Abstract
Previous reports have suggested that the protooncogene c-myb participates in T cell development in the thymus and mature T cell proliferation. We have generated two T cell-specific c-myb knockout mouse models, myb/LckCre and myb/CD4Cre. We have demonstrated that c-myb is required for the development of thymocytes at the DN3 stage, for survival and proliferation of double-positive thymocytes, for differentiation of single-positive CD4 and CD8 T cells, and for the proliferative responses of mature T cells. In addition, our data show that c-myb is directly involved in the formation of double-positive CD4+CD8+CD25+, CD4+CD25+, and CD8+CD25+ T cells, developmental processes that may imply a role for c-myb in autoimmune dysfunction.
The process of thymocyte development involves tightly regulated steps of progressive cellular differentiation, selection, and lineage commitment (1). The most immature double-negative (DN) CD4–CD8– thymocytes develop into double-positive (DP) CD4+CD8+ thymocytes, which give rise to immature single-positive (SP) CD4+ or SP CD8+ T cells. The DN population can be further subdivided into progressive levels of maturity: CD44+CD25– (DN1), CD44+CD25+ (DN2), CD44-CD25+ (DN3), and CD44-CD25– (DN4). Progression beyond the DN3 stage depends on the successful rearrangement of a T cell antigen receptor (TCR)β gene and preTCR signaling, whereas differentiation from DP to mature SP cell depends on the expression and positive selection of TCRαβ.
The nuclear protooncogene c-myb, which belongs to the myb family of transcription factors, is believed to be an important regulator of cell growth, survival, and differentiation during hematopoiesis (2). c-myb is expressed predominately in hematopoietic tissues and is crucial for fetal hepatic erythropoiesis, because homozygous c-myb null mice die in utero of severe anemia at embryonic day 15 (3). Using c-myb-null/rag-deficient blastocyst chimeric mice, it was found that c-myb is critical for maturation to the DN1 stage of thymopoiesis (4). Furthermore, by using a knock-down mouse model, it was shown that this gene is required at a high threshold for progression beyond the DN2 stage (5). In addition, transgenic expression of a dominant interfering c-Myb was found to result in the accumulation of cells at the DN3 stage of T cell development and impairment of mature T cell proliferation (6). Because constitutive loss of c-myb gene leads to embryonic lethality, and because there could be nonspecific inhibition by the dominant interfering c-Myb, it has not been possible to accurately assess the role of this gene in adult T cell development and mature T cell function. Therefore, to define the role of c-myb in T cell development and mature T cell function, we have generated two T cell-specific c-myb knockout mouse models, mybF/F/LckCre and mybF/F/CD4Cre, which delete c-myb at two specific stages of T cell development. Results presented in this report show that c-myb is essential for the development of thymocytes at the DN3 stage, for survival and proliferation of DP thymocytes, for differentiation of SP CD4 and SP CD8 T cells, and for the proper proliferative responses of mature T cells. In addition, our data indicate that c-myb is directly involved in the formation of cells expressing CD25 at both the DP and SP stages. The development of CD25+ T cells may implicate c-myb in autoimmune dysfunction.
Methods
Generation of Floxed c-myb Mice. We isolated a BamHI genomic clone of c-myb encoding exons 2–8 (7), to produce a gene-targeting vector, pCMTSK2 (Fig. 1a). A 0.9-kb fragment containing exon 6 was generated by PCR amplification and cloned into the BamHI site of the pflox vector (8, 9). The flanking 6.0-kb fragment on the 5′ end containing exons 2–5 and the flanking 2.5-kb fragment on the 3′ end containing exons 7 and 8 were generated by a combination of PCR and restriction enzyme digestion and cloned into the SalI and XhoI sites, respectively. Then the backbone of pCMTSK was modified to contain a BamHI site in place of the NheI site. After the construction of pCMTSK2, the entire clone was sequenced to ascertain that PCR technique did not produce any mutations or deletions.
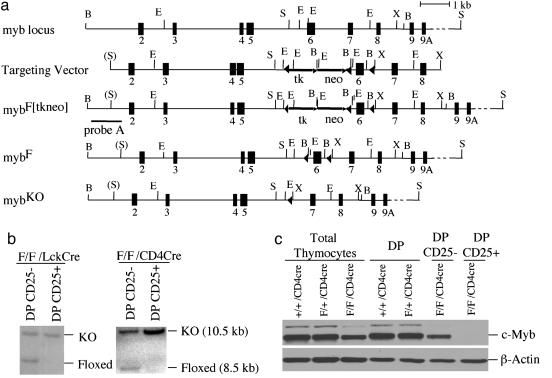
Fig. 1.
Conditional targeting of the mouse c-myb locus and expression of c-Myb protein in the thymus. (a) Strategy for flanking exon 6 of the mouse c-myb locus with loxP sites. The partial structure of the myb locus containing exons 2–9A is depicted as well as the targeting vector, which contains two selectable markers (arrows): thymidine kinase (tk) and neomycin resistant gene (neo). Thirty-four base pair loxP sites are shown as arrowheads. MybF[tkneo] is the product of homologous recombination that is used as a substrate for subsequent recombination by Cre recombinase, which deletes the tk-neo markers to generate mybF allele. The position of the probe (probe A) used to determine homologous recombination and for additional genomic Southern blotting studies of targeted c-myb allele structure is shown. Deletion of the loxP-flanked exon 6 of the mybF allele by Cre generates mybKO allele. B, BamHI; E, EcoRI; S, SalI; X, XhoI. (b) Southern blot analysis of genomic DNA from two populations DP CD25– and DP CD25+ found within the DP CD4+ CD8+ cells of the two T cell-specific c-myb knockout mouse models. (c) c-Myb protein expression in total, control DP, mutant DP CD25– and DP CD25+ thymocyte populations of myb/CD4Cre mice.
NotI-linearized pCMTSK2 was introduced into mouse R1 embryonic stem (ES) cells via electroporation. ES cells were cultured and selected as described (9). Genomic DNA from neomycin-resistant ES cells was digested with BamHI, and homologous recombinants were identified by Southern blotting by using a c-myb-specific probe upstream of exon 2 (probe A). Homologous recombinants revealed two bands: 11.5 (wild-type) and 9.8 kb (recombinant). To remove the PGKneoR-thymidine kinase marker, recombinant ES cell clones were subjected to transient expression of Cre recombinase by electroporation of pPGKcre-bpA and subsequent selection in the presence of ganciclovir (9). DNA from subclones resistant to ganciclovir were isolated, digested with BamHI, and analyzed by Southern blotting by using probe A to identify floxed and knockout alleles as 8.5 and 10.5 kb, respectively. Three mybF/+ ES clones were injected into C57BL/6 × DBA2 blastocysts by the University of Michigan Transgenic Core Facility, and chimeric male mice were crossed to C57BL/6 mice to generate two heterozygous mouse lines for the mybF allele. Both the LckCre and CD4cre transgenes were genotyped, as described on www.jax.org for LckCre transgenic mice.
Flow Cytometry, Sorting, Cell Cycle, and Western Blotting. Fluorescence-activated cell sorter (FACS) analyses were performed by using freshly prepared single-cell suspensions of thymocytes in RPMI medium 1640 without phenol red/5% heat-inactivated FBS. Stained cells were analyzed by using a FACSCalibur machine (BD Biosciences) after staining with fluorochrome-conjugated antibodies purchased from BD Biosciences: CD4 (GK1.5 and RM4-5), CD8 (53-6.7), CD3 (17A2), TCR-β (H57-597), CD69 (H1.2F3), CD25 (PC61), CD44 (Ly-24, IM7), γδ (GL3), CD24/HSA (M1/69), CD122 (TM-β1), and B220 (RA3-6B2). To avoid background staining, thymocytes were preincubated in CD16/CD32 (Fc block) according to the manufacturer's protocol (BD Biosciences). For cell sorting, single-cell suspensions were labeled and sorted on a high-speed cell sorter (MoFlo; DakoCytomation, Carpenteria, CA) to a purity of >95% at the Wistar Institute, Philadelphia. For cell cycle analysis, sorted cells were stained with DRAQ5 according to the manufacturer's instructions (Biostatus Limited, Leicestershire, U.K.). For Western analysis, 100 μg of protein extract was run on a 10% SDS–polyacrylamide gel and transferred to nitrocellulose for immunoblotting. Antibodies for c-Myb (Upstate Biotechnology, Lake Placid, NY), Proliferating Cell Nuclear Antigens, Cdk1/Cdc2 (Santa Cruz Biotechnology), and β-actin (Sigma) were used to probe the membranes.
Thymocyte Proliferation and Survival Assays. Sorted thymocytes were plated at 4 × 105 cells/well in a 96-well plate and pulsed with 1 μCi (1 Ci = 37 GBq) of [3H]thymidine for 18 h. Cells were harvested onto fiberglass filter papers (Packard), and radioactivity was quantified by using a Flexi-Vial 1209 liquid scintillation counter (Pharmacia LKB Nuclear). IL-2 treatment was carried out by using a concentration of 20 ng/ml (R & D Systems). For thymocyte survival assays, 18-h-cultured thymocytes were stained with Annexin V-FITC (BD Biosciences) and analyzed by fluorescence-activated cell sorting (FACS).
Mature T Cell Proliferation Assay and Cytokine Determination. RBC-lysed splenic cells were incubated with either anti-CD4 or anti-CD8 MACS beads (Miltenyi Biotec, Auburn, CA) and passed through a MidiMACS column to obtain 90–95% purified CD4+ and CD8+ T cells, respectively. For the proliferation assays, purified T cells were plated at 5.7 × 105 cells per well and incubated with the indicated concentrations of stimulators: 5 μg/ml of anti-CD3/anti-CD28 (BD Biosciences), 10 ng/ml of IL-2 (R & D Systems), 10 μg/ml of phytohemagglutinin (PHA) (Sigma), and 10 ng/ml of phorbol 12-myristate 13-acetate (PMA) (Sigma). After 53–55 h of incubation, supernatants were collected and the cells were pulsed with 1 μCi of [3H]thymidine and harvested at the 72-h stimulation period. Cytokine levels were measured by using the BD Biosciences Cytometric Bead Array Th1/Th2 Cytokine kit.
Statistical Analysis. Data are expressed as means ± standard deviations. Comparisons were analyzed by using Student's two-paired t test with the same variance. Differences were considered significant when P < 0.05.
Results
Generating Floxed c-myb Mutant Mice. To circumvent the embryonic lethality associated with c-myb nullizygosity, we used the Cre-LoxP recombination system to achieve tissue-specific deletion of c-myb (8, 9). We flanked exon 6 of c-myb, which encodes a high-affinity DNA-binding domain, with loxP sites (Fig. 1a). The neomycin and thymidine kinase markers were removed by transient expression of Cre recombinase in four recombinant ES cell clones. Three clones with deletion of the markers but retention of loxP sites around exon 6 were injected into blastocysts, and germ-line transmission of the floxed c-myb allele was obtained for two of the three clones. The mybF/F homozygotes had normal behavior, appearance, and development and followed a Mendelian distribution, indicating that the mybF allele is fully functional. In addition, there are no differences in the abundance of RNA and protein in the bone marrow, spleen, and thymus from the three genotypic groups (data not shown). Intercrosses between myb+/KO mice generated no mybKO/KO mice in >150 births.
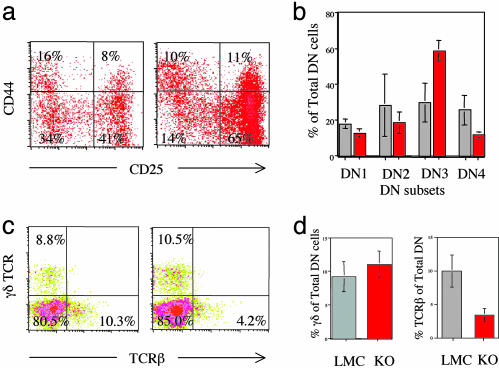
The Critical Role for c-myb at the DN3 Stage of Thymocyte Development. To determine the function of c-myb in early thymocyte development, we crossed our floxed c-myb mice with the LckCre mice, which are first active at the DN2 stage of thymocyte development (ref. 10 and data not shown). The cellularity of the thymi of mybF/F/LckCre mice is reduced to 11% of littermate controls (LMC) (Fig. 3a). Analysis of the subsets of DN thymocytes by surface staining for CD44 and CD25 showed an accumulation of thymocytes at the DN3 stage of early T cell development (Fig. 2 a and b). The accumulation of thymocytes at the DN3 stage may impede cells from progressing into the DN4 stage, which may account for the significant decrease in thymocytes at the DN4 stage in the mutant mice compared with LMC. The mutant DN cells exhibit decreased surface staining for TCRβ (Fig. 2 c and d), suggesting a defect in the TCRβ rearrangement process itself or a step before β rearrangement. Further analysis (Fig. 2 c and d) shows there is no significant difference in the percentage of γδ thymocytes within the total DN population in the mutant mice as compared with LMC, indicating that c-myb may not have a role in the development of the γδ population in these mutant mice.
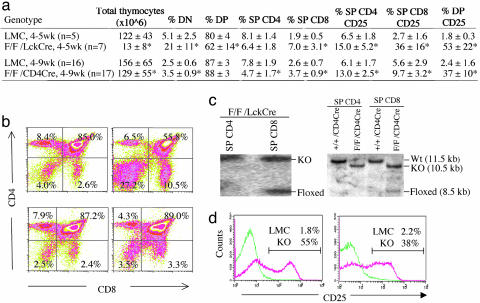
Fig. 3.
Impaired thymocyte development in mybF/F/LckCre and mybF/F/CD4Cre mice. Percentages (a) and two-color flow cytometric analysis (b) of CD4 and CD8 thymocyte subsets are shown. Percentages of SP CD4 CD25, SP CD8 CD25, and DP CD25 represent the precentages of total SP CD4, SP CD8, and DP, respectively. (c) Southern blot analysis of genomic DNA from SP thymocytes of both mutant mouse models. (d) Expression of CD25 within the DP population of c-myb mutant mice (red line) and LMC (green line). For LMC, myb+/+/Cre, mybF/+/Cre, or mybF/F, no Cre was used. *, P < 0.05. Number presented as mean ± SD.
Fig. 2.
DN thymocyte development in mybF/F/LckCre mice. Two-color flow cytometric analysis (a) and quantitation (b) of CD44 and CD25 surface expression on thymocytes. Two-color flow cytometric analysis (c) and quantitation (d) of surface γδ TCR and TCRβ within the total DN population within the live lymphoid gate of c-myb mutant (red bar) or littermate controls (gray bar).
Requirement of c-myb at the DP Stage of Thymocyte Development. In the mybF/F/LckCre mouse model, the percentage of DP population is reduced to 62% (range, 43–84%) of total thymocytes (Fig. 3a). Despite a reduction in the percentage of DP thymocytes, it is difficult to ascertain the exact contribution of c-myb to the DP stage in the c-mybF/F/LckCre model, because the DN stage is altered and there is a 100-fold expansion going from the DN to the DP stage. Therefore, we generated the mybF/F/CD4Cre mouse model, which is first active at the DP stage, to facilitate the study of the DP thymocyte population (11). Like the mybF/F/LckCre mouse model, the thymic cellularity of the mybF/F/CD4Cre is also reduced, but by a modest 17% (Fig. 3a). The percentages of DP thymocytes in the mybF/F/CD4Cre mice and their LMC are similar (Fig. 3a). However, the absolute value of the mybF/F/CD4Cre DP thymocytes (114 × 106 vs. 136 × 106 in LMC) is reduced by 17%, indicating a requirement of c-myb at the DP stage for fully normal thymocyte development.
Development of DP CD25+ Thymocytes in the Absence of c-myb. Closer examination of the DP thymocytes from the mybF/F/CD4Cre mutant mice revealed that ≈37% (range, 19–48%) of the DP thymocytes express CD25, the IL-2Rα chain, as compared with <3% in LMC (Fig. 3 a and d). This very dramatic alteration in T cell development was also observed in the DP cells of the mybF/F/LckCre mice (Fig. 3 a and d), albeit at a higher percentage (i.e., 53% compared with <2% in LMC). The DP CD25+ thymocyte population did not express CD122, the IL-2Rβ chain, suggesting that these mutant cells express only the low-affinity IL-2R on their surface (data not shown). To define the DP CD25+ population, we sorted the DP CD25+ and the DP CD25– thymocyte populations from mutant mybF/F/CD4Cre mice and DP cells from LMC and examined c-Myb protein expression (Fig. 1c). The mutant DP CD25+ had no detectable c-Myb protein expression, which correlates with DNA data that showed complete absence of the floxed c-myb allele in the mutant DP CD25+ thymocyte population, indicating that the full deletion of c-myb favors the development of DP CD25+ cells (Fig. 1b). On the contrary, the DP CD25– thymocyte population from the mutant mice had a decreased level of c-Myb protein expression, which is consistent with the partial deletion of the floxed c-myb allele (Fig. 1 b and c). Because the efficiency of Cre-mediated deletion of floxed c-myb in the DP thymocyte population is similar in both the mutant mouse models (Fig. 1b), this indicates that the dramatic decrease in the thymic cellularity seen in the mybF/F/LckCre is mainly due to events that occur during the DN stage of T cell development and not at the DP stage.
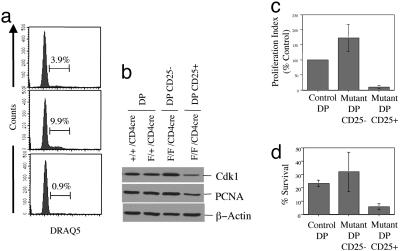
Deficient Proliferative and Survival Capacity of Mutant DP CD25+ Thymocytes. Because c-myb is implicated in proliferation in many systems, we wished to determine the proliferative activity of the unusual DP CD25+ population from the mybF/F/CD4Cre mice by measuring [3H]-thymidine incorporation (Fig. 4c). The mutant DP CD25+ thymocyte population displayed greatly impaired [3H]-thymidine uptake, exhibiting only 15% uptake compared with that of DP cells from LMC. The reduced [3H]-thymidine uptake in the mutant DP CD25+ thymocyte population is consistent with the reduced percentage of these cells in S and G2/M phases of the cell cycle when compared with DP cells from LMC (Fig. 4a). In contrast, the mutant DP CD25– thymocyte population has nearly 2-fold greater [3H]-thymidine uptake when compared with that of DP cells from LMC, suggesting that these cells may be compensating for the absence of c-Myb. This greater [3H]-thymidine uptake seen in the mutant DP cells is consistent with cell cycle data, which indicate an increased percentage of this population in the S and G2/M phases of the cell cycle as compared with DP cells from LMC (Fig. 4a). We performed RNase protection analysis (RPA) using RNA from mutant DP CD25+, mutant DP CD25–, and wild-type DP thymocyte populations and found no difference in the levels of cdk2, cyclin E, c-myc, max, bcl-w, bfl-1, bak, bax, bcl-2, bad, bcl-x, NFkB, fas, PARP, caspase3, p53, p21, and p27 among the three thymocyte populations (data not shown). However, we observed a decrease in RNA abundance of cdk1 and proliferating cell nuclear antigens (PCNA) in the mutant DP CD25+ population but not in the other two populations (data not shown). Consistent with our RPA data, Western blot analysis indicates that the abundance of Cdk1, a known c-Myb target gene, and PCNA proteins are reduced in the mutant DP CD25+ (Fig. 4b), consistent with an impairment in the proliferative capacity of DP CD25+ thymocytes (12).
Fig. 4.
Formation of DP CD25+ population in the complete absence of c-Myb. (a) Cell cycle analysis via DRAQ5 staining of sorted cells. (b) Protein expression of Cdk1 and proliferating cell nuclear antigens. (c) Proliferation via 18-h [3H]thymidine uptake expressed as a percentage of control. (d) Survival after 18 h in culture as measured by annexin V staining.
Because survival is another of the described functions of c-Myb in hematopoietic cells, we sorted mutant DP CD25+, mutant DP CD25–, and control DP cells and assessed cellular survival over 18 h by annexin V staining (Fig. 4d). Under these conditions, there was no difference in survival between the mutant DP CD25– cells or the DP cells from LMC. In contrast, the mutant DP CD25+ population lost viability more rapidly than either the mutant DP CD25– cells or the DP cells from LMC, suggesting that c-Myb provides a survival signal(s) to DP cells.
Requirement of c-myb at the SP Stage of T Cell Development. We observed a decrease in the percentage of SP CD4 T cells that is reflected in the altered ratio of CD4 to CD8 T cells in the thymi of the mybF/F/CD4Cre mice (1.3 vs. 3.0 in LMC) (Fig. 3a). The decrease in the percentage of the thymic SP CD4 T cells leads to a decrease in the percentage of mature CD4 T cells, in the ratio of CD4 to CD8 T cells, and in the ratio of CD4 T cells to B220, a pan-B cell marker, in both the spleen and the lymph nodes of both the mutant mouse models when compared with their respective LMC (Fig. 3a and Table 1). This suggests that c-myb has a role in the differentiation of the CD4 T cells. In contrast, we observed an increase in the percentage of thymic SP CD8 T cells of the mybF/F/CD4Cre mice. This observation suggests either that the deletion of c-myb directly accelerates the differentiation of SP CD8 T cells or that the impaired differentiation of SP CD4 T cells enhances the formation of SP CD8 T cells (Fig. 3a and Table 1). However, there is no change in the percentage of CD8 T cells in either the spleen or lymph nodes of the mybF/F/CD4Cre mice when compared with LMC (Table 1).
Table 1. Distribution of mature CD4 and CD8 T cells in spleen and lymph nodes of mutant mice.
| Genotype | Total cells (× 106) | % CD4 | % CD8 | CD4/CD8 | % CD4 CD25+ | %CD8 CD25+ |
|---|---|---|---|---|---|---|
| Spleen | ||||||
| LMC, 4-5 wk | 65 ± 13 | 16.0 ± 5.2 | 7.0 ± 3.5 | 2.3 | 12.4 ± 3.0 | 2.6 ± 2.0 |
| F/F/LckCre, 4-5 wk (n = 7) | 68 ± 31 | 9.7 ± 3.7* | 5.1 ± 2.3 | 1.9 | 21.5 ± 2.0* | 8.4 ± 3.3* |
| LMC, 4-9 wk | 102 ± 41 | 19.9 ± 2.2 | 11.3 ± 1.4 | 1.8 | 11.3 ± 1.4 | 7.7 ± 6.0 |
| F/F/CD4Cre, 4-9 wk | 74 ± 32 | 11.8 ± 1.7* | 9.1 ± 1.6* | 1.3 | 13.6 ± 2.9 | 10.0 ± 6.7 |
| Lymph node | ||||||
| LMC, 4-5 wk | — | 43 ± 3 | 30 ± 4 | 1.4 | 13.6 ± 2.1 | 2.4 ± 2 |
| F/F/LckCre, 4-5 wk (n = 3) | — | 23 ± 3* | 23 ± 4 | 1.0 | 29.1 ± 2.0* | 8.6 ± 3* |
| LMC, 4-9 wk | — | 37 ± 4 | 24 ± 3 | 1.5 | 13.1 ± 2.1 | 1.7 ± 1.2 |
| F/F/CD4Cre, 4-9 wk | — | 28 ± 2* | 24 ± 4 | 1.2 | 17.7 ± 2.0* | 3.0 ± 1.9* |
Percentages of CD4 CD25+ and CD8 CD25+ represent the total CD4 and CD8 T cells, respectively. *, P < 0.05.
Increased Presence of Immature and Mature CD4 CD25+ and CD8 CD25+ in the Mutant Mouse Models. In both the mybF/F/CD4Cre and mybF/F/LckCre mouse models, we detected a slight but consistent increase in the percentage of thymic SP CD4 CD25+ cells that are CD3hi (Fig. 3a and data not shown). We observed that this increase in thymic SP CD4 CD25+ cells results in a slight but significant percentage increase in the mature CD4 CD25+ T cells in the lymph nodes of the mybF/F/CD4Cre mice and in both the spleen and lymph nodes of the mybF/F/LckCre mice. We also found that there are 2- and 13-fold increases in thymic CD3hi SP CD8 CD25+ cells in the mybF/F/CD4Cre and mybF/F/LckCre mice, respectively. This increase in thymic SP CD8 CD25+ translates into a slight percentage increase of peripheral CD8 CD25+ T cells in both the spleen and lymph nodes of the mybF/F/CD4Cre mice. Similarly, the 13-fold increase in the SP CD8 CD25+ thymocytes of the mybF/F/LckCre mice accounts for a 3-fold increase in mature CD8 CD25+ T cells in the spleen and lymph nodes. Our data suggest that formation of these mutant CD4 CD25+ and CD8 CD25+ immature and mature T cells is due to altered events at the DP stage as a consequence of depleted abundance of c-Myb.
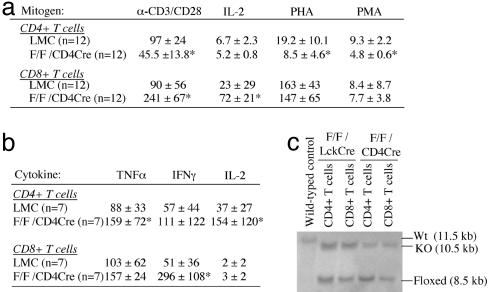
Requirement of c-myb for Proper Proliferative Response of Mature T Cells to Mitogens. We next tested the proliferative response of purified splenic CD4 and CD8 T cells from mybF/F/CD4Cre mice to stimulation by either anti-CD3/anti-CD28, IL-2, PHA, or PMA (Fig. 5a). Results show that purified splenic CD4 T cells from mutant mice have an ≈50% lower response as measured by [3H]-thymidine uptake after stimulation with either anti-CD3/anti-CD28, PHA, or PMA. However, there was no difference in the response to IL-2 when compared with LMC, which is in contrast to previously published data (13, 14). In contrast, purified splenic CD8 T cells show a 3-fold greater response to either anti-CD3/anti-CD28 or IL-2, but not PHA or PMA (Fig. 5a). These results are consistent with data showing a 6-fold increase in IFNγ secretion by the mutant CD8 T cells in response to anti-CD3/anti-CD28 (Fig. 5b).
Fig. 5.
Impaired proliferation response of mature splenic T cells from mybF/F/CD4Cre mice to mitogens. (a) Proliferation index of purified CD4 and CD8 T cells. (b) Cytokine concentration (×10–8 g/ml) in supernatant from CD4 and CD8 T cells treated with αCD3/CD28. There were no differences in the levels of tumor necrosis factor α, IFNγ, and IL-2 cytokines for any of the other treatments listed in a.(c) Southern blot analysis of genomic DNA from purified splenic CD4 and CD8 T cells of both mutant mouse models. *, P < 0.05.
Discussion
We have created two T cell-specific c-myb knockout mouse models, mybF/F/CD4Cre and mybF/F/LckCre mice, by using the Cre-Lox recombination system, which enabled us to examine various stages of T cell development as well as mature T cell function. We observed an accumulation of thymocytes at the DN3 stage that indicates an important role for c-myb at this point in early T cell development. While this manuscript was in preparation, we became aware of a report by Bender et al. (15) showing that c-myb is essential at the DN3 stage of T cell development. Although Bender's group and we arrived at the same conclusion, the accumulation of thymocytes at the DN3 stage is less striking in our model. This probably reflects the different efficiencies of Cremediated deletion of the floxed c-myb in our LckCre (10) mice vs. the cwLckCre (11) mice used by Bender et al. (15). However, at odds with this report, we detected no significant difference in the percentage of the γδ population as a total of the DN population in the mybF/F/LckCre mice compared with LMC. The disparate results may reflect the different Cre mice used and the different efficiencies of Cre-mediated deletion of floxed c-myb in the DN stage.
Our data from the mybF/F/CD4Cre mouse model indicate that there is a requirement for c-myb in DP thymocyte development. Our data suggest that the function of c-myb in these cells is to confer both proliferative and survival capabilities. When c-Myb is lost in DP thymocytes, the proliferative and survival capacities of these cells are impaired, and this is associated with the expression of CD25. We do not believe that CD25 is a bona fide c-Myb target gene, at least at the DP stage. For instance, in the mutant DP CD25– cells where floxed c-myb allele is partially deleted, CD25 is not expressed. We therefore believe that the expression of CD25 by the c-Myb-deficient DP thymocytes serves only as a marker of defective cells that need to be eliminated from the thymic environment. Consistent with this notion, addition of IL-2 has no effect on 3H-thymidine uptake or apoptosis in the mutant DP CD25+ thymocytes (data not shown). Furthermore, our interpretation is consistent with the phenotype of IL-2-deficient mice that develop various forms of autoimmunity, because autoreactive T cells are not deleted to provide central tolerance (16, 17). Interestingly, it has been reported that DP thymocytes treated with anti-CD3 to induce cell death down-regulate CD4 and CD8 but up-regulate TCRβ, CD3, CD69, and CD25 (18). Although c-myb-deficient DP CD25+ cells do not down-regulate CD4 and CD8 and are CD69–, CD3lo, and TCRβlo (data not shown), expression of CD25 on DP thymocytes may be indicative of defective cells that would normally be purged from the thymic environment.
In the mutant DP CD25– thymocytes, there is nearly twice the proliferative index of the DP thymocytes of LMC. Because our mice are c-Myb depletion models and the half-life of c-Myb protein is ≈60 min (19), we believe that virtually all of the remaining c-Myb protein in the DP CD25– thymocytes (Fig. 1c) comes from the undeleted floxed c-myb allele (Fig. 1b). We believe that the mutant cells have compensated for the absence of c-Myb by using a c-Myb-independent mechanism for proliferation, which is also A-Myb independent because there are no changes in the levels of A-Myb in mutant DP CD25+, DP CD25–, and DP of LMC (data not shown). Because the mutant DP CD25– thymocytes can compensate for the proliferative capacity as well as the survival capacity in the absence of c-Myb, these cells are able to progress to the SP stage. Consistent with this interpretation, we observed the presence of SP thymocytes with a partially deleted c-myb allele in the thymi of both c-myb mutant mouse models.
Our data suggest that c-myb is also required in the differentiation of CD4 SP T cells, which is consistent with the study by Bender et al. (15). Interestingly, we noticed that only ≈50% of the floxed c-myb alleles is deleted in the purified splenic CD4 T cells, whereas a near-complete deletion of our floxed c-myb alleles is seen in the SP CD4 T cells in the thymus unlike Bender et al. (15). This observation suggests that the c-Myb-deficient SP CD4 T cells have a slower maturation rate at the SP stage, a defect in their ability to home to the spleen, and a shorter life span in the periphery and/or compete poorly for the available niches in the spleen than c-Myb-sufficient CD4 T cells. Further experiments should clarify these possibilities.
We observed an increase in the percentage of SP CD8 in our mybF/F/CD4Cre mouse model, suggesting that c-myb is required for the differentiation of SP CD8 T cells, which is at odds with the report of Bender et al. (15). The reason for this discrepancy may lie in the myb/Cre models used. There appears to be a correlation between the degree of perturbation to the DN stage as measured by the accumulation of thymocytes in the DN stage and in the DN3 stage and the increase in the SP CD8 thymocyte subset. For instance, among the four myb/Cre models used in our study and that of Bender et al. (15), the perturbation to the DN stage in order of greatest to least is as follows: Bender's mybf/f/cwLckCre, our mybF/F/LckCre, Bender's mybf/f/LckCre, and our mybF/F/CD4Cre mice. In the mybF/F/CD4Cre mouse model, no accumulation of cells at the DN3 stage was observed (data not shown). If we now compare the absolute number of SP CD8 thymocytes as a percentage of LMC, the results for the four mouse models in the same order as above are as follow: 12% (6 × 105 vs. 5 × 106 in LMC), 40% (9.1 × 105 vs. 2.3 × 106 in LMC), 80% (4 × 106 vs. 5 × 106 in LMC), and 117% (4.8 × 106 vs. 4.1 × 106 in LMC), respectively. Because the mybF/F/CD4Cre mouse model has the least perturbation to the DN stage and because events at the DN stage do influence the DP compartment, we believe that the mybF/F/CD4Cre mouse model is the model that best portrays the events at the DP and SP stages of T cell development.
Previous attempts to study the role of c-Myb in the function of mature T cells have mainly used antisense oligonucleotides or a dominant interfering c-Myb protein, which can have nonspecific effects (6, 20, 21). We determined that purified mutant CD4 T cells have a weaker response to anti-CD3/anti-CD28, PHA, or PMA. On the contrary, mutant CD8 T cells have a greater proliferative response to anti-CD3/CD28 and IL-2 that may reflect an altered proliferative mechanism as we have observed in the mutant DP CD25– thymocytes. Hence, our data indicate that c-myb is required for normal proliferative activity of mature CD4 and CD8 T cells.
Interestingly, in both our models, we observed an increase of SP CD4+CD25+, SP CD8+CD25+, mature CD4 CD25+, and mature CD4 CD25+ T cells. It is not clear whether these mutant cells have suppressor functions. However, it appears that these CD25+ T cells may have arisen from the DP CD25+ cell population. For instance, there is a positive correlation between DP CD25+ and SP CD8+CD25+ in the mybF/F/LckCre mice (data not shown). Interestingly, although we observed that 36% of SP CD8 T cells express CD25 in the thymi of mybF/F/LckCre mice, CD8 CD25+ T cells represent only ≈8% of the splenic CD8 T cells and 9% of lymph node CD8 T cells. This observation may reflect that some of these cells are deleted in the thymi, or these cells migrate slowly to the periphery, have a shorter life span in the periphery, or home to nonlymphoid organs. Further studies should clarify these possibilities.
Conclusion
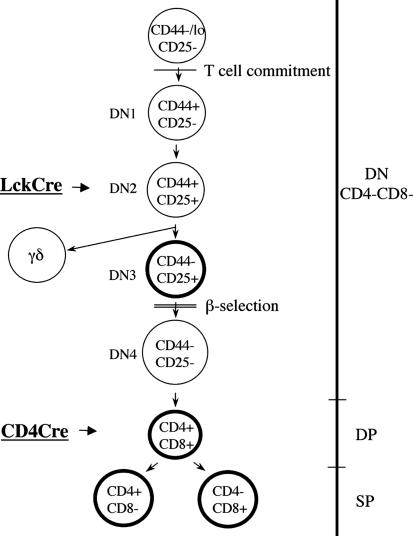
Our data indicate that c-myb is required for T cell development at the stages depicted in Fig. 6: DN3, DP, SP CD4, and SP CD8. Because c-myb is required for T cell development and normal proliferative response in the periphery, it will be necessary to decipher whether c-myb has a role not only in autoimmune dysfunction but also in T cell leukemia. If we extrapolated our findings here to T leukemic cells, we would predict that depletion of c-Myb would greatly impair the proliferative and survival capacities of most T leukemic cells similarly to what we observed in the DP CD25+ cells. However, the presence of DP CD25– cells suggests that in some instances, replacement of c-Myb function may occur. This assumption would be consistent with the observation of Venturelli et al. (22), who have shown that down-regulation of c-myb by antisense oligodeoxynucleotides inhibited DNA synthesis of T leukemic cells in most but not all patients.
Fig. 6.
c-myb and T cell development. Highlighted in bold are the stages where we observed that loss of c-myb resulted in altered phenotype.
Acknowledgments
We thank Dr. Jamey D. Marth (The Burnham Institute, La Jolla, CA) for the gift of the pFlox plasmid and Dr. Mariano Barbacid (Centro Nacional de Investigaciones Oncologias, Madrid) for the pPGK-Cre-bpA plasmid. In addition, we thank Joe Meissler for help with the radioactive harvester and Filip Bednar, Alaina Borden, and Mayumi Kataoka for help with flow cytometric acquisition. Y.K.L. was supported by a predoctoral fellowship from the Department of Defense Breast Cancer Research Program. This work was supported by National Institutes of Health Grants CA79086 (to E.P.R.), DA-06650, DA-11130, DA-1423, DA-16544, and P30 DA-13429 (to T.J.R.), and the Fels Foundation. Core facilities used in this study were supported by National Institutes of Health Grant R24 CA88261 (to E.P.R.).
Author contributions: Y.K.L., A.K., T.J.R., and E.P.R. designed research; Y.K.L., A.K., and A.G.P. performed research; Y.K.L. and A.K. contributed new reagents/analytic tools; Y.K.L. and E.P.R. analyzed data; and Y.K.L., T.J.R., and E.P.R. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DP, double positive; SP, single positive; DN, double negative; ES, embryonic stem; PMA, phorbol 12-myristate 13-acetate; PHA, phytohemagglutinin; LMC, littermate controls; TCR, T cell antigen receptor.
References
- 1.Bommhardt, U., Beyer, M., Hunig, T. & Reichardt, H. M. (2004) Cell Mol. Life Sci. 61, 263–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oh, I. H. & Reddy, E. P. (1999) Oncogene 18, 3017–3033. [DOI] [PubMed] [Google Scholar]
- 3.Mucenski, M. L., McLain, K., Kier, A. B., Swerdlow, S. H., Schreiner, C. M., Miller, T. A., Pietryga, D. W., Scott, W. J., Jr., & Potter, S. S. (1991) Cell 65, 677–689. [DOI] [PubMed] [Google Scholar]
- 4.Allen, R. D., III, Bender, T. P. & Siu, G. (1999) Genes Dev. 13, 1073–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emambokus, N., Vegiopoulos, A., Harman, B., Jenkinson, E., Anderson, G. & Frampton, J. (2003) EMBO J. 22, 4478–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badiani, P., Corbella, P., Kioussis, D., Marvel, J. & Weston, K. (1994) Genes Dev. 8, 770–782. [DOI] [PubMed] [Google Scholar]
- 7.Tantravahi, R., Dudek, H., Patel, G. & Reddy, E. P. (1996) Oncogene 13, 1187–1196. [PubMed] [Google Scholar]
- 8.Gu, H., Marth, J. D., Orban, P. C., Mossmann, H. & Rajewsky, K. (1994) Science 265, 103–106. [DOI] [PubMed] [Google Scholar]
- 9.Priatel, J. J., Sarkar, M., Schachter, H. & Marth, J. D. (1997) Glycobiology 7, 45–56. [DOI] [PubMed] [Google Scholar]
- 10.Hennet, T., Hagen, F. K., Tabak, L. A. & Marth, J. D. (1995) Proc. Natl. Acad. Sci. USA 92, 12070–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, P. P., Fitzpatrick, D. R., Beard, C., Jessup, H. K., Lehar, S., Makar, K. W., Perez-Melgosa, M., Sweetser, M. T., Schlissel, M. S., Nguyen, S., et al. (2001) Immunity 15, 763–774. [DOI] [PubMed] [Google Scholar]
- 12.Furukawa, Y., Piwnica-Worms, H., Ernst, T. J., Kanakura, Y. & Griffin, J. D. (1990) Science 250, 805–808. [DOI] [PubMed] [Google Scholar]
- 13.Stern, J. B. & Smith, K. A. (1986) Science 233, 203–206. [DOI] [PubMed] [Google Scholar]
- 14.Rohwer, F., Todd, S. & McGuire, K. L. (1996) J. Immunol. 157, 643–649. [PubMed] [Google Scholar]
- 15.Bender, T. P., Kremer, C. S., Kraus, M., Buch, T. & Rajewsky, K. (2004) Nat. Immunol. 5, 721–729. [DOI] [PubMed] [Google Scholar]
- 16.Willerford, D. M., Chen, J., Ferry, J. A., Davidson, L., Ma, A. & Alt, F. W. (1995) Immunity 3, 521–530. [DOI] [PubMed] [Google Scholar]
- 17.Sadlack, B., Merz, H., Schorle, H., Schimpl, A., Feller, A. C. & Horak, I. (1993) Cell 75, 253–261. [DOI] [PubMed] [Google Scholar]
- 18.Kishimoto, H., Surh, C. D. & Sprent, J. (1995) J. Exp. Med. 181, 649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klempnauer, K.-H., Bonifer, C. & Sippel, A. E. (1986) EMBO J. 5, 1903–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Churilla, A. M., Braciale, T. J. & Braciale, V. L. (1989) J. Exp. Med. 170, 105–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gewirtz, A. M. & Calabretta, B. (1988) Science 242, 1303–1306. [DOI] [PubMed] [Google Scholar]
- 22.Venturelli, D., Mariano, M. T., Szczylik, C., Valtieri, M., Lange, B., Crist, W., Link, M. & Calabretta, B. (1990) Cancer Res. 50, 7371–7375. [PubMed] [Google Scholar]