of fusion proteins were incubated at 37°C for 24 h and then transferred to 30°C for 4–16 h to promote fluorophore maturation. The fluorescence emissions of the cells were imaged as described (24, 25).
Immunofluorescence and Confocal Analysis. Cells transfected with plasmids encoding the indicated combination of fusion proteins. were fixed with 3.7% paraformaldehyde after 36 h. Fixed cells were washed with 0.1% Triton X-100 and incubated with the antibodies indicated followed by secondary antibody labeled with Alexa Fluor 594. Fluorescence images were collected by confocal microscopy.
Immunoprecipitation, Immunoblotting, and Metabolic Labeling. Extracts were prepared from COS-1 cells transfected with the plasmids indicated. The extracts were analyzed by immunoprecipitation and immunoblotting as described (26). Metabolic labeling with [35S]methionine/[35S]cysteine and pulse–chase analysis were performed as described (26).
Cell Fractionation and Lysosome Isolation. Cells were lysed in hypotonic buffer as described (26). The nuclei were pelleted by centrifugation at 3,000 × g for 5 min. The nuclear pellet was resuspended in 1% Nonidet P-40 cell lysis buffer, and insoluble material was removed by centrifugation at 16,000 × g for 10 min. Lysosomes were purified by Percoll density gradient sedimentation, and β-hexosaminidase activity was measured as described (27).
Results
Visualization of Ubiquitination in Living Cells. Ubiquitination is generally detected by immunoprecipitation followed by Western blot analysis using antibodies directed against epitope tags linked to ubiquitin and to a putative substrate protein (28). This approach has the inherent limitation that it does not allow analysis of ubiquitination in living cells. The study of ubiquitination in cells is further impeded by the small subpopulation of each protein that is ubiquitinated at any one time.
To develop an approach for the visualization of specific ubiquitinated proteins in living cells, we took advantage of the ability of fragments of selected fluorescent proteins to form a fluorescent complex when brought together by the association of proteins fused to the fragments (24, 25). We hypothesized that fusion of ubiquitin to one fragment of a fluorescent protein, as well as a putative substrate to the complementary fragment, would allow selective visualization of the ubiquitin conjugate in living cells. We designate this approach ubiquitin-mediated fluorescence complementation (UbFC).
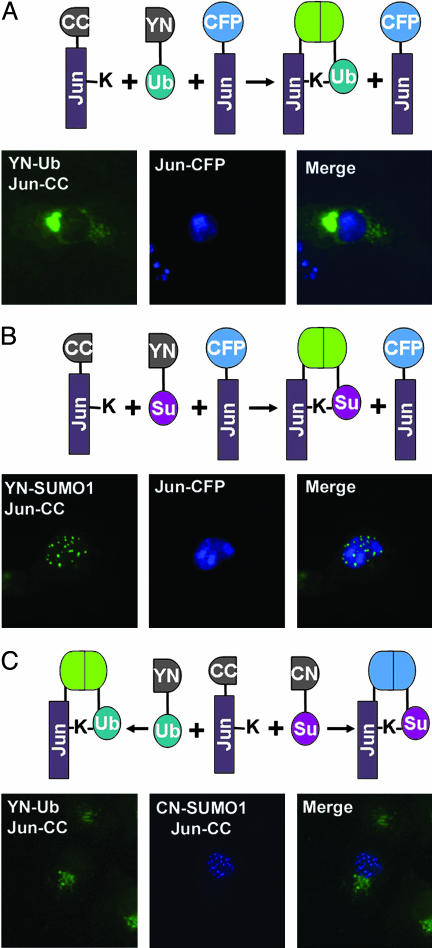
To test the feasibility of the UbFC approach, we fused the N-terminal fragment of the yellow fluorescent protein to ubiquitin and the complementary fragment to Jun. Plasmids encoding both fusion proteins were transfected into COS-1 cells, and the cells were observed by fluorescence microscopy. Cells transfected with both plasmids, but not cells transfected with either plasmid alone, were fluorescent (Fig. 1A Left). Surprisingly, the fluorescence was located mainly outside the nucleus and was concentrated in small spherical structures in the cytoplasm, including an aggregation of these structures that was frequently adjacent to the nucleus. Similar patterns of fluorescence complementation were observed when the fusion proteins were expressed in HEK293T or NIH 3T3 cells. To ascertain that expression of the fusion proteins did not disrupt the normal mechanisms for Jun localization, we coexpressed Jun fused to full length CFP in the same cells. Jun-CFP was localized mainly to the nucleus (Fig. 1 A Center). These results suggest that ubiquitination of Jun caused export from the nucleus into discrete cytoplasmic structures.
Fig. 1.
Visualization of Jun conjugated to ubiquitin and SUMO1 in living cells. YN-Ub, Jun-CC, and Jun-CFP (A); YN-SUMO1, Jun-CC, and Jun-CFP (B); and YN-Ub, CN-SUMO1, and Jun-CC (C) were expressed in COS-1 cells. Fluorescence images were acquired 36 h after transfection by using filters selective for YN-CC (Left) and CFP (Center). The two images were merged (Right). The images shown are representative of >90% of the fluorescent cells in each population. The diagrams above the images describe each experimental design.
Different ubiquitin family peptides have distinct biological effects when conjugated to substrate proteins. We examined the effect of the SUMO1 on Jun in cells using the UbFC assay. Expression of SUMO1 and Jun fused to complementary fluorescent protein fragments produced fluorescence that was localized to subnuclear foci (Fig. 1B Left). Coexpressed Jun-CFP was distributed throughout the nucleoplasm and enriched in nucleoli (Fig. 1B Center). The results of these experiments indicate that SUMO1 conjugation could induce Jun localization to specific subnuclear structures.
The distinct subcellular locations of the fluorescent complexes formed by Jun with ubiquitin versus SUMO1 suggest that different modifications have distinct effects on Jun localization. To compare the effects of these modifications in the same cell, we used a multicolor adaptation of the UbFC assay (25). We coexpressed ubiquitin and SUMO1 fused to fragments of different fluorescent proteins together with Jun fused to a complementary fragment. The ubiquitin and SUMO1 conjugates of Jun exhibited nonoverlapping distributions similar to those that were observed when the conjugates were produced in separate cells (Fig. 1C). The distinct subcellular distributions of these conjugates were therefore not caused by distinct cellular responses to the expression of ubiquitin versus SUMO1 but were determined by intrinsic localization determinants on these conjugates.
The UbFC approach requires fusion of fluorescent protein fragments to the ubiquitin family peptide as well as to the putative substrate. We examined whether fusion of the fluorescent protein fragments to ubiquitin and SUMO1 affected their conjugation to substrate proteins. Western blot analysis of extracts from cells that expressed ubiquitin fused to a fluorescent protein fragment produced a high-molecular-weight smear of conjugates that was of similar intensity but of higher apparent molecular weight compared with that observed when extracts from cells that expressed epitope-tagged ubiquitin were analyzed (Fig. 7A, which is published as supporting information on the PNAS web site). Likewise, Western blot analysis of anti-Jun immunoprecipitates from cells expressing SUMO1 fused to a fluorescent protein fragment produced two bands of similar intensities to those observed when SUMO1 lacking the fusion was expressed (Fig. 7B). The two bands likely correspond to Jun modified at one or two lysine residues. The fraction of Jun modified at two sites was much higher than predicted if the sites were modified independently. Thus, ubiquitin and SUMO1 fused to the fluorescent protein fragment were conjugated to substrates as efficiently as the peptides lacking the fusions.
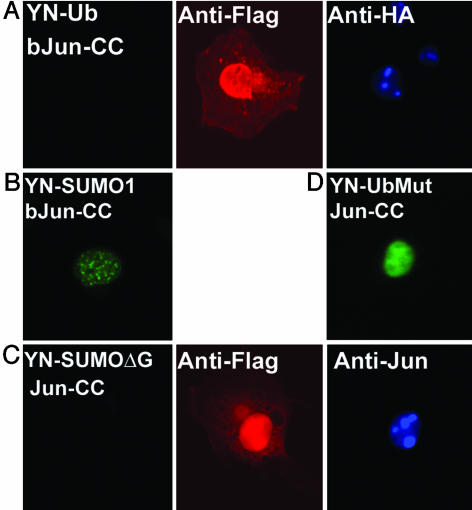
The distinct distributions of the fluorescent complexes formed by Jun with ubiquitin versus SUMO1 suggested that the complexes represented specific conjugates. To ascertain the specificity of the UbFC assay, we examined the effects of mutations on fluorescence complementation. Deletion of regions of Jun outside the bZIP domain eliminated fluorescence complementation with ubiquitin but did not prevent fluorescence complementation with SUMO1 (Fig. 2 A and B). The level of ubiquitination of the bZIP domain of Jun detected by Western blot analysis of cell extracts was >10-fold lower than that of full length Jun (data not shown). The selective loss of fluorescence complementation caused by deletion of regions outside the bZIP domain of Jun corroborates the specificity of the UbFC assay of Jun ubiquitination.
Fig. 2.
Specificity of the UbFC assay. YN-Ub and bJun-CC (A), YN-SUMO1 and bJun-CC (B), YN-SUMO1ΔG and Jun-CC (C), and YN-UbMut and Jun-CC (D) were expressed in COS-1 cells. Fluorescence images were acquired by using cells fixed 36 h after transfection. The expression of the fusion proteins in cells that did not display fluorescence complementation was visualized by using the antibodies indicated (Center and Right).
Fluorescence complementation can be facilitated by the covalent or noncovalent association of proteins fused to the fluorescent protein fragments. We examined the effects of mutations that prevented conjugation on fluorescence complementation. Deletion of the C-terminal glycines of SUMO1 eliminated the fluorescence complementation with Jun (Fig. 2C). This mutation also eliminated the SUMO1 conjugates of Jun detected in cell extracts (Fig. 7B). The fusions were expressed in overlapping regions of the cell, and the level of expression of mutated SUMO1 was comparable to that observed for the wild-type peptide. The lack of fluorescence complementation therefore indicates that the complementation observed in the UbFC assay reflected the covalent conjugation of SUMO1 to Jun. In contrast, when mutated ubiquitin fused to the N-terminal fragment of yellow fluorescent protein was expressed with Jun fused to the complementary fragment, fluorescence complementation was detected (Fig. 2D). This fluorescence was localized predominantly to the nucleus and exhibited a distribution identical to that of unmodified Jun. No covalent conjugates of the mutated ubiquitin were detected in cell extracts, indicating that the complementation resulted from noncovalent association of the fusion proteins. The nonconvalent bimolecular fluorescent complexes formed by mutated ubiquitin with Jun were exclusively nuclear, indicating that the cytoplasmic localization of complexes formed by wild-type ubiquitin required covalent conjugation to Jun.
Fluorescence complementation requires that the fragments of the fluorescent protein can associate with each other in the complex. To examine whether conjugates with different steric arrangements of the fluorescent protein fragments had different subcellular distributions, we examined complementation using proteins in which the fluorescent protein fragments were fused to different ends of Jun. These proteins formed fluorescent complexes with ubiquitin that exhibited identical subcellular distributions (Fig. 8 A and B, which is published as supporting information on the PNAS web site). In contrast, no fluorescence complementation was observed when Jun fused to a fluorescent protein fragment on the N-terminal end was expressed together with SUMO1 fused to the complementary fragment (Fig. 8D). The difference between the effects of the position of the fusion on fluorescence complementation with ubiquitin versus SUMO1 is consistent with the greater reach and flexibility predicted for a polyubiquitin chain relative to a mono-SUMO1 adduct.
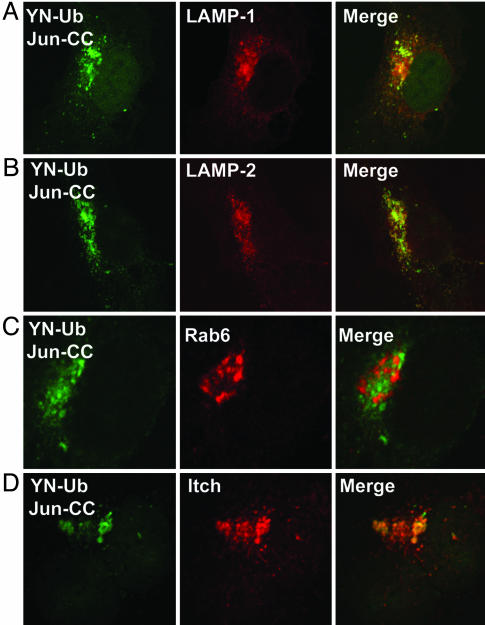
Subcellular Localization of Ubiquitinated Jun. To identify the subcellular compartment where ubiquitinated Jun was localized, we compared the locations of fluorescence complementation with those of cellular proteins with known subcellular distributions. Because ubiquitin-conjugated Jun was distributed in a pattern reminiscent of transport vesicles, we compared its localization with markers for various endosomal compartments. We found that ubiquitinated Jun closely colocalized with the LAMP1 and LAMP2 lysosomal membrane proteins (Fig. 3 A and B) (29). In contrast, there was no colocalization of ubiquitin-conjugated Jun with the small GTPase Rab6, which participates in vesicular sorting in the Golgi and endoplasmic reticulum (30) (Fig. 3C). These results suggest that ubiquitin-conjugated Jun is localized to lysosomes.
Fig. 3.
Subcellular localization of ubiquitinated Jun. YN-Ub and Jun-CC were expressed in COS-1 cells, and cells fixed 36 h after transfection were immunostained with antibodies directed against LAMP1 (A), LAMP2 (B), and Rab6 (C). (D) Xpress-tagged Itch was coexpressed, and the cells were immunostained with anti-Xpress antibody. Fluorescence images were collected by confocal microscopy by using filters selective for YN-CC (Left) and Alexa Fluor 594 (Center) fluorescence. The merged images are shown (Right).
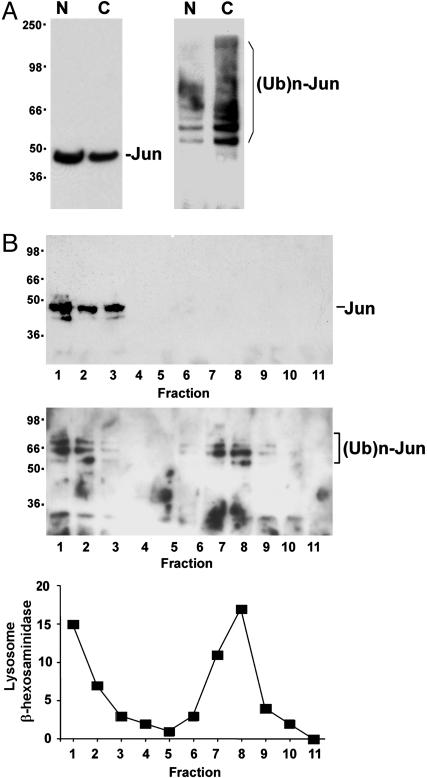
To complement the visualization of ubiquitinated Jun in living cells, we examined the fractionation of ubiquitinated Jun in cell lysates. Lysates from cells that expressed epitope-tagged Jun and ubiquitin were separated into nuclear and cytoplasmic fractions. The majority of unmodified Jun was found in the nuclear fraction (Fig. 4A Left). In contrast, most of the ubiquitinated Jun was found in the cytoplasmic fraction (Fig. 4A Right). Because these experiments were performed by using Jun and ubiquitin that were not fused to fluorescent protein fragments, the results demonstrate that the cytoplasmic localization of the conjugate was independent of fluorescent complex formation. The majority of ubiquitinated endogenous Jun was also found in the cytoplasmic fraction (data not shown). The partitioning of ubiquitinated Jun into the cytoplasmic fraction is consistent with the cytoplasmic localization of the UbFC conjugate in living cells.
Fig. 4.
Subcellular fractionation of ubiquitinated Jun. (A) Nuclear (N) and cytoplasmic (C) fractions were prepared from cells that expressed Jun and ubiquitin tagged with the Xpress and hemagglutinin (HA) epitope tags, respectively. Aliquots of the fractions were analyzed by Western blotting with anti-Xpress antibody to detect total Jun (Left). The fractions were immunoprecipitated by using anti-Xpress antibody, and the immunoprecipitates were analyzed by Western blotting by using anti-HA antibody to detect ubiquitinated Jun (Right). (B) The cytoplasmic extract was fractionated by density gradient sedimentation. Total Jun (Top) and ubiquitinated Jun (Middle) in each fraction was analyzed as described in A. (Bottom) Lysosomal β-hexosaminidase activity in each fraction is plotted as a percentage of the total activity applied to the gradient.
To examine whether the ubiquitinated Jun in cell lysates was associated with lysosomes, we analyzed the cytoplasmic fraction by density gradient sedimentation. Lysosomes have a high density and sediment toward the bottom of the gradient, whereas other membranes and soluble proteins have a lower density and sediment near the top of the gradient. Virtually all unmodified Jun was recovered in the top three fractions (Fig. 4B Top). Ubiquitinated Jun separated into two populations, one at the top and a second toward the bottom of the gradient (Fig. 4B Middle). The location of lysosomes in the gradient was determined by measuring lysosomal β-hexosaminidase activity (Fig. 4B Bottom). β-hexosaminidase activity cosedimented in all fractions with ubiquitinated Jun. The ubiquitinated Jun and β-hexosaminidase activity at the top of the gradient may be due to lysis of part of the lysosomes during cell extraction. The cofractionation of ubiquitinated Jun with lysosomal β-hexosaminidase is consistent with the lysosomal localization of the conjugate detected by UbFC analysis.
Identification of the E3 Ligase That Ubiquitinated Jun in Cells. Several different E3 ligases can facilitate Jun ubiquitination in reconstituted reactions in vitro and subunits of these E3 ligases can interact with Jun in cell extracts (15, 16). We attempted to identify the E3 ligase(s) that mediated the ubiquitination of Jun detected in living cells. Both Jun and JunB can interact with the E3 ligase Itch in cell extracts, and Itch overexpression increases the level of JunB ubiquitination detected by Western analysis (26). Itch is localized to lysosomes in some cells (31). We compared the distributions of ubiquitin-conjugated Jun and Itch in COS-1 cells (Fig. 3D). The majority of ubiquitinated Jun was localized to compartments that also contained high levels of Itch.
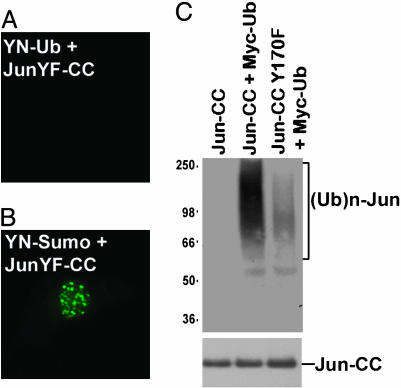
To determine whether Itch was responsible for the ubiquitination of Jun in cells, we sought to identify mutations in Jun that prevented its recognition by Itch. Jun contains a sequence resembling the Itch recognition motif (167PPVY170) within the N-terminal region required for ubiquitination. A single amino acid substitution in this sequence (Y170F) eliminated fluorescence complementation with ubiquitin (Fig. 5A). The fusion proteins were coexpressed in the same cells and exhibited overlapping distributions (data not shown). Coexpression of JunY170F with SUMO1 fused to the same fragments produced fluorescence complementation identical to that observed for wild-type Jun (Fig. 5B). Similar results were obtained in both COS-1 and HEK293T cells. Elimination of the Itch recognition sequence in Jun selectively abolished fluorescence complementation with ubiquitin in living cells. We also examined the role of Itch in Jun ubiquitination by Western blot analysis. The Y170F mutation dramatically reduced the level of ubiquitinated Jun detected in cell extracts (Fig. 5C). Taken together, the results of these experiments suggest that Jun ubiquitination in these cells is catalyzed primarily by Itch and/or E3 ligases that recognize the same sequence motif in Jun.
Fig. 5.
Specificity of Jun ubiquitination by Itch in living cells and cell extracts. (A and B) YN-Ub and JunY170F-CC (A) or YN-SUMO1 and JunY170F-CC (B) were expressed in COS-1 cells, and fluorescence images were acquired 36 h after transfection. (C) The proteins indicated above each lane were expressed in HEK293T cells. Cell extracts were precipitated with anti-Jun antibody, and equal amounts of the proteins were analyzed by Western blot analysis with anti-Myc antibody to detect ubiquitinated proteins (Upper). The membrane was reprobed with anti-Jun antibody to confirm that equal amounts of Jun were loaded in each lane (Lower).
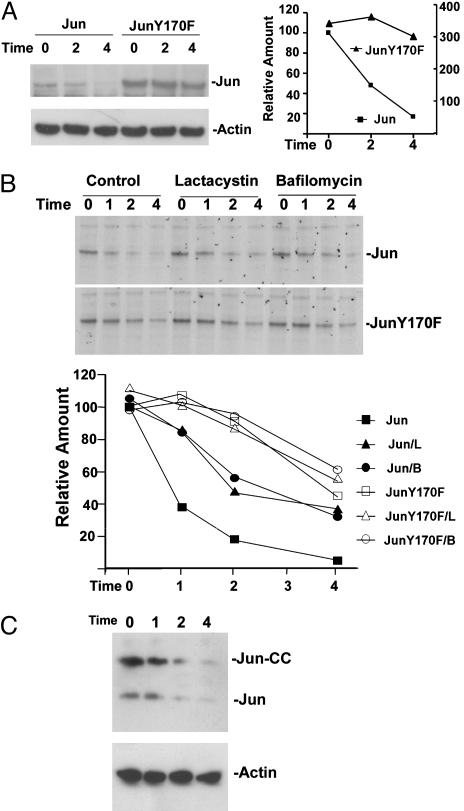
Degradation of Ubiquitinated Jun. To investigate the role of ubiquitination by Itch in Jun turnover, we examined the effects of the Y170F mutation on the stability of Jun. The steady-state level of JunY170F expression was higher than that observed for wild-type Jun when equal amounts of plasmids encoding the proteins were transfected into cells (Fig. 6A). When protein synthesis was blocked by the addition of cycloheximide, >80% of wild-type Jun was degraded in 4 h. In contrast, less than half of JunY170F was degraded in 4 h (longer incubation caused morphological changes and cell death). To ascertain whether the difference between the levels of expression of wild-type and Y170F Jun was caused by a difference between their rates of degradation, we measured their half-lives by pulse–chase analysis (Fig. 6B). Wild-type Jun had a half-life of 1 h, whereas JunY170F has a half-life of 4 h. Thus, the Y170F mutation markedly stabilized Jun in transfected cells.
Fig. 6.
Stabilization of Jun by the Y170F mutation and by inhibitors of proteasomes and lysosomal H+-ATPase. (A) Jun and JunY170F with Xpress epitope tags were expressed in HEK293T cells. Fifty micromolar cycloheximide was added 24 h after transfection (Time 0). The cells were harvested at the times (h) indicated above the lanes, and cell extracts were analyzed by Western blotting with antibodies directed against the Xpress tag (Left Upper) and actin (Left Lower). The relative amounts of Jun were quantified and are plotted in the graph (Right) (Jun levels are plotted on the left axis, and JunY170F levels are plotted on the right axis). (B) COS-1 cells that expressed Jun (Top) or JunY170F (Middle) with Xpress epitope tags were labeled with 35S amino acids. The radiolabel was washed out (Time 0), and the cells were cultured in the absence (Control) or in the presence of 20 μM lactacystin or 0.5 μM bafilomycin A. The cells were harvested at the times indicated above the lanes, and proteins were precipitated by using anti-Xpress antibodies and resolved by SDS/PAGE. The amounts of Jun (filled symbols) and JunY170F (open symbols) were quantified and plotted in Bottom (L, lactacystin; B, bafilomycin). (C) Jun-CC was expressed in COS-1 cells. Cycloheximide was added 24 h after transfection (Time 0). The cells were harvested at the times indicated above the lanes (h), and cell extracts were analyzed by Western blotting by using antibodies directed against Jun (Upper) and actin (Lower). The bands corresponding to transiently expressed Jun-CC and endogenous Jun are indicated.
Because ubiquitinated Jun was localized to lysosomal vesicles, and the Y170F mutation that eliminated Jun ubiquitination also reduced its rate of degradation, we examined the role of lysosomes in Jun degradation. Treatment of cells with chloroquine or bafilomycin A that inhibit lysosomal H+-ATPase extended the half-life of Jun to 2 h (Fig. 6B and data not shown). Treatment of cells with the MG132 or lactacystin proteasome inhibitors also increased the half-life of Jun to 2 h (Fig. 6B and data not shown). None of these inhibitors affected the degradation of JunY170F. Thus, either the degradation of Jun involved both the proteasomal and lysosomal pathways, or these inhibitors had indirect effects on multiple pathways.
To examine whether transiently transfected Jun fused to a fluorescent protein fragment and epitope-tagged Jun were valid models for the study of Jun degradation, we compared their half-lives with that of endogenous Jun in the same cells. All of the proteins exhibited virtually identical rates of degradation (Fig. 6C). The average level of exogenous Jun in transfected cells was 6-fold higher than the level of endogenous Jun. This modest level of overexpression did not alter the rate of degradation, suggesting that the transiently expressed fusion proteins were degraded via the same pathway(s) as endogenous Jun.
Discussion
The direct visualization of ubiquitinated Jun in living cells using the UbFC approach has uncovered a new pathway for Jun degradation mediated by ubiquitination by Itch and translocation to lysosomal vesicles. The exclusion of ubiquitinated Jun from the nucleus provides a mechanism for rapid abolition of its transcriptional activity. Sequestration of ubiquitinated Jun to lysosomal vesicles can prevent reactivation of the protein by ubiquitin hydrolases and may be important under conditions when protein degradation is inefficient due to limiting ATP or other factors.
Analysis of the effects of ubiquitin family peptide conjugation on protein localization using the UbFC approach requires that the fluorescent protein fragment fusions do not affect localization of the conjugate. In the case of Jun, the results of UbFC analysis and subcellular fractionation using proteins without fluorescent protein fragment fusions produced concordant results. Furthermore, the fusion proteins exhibited subcellular distributions indistinguishable from those of proteins lacking the fusions, and the Jun fusion had a turnover rate comparable to that of endogenous Jun.
The Y170F mutation that prevented Jun ubiquitination by Itch reduced the rate of Jun degradation by >70%. The Itch recognition motif was therefore required for the major pathway of Jun degradation in COS-1 cells. The 167PPVY170 recognition sequence for Itch overlaps a putative phosphorylation site for Abl kinase in Jun (32). Phosphorylation of this tyrosine by Abl or other tyrosine kinases could regulate recognition of Jun by Itch/AIP4 or other HECT family E3 ligases.
Two other E3 ligases that can ubiquitinate Jun have been identified (15, 16). The SCFFbw7 E3 ligase specifically recognizes Jun phosphorylated by JNK (16). Overexpression of the DCXhDET1-hCOP1 E3 ligase reduced the level of Jun, and depletion of individual components of the complex increased the half-life of Jun by ≈30% (15). The multisubunit SCFFbw7 and DCXhDET1-hCOP1 RING-type E3 ligases are structurally unrelated to the single-polypeptide HECT-type E3 ligase Itch. They recognize Jun sequences that are more than 50 amino acid residues away from the 167PPVY170 motif. It is therefore unlikely that the Y170F mutation in Jun affected ubiquitination by these RING-type E3 ligases. Jun ubiquitinated by DCXhDET1-hCOP1 is thought to be degraded by proteasomes, because degradation in cells that overexpressed this E3 ligase is inhibited by MG132 (15). The use of several ubiquitin ligases and degradation pathways for the regulation of Jun stability provides the potential for control of Jun turnover through multiple independent mechanisms. Visualization of the subcellular localization of Jun ubiquitinated by different E3 ligases and analysis of the mechanisms of degradation of these conjugates may allow comparison of these pathways of Jun degradation.
Supplementary Material
Acknowledgments
We thank Jack Dixon (University of California at San Diego, La Jolla) for plasmids encoding ubiquitin and SUMO1, Yun-Cai Liu (La Jolla Institute for Allergy and Immunology, La Jolla, CA) for the Itch expression vector and discussions regarding the role of Itch in the ubiquitination of Jun, Limin Zhang for the preparation of reagents and assistance with some experiments, and members of the Kerppola laboratory for helpful discussions. This work was supported by grants from the Cancer Cell Biology Training Program (to D.F.) and the Human Frontier Science Program (to T.K.K.) and by the Howard Hughes Medical Institute.
Author contributions: D.F. and T.K.K. designed research, performed research, contributed new reagents/analytic tools, analyzed data, and wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: UbFC, ubiquitin-mediated fluorescence complementation; SUMO1, small ubiquitin-related modifier 1; CFP, cyan fluorescent protein.
References
- 1.Goldknopf, I. L., Wilson, G., Ballal, N.R. & Busch, H. (1980) J. Biol. Chem. 255, 10555–10558. [PubMed] [Google Scholar]
- 2.Pickart, C. M. (2004) Cell 116, 181–190. [DOI] [PubMed] [Google Scholar]
- 3.McGuire, M. J., Croall, D. E. & DeMartino, G. N. (1988) Arch. Biochem. Biophys. 262, 273–285. [DOI] [PubMed] [Google Scholar]
- 4.Driscoll, J. & Goldberg, A. L. (1990) J. Biol. Chem. 265, 4789–4792. [PubMed] [Google Scholar]
- 5.Hicke, L. & Riezman, H. (1996) Cell 84, 277–287. [DOI] [PubMed] [Google Scholar]
- 6.Salghetti, S. E., Caudy, A. A., Chenoweth, J. G. & Tansey, W. P. (2001) Science 293, 1651–1653. [DOI] [PubMed] [Google Scholar]
- 7.Babst, M., Odorizzi, G., Estepa, E. J. & Emr, S. D. (2000) Traffic 1, 248–258. [DOI] [PubMed] [Google Scholar]
- 8.Conaway, R. C., Brower, C. S. & Conaway, J. W. (2002) Science 296, 1254–1258. [DOI] [PubMed] [Google Scholar]
- 9.von der Lehr, N., Johansson, S., Wu, S., Bahram, F., Castell, A., Cetinkaya, C., Hydbring, P., Weidung, I., Nakayama, K., Nakayama, K. I., et al. (2003) Mol. Cell 11, 1189–1200. [DOI] [PubMed] [Google Scholar]
- 10.Kumar, S., Yoshida, Y. & Noda, M. (1993) Biochem. Biophys. Res. Commun. 195, 393–399. [DOI] [PubMed] [Google Scholar]
- 11.Mahajan, R., Delphin, C., Guan, T., Gerace, L. & Melchior, F. (1997) Cell 88, 97–107. [DOI] [PubMed] [Google Scholar]
- 12.Schwartz, D. C. & Hochstrasser, M. (2003) Trends Biochem. Sci. 28, 321–328. [DOI] [PubMed] [Google Scholar]
- 13.Treier, M., Staszewski, L. M. & Bohmann, D. (1994) Cell 78, 787–798. [DOI] [PubMed] [Google Scholar]
- 14.Musti, A. M., Treier, M. & Bohmann, D. (1997) Science 275, 400–402. [DOI] [PubMed] [Google Scholar]
- 15.Wertz, I. E., O'Rourke, K. M., Zhang, Z., Dornan, D., Arnott, D., Deshaies, R. J. & Dixit, V. M. (2004) Science 303, 1371–1374. [DOI] [PubMed] [Google Scholar]
- 16.Nateri, A. S., Riera-Sans, L., Da Costa, C. & Behrens, A. (2004) Science 303, 1374–1378. [DOI] [PubMed] [Google Scholar]
- 17.Seeler, J. S. & Dejean, A. (2003) Nat. Rev. Mol. Cell. Biol. 4, 690–699. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser, P., Flick, K., Wittenberg, C. & Reed, S. I. (2000) Cell 102, 303–314. [DOI] [PubMed] [Google Scholar]
- 19.Muller, S., Matunis, M. J. & Dejean, A. (1998) EMBO J. 17, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross, S., Best, J. L., Zon, L. I. & Gill, G. (2002) Mol. Cell 10, 831–842. [DOI] [PubMed] [Google Scholar]
- 21.Fogal, V., Gostissa, M., Sandy, P., Zacchi, P., Sternsdorf, T., Jensen, K., Pandolfi, P. P., Will, H., Schneider, C. & Del Sal, G. (2000) EMBO J. 19, 6185–6195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez, M. S., Desterro, J. M., Lain, S., Midgley, C. A., Lane, D. P. & Hay, R. T. (1999) EMBO J. 18, 6455–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogt, P. K. (2002) Nat. Rev. Cancer 2, 465–469. [DOI] [PubMed] [Google Scholar]
- 24.Hu, C. D., Chinenov, Y. & Kerppola, T. K. (2002) Mol. Cell 9, 789–798. [DOI] [PubMed] [Google Scholar]
- 25.Hu, C. D. & Kerppola, T. K. (2003) Nat. Biotechnol. 21, 539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fang, D., Elly, C., Gao, B., Fang, N., Altman, Y., Joazeiro, C., Hunter, T., Copeland, N., Jenkins, N. & Liu, Y. C. (2002) Nat. Immunol. 3, 281–287. [DOI] [PubMed] [Google Scholar]
- 27.Green, S. A., Zimmer, K. P., Griffiths, G. & Mellman, I. (1987) J. Cell Biol. 105, 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Finley, D., Ciechanover, A. & Varshavsky, A. (1984) Cell 37, 43–55. [DOI] [PubMed] [Google Scholar]
- 29.Chen, J. W., Murphy, T. L., Willingham, M. C., Pastan, I. & August, J. T. (1985) J. Cell Biol. 101, 85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tisdale, E. J., Bourne, J. R., Khosravi-Far, R., Der, C. J. & Balch, W. E. (1992) J. Cell Biol. 119, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angers, A., Ramjaun, A. R. & McPherson, P. S. (2004) J. Biol. Chem. 279, 11471–11479. [DOI] [PubMed] [Google Scholar]
- 32.Barila, D., Mangano, R., Gonfloni, S., Kretzschmar, J., Moro, M., Bohmann, D. & Superti-Furga, G. (2000) EMBO J. 19, 273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.