Abstract
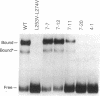
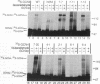
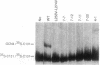
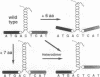
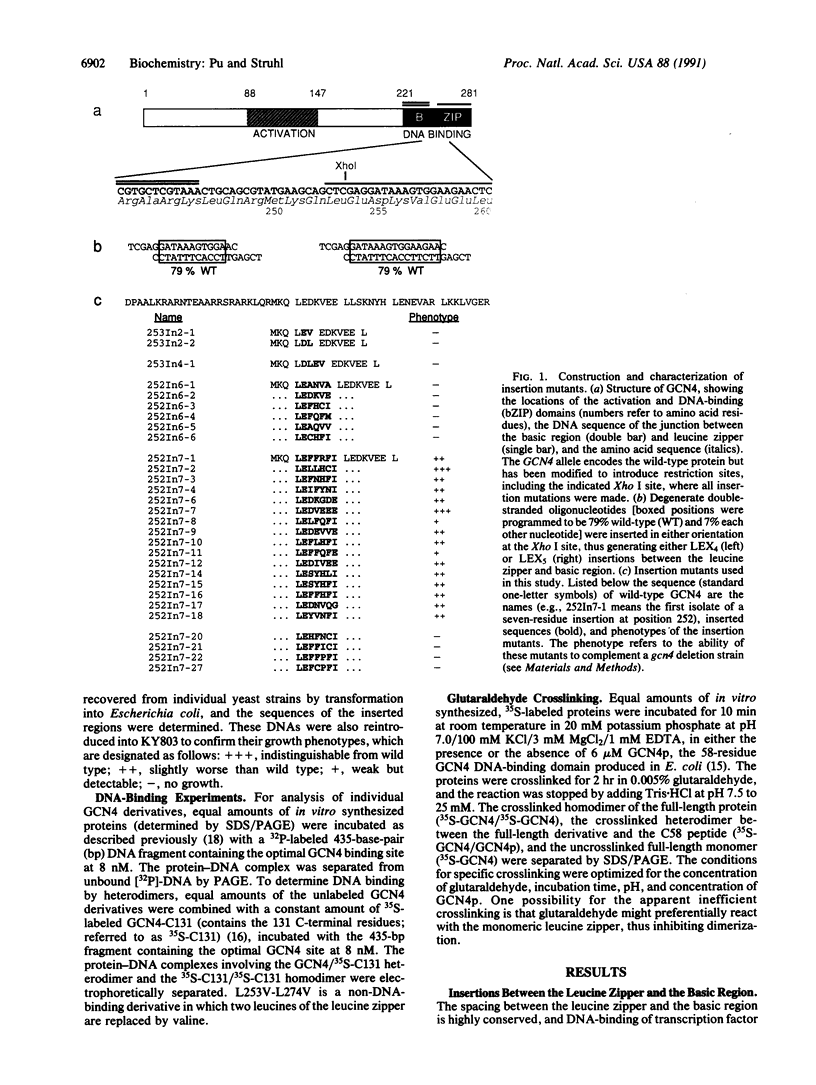
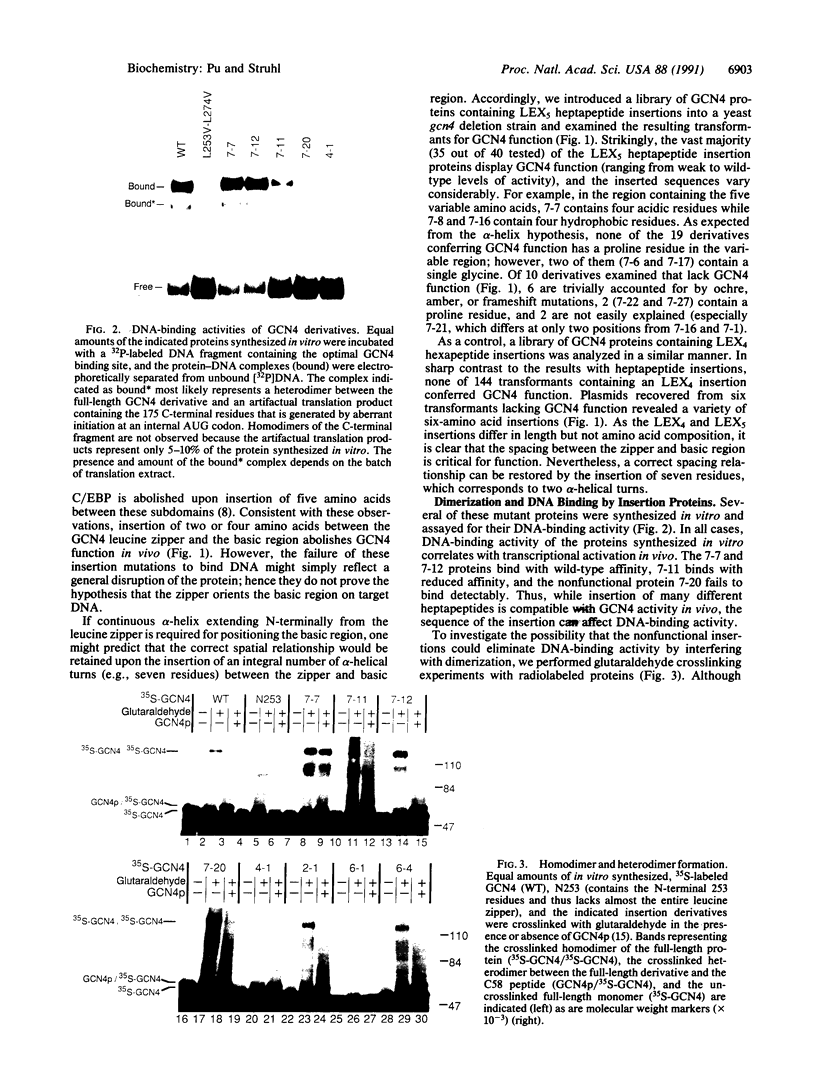
The bZIP structural motif present in several eukaryotic transcription factors is defined by the leucine zipper, a coiled-coil dimerization interface, and an adjacent basic region that directly interacts with DNA. To examine the functional importance of the highly conserved spacing between the leucine zipper and the basic region, we have analyzed the DNA-binding ability of yeast GCN4 proteins containing amino acid insertions between these two subdomains. Proteins containing a surprisingly wide variety of seven-amino acid insertions, but none containing two-, four-, or six-amino acid insertions, are functional. However, heterodimers between wild-type GCN4 and functional derivatives containing seven amino acid insertions are unable to bind DNA. These observations provide strong experimental support for several aspects of the scissors grip and induced fork models for DNA-binding by bZIP proteins. Specifically, they demonstrate that continuous alpha-helices symmetrically diverging from the leucine zipper correctly position the two basic regions for specific binding to abutting DNA half-sites. In addition, the results indicate that GCN4 homodimers are primarily responsible for transcriptional activation in yeast cells.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agre P., Johnson P. F., McKnight S. L. Cognate DNA binding specificity retained after leucine zipper exchange between GCN4 and C/EBP. Science. 1989 Nov 17;246(4932):922–926. doi: 10.1126/science.2530632. [DOI] [PubMed] [Google Scholar]
- Halazonetis T. D., Georgopoulos K., Greenberg M. E., Leder P. c-Jun dimerizes with itself and with c-Fos, forming complexes of different DNA binding affinities. Cell. 1988 Dec 2;55(5):917–924. doi: 10.1016/0092-8674(88)90147-x. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T., Ziff E. Leucine zippers of fos, jun and GCN4 dictate dimerization specificity and thereby control DNA binding. Nature. 1989 Aug 17;340(6234):568–571. doi: 10.1038/340568a0. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989 Mar 31;243(4899):1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- Landschulz W. H., Johnson P. F., McKnight S. L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988 Jun 24;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Ryder K., Nathans D. DNA binding activities of three murine Jun proteins: stimulation by Fos. Cell. 1988 Dec 2;55(5):907–915. doi: 10.1016/0092-8674(88)90146-8. [DOI] [PubMed] [Google Scholar]
- Neuberg M., Schuermann M., Hunter J. B., Müller R. Two functionally different regions in Fos are required for the sequence-specific DNA interaction of the Fos/Jun protein complex. Nature. 1989 Apr 13;338(6216):589–590. doi: 10.1038/338589a0. [DOI] [PubMed] [Google Scholar]
- O'Neil K. T., Hoess R. H., DeGrado W. F. Design of DNA-binding peptides based on the leucine zipper motif. Science. 1990 Aug 17;249(4970):774–778. doi: 10.1126/science.2389143. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Kim P. S. Evidence that the leucine zipper is a coiled coil. Science. 1989 Jan 27;243(4890):538–542. doi: 10.1126/science.2911757. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Rutkowski R., Stafford W. F., 3rd, Kim P. S. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science. 1989 Aug 11;245(4918):646–648. doi: 10.1126/science.2503872. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R., Nussbaum A. L., Struhl K. Cloning of random-sequence oligodeoxynucleotides. Gene. 1986;44(2-3):177–183. doi: 10.1016/0378-1119(86)90180-0. [DOI] [PubMed] [Google Scholar]
- Sellers J. W., Struhl K. Changing fos oncoprotein to a jun-independent DNA binding protein with GCN4 dimerization specificity by swapping "leucine zippers". Nature. 1989 Sep 7;341(6237):74–76. doi: 10.1038/341074a0. [DOI] [PubMed] [Google Scholar]
- Talanian R. V., McKnight C. J., Kim P. S. Sequence-specific DNA binding by a short peptide dimer. Science. 1990 Aug 17;249(4970):769–771. doi: 10.1126/science.2389142. [DOI] [PubMed] [Google Scholar]
- Vinson C. R., Sigler P. B., McKnight S. L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989 Nov 17;246(4932):911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- Weiss M. A., Ellenberger T., Wobbe C. R., Lee J. P., Harrison S. C., Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990 Oct 11;347(6293):575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]