Abstract
Data are presented on quantum speciation in the Sitopsis section of the genus Aegilops (Poaceae, Monocotyledones). Two small, peripheral, isolated, wild populations of annual cross-pollinated Ae. speltoides and annual self-pollinated Ae. sharonensis are located 30 m apart on different soil types. Despite the close proximity of the two species and their close relatedness, no mixed groups are known. Comparative molecular cytogenetic analysis based on the intrapopulation variability of rRNA-encoding DNA (rDNA) chromosomal patterns of individual Ae. speltoides geno-types revealed an ongoing dynamic process of permanent chromosomal rearrangements. Chromosomal mutations can arise de novo and can be eliminated. Analysis of the progeny of the investigated genotypes testifies that inheritance of de novo rDNA sites happens frequently. Heterologous recombination and/or transposable elements-mediated rDNA transfer seem to be the mechanisms for observed chromosomal repatterning. Consequently, several modified genomic forms, intermediate between Ae. speltoides and Ae. sharonensis, permanently arise in the studied wild population of Ae. speltoides, which make it possible to recognize Ae. sharonensis as a derivative species of Ae. speltoides, as well as to propose rapidness and canalization of quantum speciation in Sitopsis species.
Keywords: in situ hybridization, En/Spm transposons, peripheral populations, ecological speciation
Speciation is the most intriguing enigma of biology. During the last century, our knowledge of fundamental components of the speciation process became better defined, but we are still nowhere nearer to a complete understanding. Currently, a number of speciation models and mechanisms are proposed. The “genic” and “chromosomal” hypothesis of the predominant speciation mechanisms is still hotly debated (reviewed in refs. 1–6). Here, we will present molecular cytogenetic evidence supporting the model of quantum speciation by chromosomal repatterning in the genus Aegilops. Quantum speciation implies rapid origination of a new species in a small, ecologically marginal population on the periphery of the parent species area (3, 7, 8).
Carl Linnaeus described Aegilops in 1737. Systematically, this genus takes an intermediate position between the genus Triticum and the genus Agropyron (9). The section Sitopsis of the genus Aegilops is closest to the genus Triticum. This section includes five diploid species: Ae. speltoides Tausch., Ae. longissima Sweinf. et Muschl., Ae. searsii Feldman et Kislev, Ae. sharonensis Eig, and Ae. bicornis Forssk. Within the Sitopsis group, Ae. speltoides stands out from the four other species in several important features: (i) it is the only cross-pollinated species in the section; (ii) it grows on terra rossa or alluvial soils, although the rest of the Sitopsis species (except Ae. searsii) are constrained to light sand soils; (iii) it exhibits significant differences in molecular composition of repetitive DNA fraction (10); and (iv) geographically, it is distributed in the central part of the genus area, i.e., in and around the Fertile Crescent, whereas the rest of the Sitopsis species are local peripheral endemics of the southern part of the western wing of the Fertile Crescent (11).
The Fertile Crescent center is considered to be the primary center of Triticum/Aegilops species variability where local populations of wild progenitors of cultivated wheats exhibit significant genetic diversity, and until now, this center preserves its speciation potential (9). Notably, the area of Ae. longissima, Ae. searsii, Ae. sharonensis, and Ae. bicornis marginally overlaps with the southwestern part of the area of Ae. speltoides, and it seems as if these species replace Ae. speltoides in the southern climatically and edaphically special environments. All recorded features make it possible to hypothesize Ae. speltoides as an ancestral species for the Sitopsis section.
To check the last statement and to investigate the evolutionary interrelations between two diploid members of the Sitopsis section, namely, between annual cross-pollinated Ae. speltoides (2n = 2x = 14) and annual self-pollinated Ae. sharonensis (2n = 2x = 14), we conducted comparative molecular cytogenetic analysis of individual genotypes from two isolated wild populations of Ae. speltoides and Ae. sharonensis. These populations are located on the western banks of the Kishon River (Haifa Bay area, Israel) 30 m apart (Fig. 1). It is essential to note that, despite the close proximity of the two species, no mixed groups are known. The Kishon populations of Ae. speltoides and Ae. sharonensis were selected as a speciation model for several reasons. (i) This is the south border range of Ae. speltoides, the population is small (≈100 m2), and it has long been established that on the species border range in small populations intrapopulation differentiation processes are intensified (3, 12). (ii) According to our previous data (13), the Kishon population of Ae. speltoides is highly heteromorphic, exhibits a high level of Enhancer/Suppressor-mutator (En/Spm) transposons activity during male gametogenesis, and possesses a wide spectrum of chromosomal abnormalities, including supernumerary chromosomes, heterozygosity for translocations, and variability in chromosomal position/number of 45S and 5S rDNA sites. It has been known that chromosomal repatterning is an integral part of the speciation process (2, 7), and, as a consequence, speciation in wild diploid and polyploid wheats is associated with significant repatterning of rDNA sites (14–17). It is obvious that speciation-related chromosomal repatterning extended further rDNA sites, but their dynamics can be regarded as a strong indicator for significant intragenomic processes (18). Thus, we used changes in rDNA chromosomal pattern as a marker for potential micro-evolution in the Kishon populations of Ae. speltoides and Ae. sharonensis. We assumed that Ae. sharonensis is a derivative of Ae. speltoides adapted for special edaphic ecology (light sand soil).
Fig. 1.
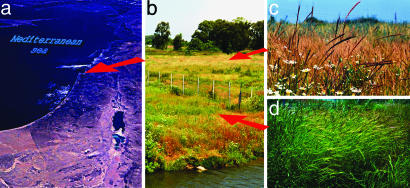
Geographical location and photos of the investigated populations. (a) Satellite image of eastern Mediterranean. (b) Field position of the studied populations. (c) Ae. sharonensis. (d) Ae. speltoides. The photographs of Ae. sharonensis and Ae. speltoides were taken in the same day. Although Ae. speltoides is blossoming, Ae. sharonensis is already mature.
Materials and Methods
Plant Material. Individual spikes of Ae. speltoides (ssp. aucheri and ssp. ligustica) and Ae. sharonensis were collected in 2001 in the Kishon populations (Fig. 1). The populations are located in the Haifa Bay area: the population of Ae. sharonensis inside, and the population of Ae. speltoides close to the Akko plain terminal of desert plants (19). The Kishon population of Ae. sharonensis is much larger (≈9,000 m2) than the population of Ae. speltoides (≈100 m2).
Chromosome counting of 57 individual plants of Ae. speltoides from the Kishon population was conducted. The following genotypes were analyzed: 26 of Ae. speltoides (15 original and 11 progeny genotypes) and 7 original genotypes of Ae. sharonensis from the Kishon populations, and 5 original genotypes of Ae. sharonensis from Caesarea population 60 km south of the Kishon population in the coastal plain. Plants of Ae. speltoides ssp. aucheri Ts-84 (Latakia, Syria), Ae. speltoides ssp. ligustica Ts-24 (Eregli, Turkey), and Ae. sharonensis (Caesarea, Israel) were used as controls (Table 1 and Fig. 2).
Table 1. Morphological characters of Ae. speltoides and Ae. sharonensis.
| Morphological characters | Spike, cm | Lemma: lateral awns | Glume: keel tooth (teeth) |
|---|---|---|---|
| Ae. speltoides ssp. aucheri Ts-84* | 8-9 | Absence | Absence |
| Ae. speltoides ssp. ligustica Ts-24† | 8-9 | Presence | Presence |
| Ae. speltoides ssp. aucheri (Kishon)‡ | 8-20 | Absence | Presence/absence (polymorphic) |
| Ae. speltoides ssp. ligustica (Kishon)‡ | 8-20 | Presence/absence (polymorphic) | Presence/absence (polymorphic) |
| Ae. sharonensis (Kishon)‡ | 6-14 | Presence | Presence |
| Ae. sharonensis (Caesarea)‡ | 6-14 | Presence | Presence |
From Latakia Province, Syria (PI 487235, USDA).
From Eregli, Turkey (University of California, Riverside, G-1038).
From Israel (Gene bank of the Institute of Evolution, University of Haifa).
Fig. 2.
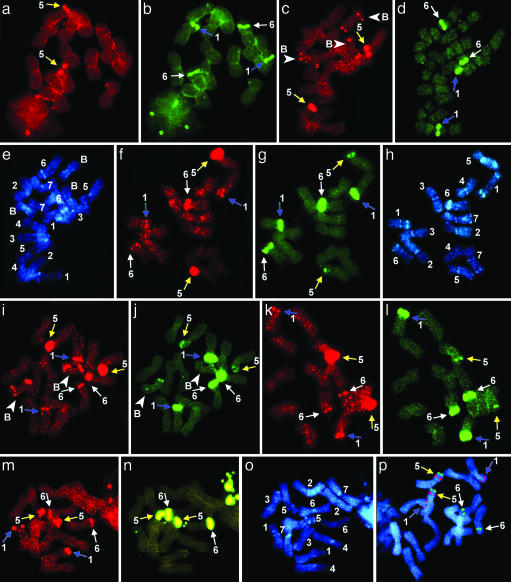
In situ hybridization of 5S (detected red) and 45S rDNA (detected green) on metaphase chromosomes of Aegilops speltoides and Ae. sharonensis and differential staining with Hoechst 33258. (a and b) Ae. speltoides Ts-24, standard karyotype. (c–e) Ae. speltoides (Kishon), genotype 4-1. Original genotypes presented in (f–h) and their progeny (k and l) carry additional 5S and/or 45S rDNA sites on chromosomes 1, 5, and 6. (f–h) Genotype 1-1, heterozygote for deletion of chromosome 1 long arm. (i and j) Genotype 1-2, from the same spike as genotype 1-1. (k and l) Progeny of the genotype 1-1 after self-pollination, homozygote for deletion of chromosome 1 long arm. (m–o) Ae. sharonensis (Kishon). Chromosomes 5 and 6 carry additional rDNA sites. (p) Ae. sharonensis (Caesarea), standard karyotype. Chromosomes 1, 5, and 6 are shown by blue, yellow, and white arrows, respectively. B-chromosomes are shown by white arrowheads.
Probe Labeling, in Situ Hybridization, and Detection. For fluorescent in situ hybridization (FISH) localization of 45S rRNA and 5S rRNA gene regions, we used pTa71 (20) and As5SDNAE (21) probes, respectively. Probe pTa71 was labeled with biotin-16-dUTP (Roche) and detected with Alexa 488 (Molecular Probes). Probe As5SDNAE was directly labeled with Cy-3 (Amersham Pharmacia). Clone ESas-2 (AY265311), the internal part of the transposase (TPase) region of En/Spm-like transposons (13), was labeled with digoxigenin-11-dUTP (Roche) and detected with rhodamine-conjugated sheep anti-digoxigenin Fab fragment (Roche). Telomeres were localized by hybridization with a PCR-generated synthetic probe consisting of a tandem repeat array of the short sequence TTTAGGG (22) labeled with digoxigenin-11-dUTP (Roche) and detected with Alexa 350 (Molecular Probes). The FISH procedure has been described in detail (13, 23, 24).
Chromosomal Identification. Ae. speltoides and Ae. sharonensis chromosomes were identified according to rDNA patterns (15) and differential staining with AT-specific fluorochrome Hoechst 33258 on the same slides used for fluorescent in situ hybridization (Fig. 2 e, h, and o) (23). Slides were examined on a Leica (Deerfield, IL) DMR microscope by using black and white Kodak T 400 CN and color Fujicolor Superia X-TRA 400 microfilms. Images were scanned with a Nikon LS-400.
Results
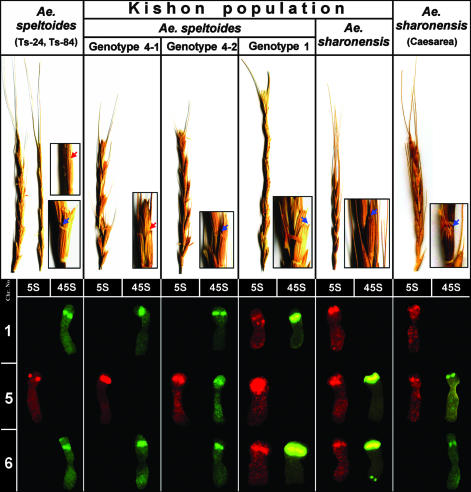
Ae. speltoides Morphological and Chromosomal Diversity. There are two morphological types of Ae. speltoides (11). The aucheri-type is characterized by cylindrical spikes with widely interspersed spikelets. Only the apical spikelet is awned, and the rachis is tough. The ligustica-type is characterized by denser, two-rowed ears in which the lateral spikelets are also awned and the rachis is brittle (Fig. 3). Analysis of the three important morphological characters [spike size, glume morphology (presence/absence of keel tooth), and lateral awns expressivity (latter for ssp. ligustica) (Fig. 3) (25)] revealed significant intrapopulation morphological variability of Ae. speltoides that guided the successive cytogenetical screening. Variability of morphological characters for different genotypes is shown in Table 1. Chromosome counting of 57 (among these, 15 genotypes used for in situ hybridization) original individual plants of Ae. speltoides, from the Kishon population grown in the greenhouse from 10 spikes with different phenotypes, revealed from one to eight supernumerary B-chromosomes and a wide spectrum of chromosomal rearrangements, including variability in chromosomal position/number of 45S and 5S rDNA sites (see also ref. 13). Nevertheless, all plants were fertile after self- and cross-pollination in the greenhouse.
Fig. 3.
Morphological and karyotypic variability of selected genotypes of Aegilops speltoides and Ae. sharonensis. Spikes of different genotypes are presented. Ae. speltoides ssp. aucheri Ts-84 and genotype 1: only terminal spikelet awned. Ae. speltoides ssp. ligustica Ts-24 and genotypes 4-1, 4-2, Ae. sharonensis: terminal spikelet and lemmas of lateral spikelets awned. Presence or absence of keel tooth on the glume is shown, respectively, by red and blue arrows in the enlargement fragments of the spikes (Ae. speltoides ssp. aucheri Ts-84 and genotype 4-1 do not have the tooth on the glume, red arrows). Simultaneous in situ hybridization of 5S (detected red) and 45S rDNA (detected green) on the somatic chromosomes. Only chromosomes 1, 5, and 6 with signals are shown. Polymorphism in the morphological characters and chromosomal rDNA patterns are observed in plants of Ae. speltoides and Ae. sharonensis from the Kishon.
Ae. speltoides rDNA Chromosomal Pattern. Chromosomes 1, 5, and 6 (specifically investigated in this study) could be unambiguously identified in the Ae. speltoides genome according to rDNAs and AT-enriched heterochromatin patterns (Fig. 2 e and h). Normally, Ae. speltoides carries 5S rDNA sites on the short arm of chromosome 5, and chromosomes 1 and 6 carry 45S rDNA sites on the short arms in the secondary constriction regions (Figs. 2 a and b and 3) (15).
Several types of rDNA patterns have been observed.
The normal pattern similar to that of Ts-24 and Ts-84 (Figs. 2 c and d and 3, genotype 4-1).
-
Patterns with additional rDNA sites on chromosomes 1, 5, and 6.
- Additional 45S rDNA sites on chromosome 5 plus 5S rDNA sites on chromosomes 1 and 6 (Fig. 2 f–l).
Fig. 4.
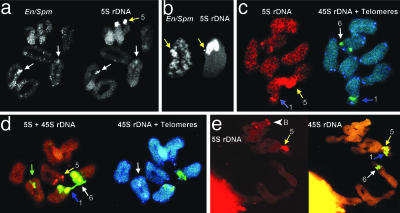
Intragenomic dynamics of rDNA sites in male gametogenesis of Ae. speltoides.(a) Intragenomic En/Spm-mediated transfers of 5S rDNA. Probe As5SDNAE (detected green) for detection of 5S rDNA and clone ESas-2 (detected red) for detection of the internal part of TPase region of En/Spm-like transposons were hybridized simultaneously on meiotic chromosomes. Extra 5S rDNA sites coupled to En/Spm transposons are shown by white arrows. (b) Chromosome 5, concurrence of En/Spm and additional 5S rDNA clusters are shown by yellow arrows. (c) Meiotic chromosomes (diakinesis stage) of the progeny of the same genotype after selfing. Chromosomes 1 and 6 carry additional 5S rDNA blocks concurrent with 45S rDNA. Moreover, one of the conjugated homologues of chromosome 1 carries one more additional intercalary 5S rDNA block on the short arm. One of the conjugated homologues of chromosome 5 carries an extra 5S rDNA block (3 blocks on bivalent totally). Chromosome 6 demonstrates a reduced satellite. (d) Meiotic chromosomes from the same anther as c. The consequences of heterologous recombination are observed. The bridge between 45S rDNA blocks of the chromosomes 1 and 6 still exist. Third (additional) 5S rDNA block on chromosome 5 is lost, whereas the other bivalent (green arrow) simultaneously carries 5S, 45S rDNA blocks and telomeric repeats (detected blue). (e) Meiotic chromosomes, diplotene stage. Heterologous synapses of chromosomes 1 and 5. Chromosome 5 carries an extra 45S rDNA block. Chromosomes 1, 5, and 6 are shown by blue, yellow, and white arrows, respectively. B-chromosomes are shown by white arrowheads.
Moreover, in a number of original genotypes, chromosome 1 carries a second additional intercalary 5S rDNA block in the short arm (Fig. 3, genotype 1) in homo- (Fig. 2f) or heterozygous conditions (Fig. 2i).
The progeny of plants carrying extra-rDNA (Fig. 2 f–h) mainly followed parental genotypes after selfing (Fig. 2 k and l). However, we regularly observed the emergence of new rDNA sites as well as genome “normalization,” i.e., elimination of additional rDNA sites from chromosome 5 and/or chromosomes 1 and 6. For example, in the progeny of genotype 2 (similar to genotype 1), one plant followed the parental genotype, whereas chromosomes 1 and 6 in another plant simultaneously lost additional 5S rDNA blocks and became “regular” (not shown).
Satellites on chromosomes 1 and 6 (Figs. 2 f and g and 3, genotype 1) were significantly reduced in a number of genotypes. Supernumerary B-chromosomes, if they exist, carry distal 5S rDNA blocks on both arms (Figs. 2 c and i and 4e) and sometimes also 45S blocks (Fig. 2j).
Ae. sharonensis rDNA Chromosomal Pattern. Normally, chromosome 1 of Ae. sharonensis carries a distal 5S rDNA site on the short arm, chromosome 5 carries one distal 5S rDNA and one telomeric 45S rDNA block on the short arm, and chromosome 6 carries a 45S rDNA block in the secondary constriction region on the short arm (Figs. 2p and 3) (15). This pattern was observed in the plants from the control Caesarea population.
Besides the norm, plants with modified rDNA pattern have often been detected in the Kishon population of Ae. sharonensis (Figs. 2 m–o and 3): chromosome 5 carries an additional 5S rDNA block concurrent with a regular terminal 45S rDNA site. Chromosome 6 carries an additional 5S rDNA block in the short arm (Figs. 2 m and n and 3). An additional terminal 45S rDNA block is observed on the long arm of chromosome 6, and the satellite on the short arm is significantly reduced in comparison with chromosome 6 from Caesarea plants (Fig. 3).
Thus, remarkably, the modified genotypes of Ae. speltoides (Figs. 2 f–l and 3, genotypes 4-2 and -1) and Ae. sharonensis (Figs. 2 m–o and 3) from the Kishon populations have additional rDNA sites on chromosomes 1, 5, and 6 and reduced satellites on chromosome 6.
Additional rDNA Clusters in Connection with En/Spm Transposons Activity in Meiosis. Simultaneous in situ hybridization with TPase fragments of the En/Spm transposons and 5S rDNA was conducted on the meiotic chromosomes of Ae. speltoides plants from the Kishon population (Fig. 4 a and b) (see also ref. 13). The concurrence of chromosomal localization of the En/Spm clusters with additional clusters of 5S rDNA has also been revealed (Fig. 4 a and b).
rDNA Sites Can Provoke Heterologous Synapses and Recombination. Chromosomes that carry extended rDNA arrays, such as rDNA sites, telomeric repeats, etc., are involved in heterologous synapses and recombination (Fig. 4 d and e). Thus, we observed the bridge between 45S rDNA blocks of chromosomes 1 and 6 (Fig. 4d). A combined block of 5S rDNA, 45S rDNA, and telomeric repeat on the other bivalent is also the consequence of such an event (Fig. 4d). Another example of heterologous synapses between chromosome 5, which carries an additional 45S rDNA site, and chromosome 1 is shown in Fig. 4e.
Discussion
Intrapopulation Variability of rDNA Sites. Chromosomal distribution of 5S and 45S rDNAs is different in the two analyzed species. Given that our assumption is true, and Ae. sharonensis is indeed a derivative of Ae. speltoides adapted for special sandstone edaphic ecology, there must formally be at least two speciation events concerning the remodeling of 5S and 45S ribosomal sites: (i) emergence of a new 5S rDNA cluster on chromosome 1, and (ii) transfer of the 45S rDNA cluster from chromosome 1 to a distal position on the short arm of chromosome 5 (Fig. 3). It is unlikely that these events occur simultaneously, and a priori we expect intermediate genotypes. Indeed, this is what we found. Although several genotypes exhibit a speltoides-type of rDNA chromosomal distribution (Figs. 2 c and d and 3, genotype 4-1), other genotypes (Figs. 2 f–l and 3, genotypes 4-2 and -1) show intermediate rDNA patterning between speltoides and sharonensis types (Figs. 2 m–p and 3). These intermediate genotypes already possess intercalary 5S rDNA sites on chromosome 1, but do not yet have the distal 45S cluster on chromosome 5. Genotype 4-2 possesses chromosome 5 with a sharonensis-type of rDNA distribution, i.e., mobile cluster of 45S rDNA colocated with a regular 5S rDNA site, but chromosomes 1 and 6 are still of the speltoides-type (Fig. 3). It is important to emphasize that genotypes 4-1 with normal rDNA patterning and 4-2 with a modified pattern were grown from the same spike, a strong argument in favor of rDNA remodeling permanency. Plants of Ae. sharonensis from the Kishon population still possess some features of rDNA chromosomal distribution of intermediate genotypes: the presence of the additional 5S rDNA cluster on chromosome 6 of genotype 1, for instance (Figs. 2 and 3).
Formally, the presence of intermediate genotypes may be explained by introgressive hybridization. It should be stated, however, that, from the very beginning, we excluded this possibility for three reasons. First, natural hybridization among the diploid Aegilops species is a very rare phenomenon despite the fact that several diploids have massive spatial contact (11). Second, due to our field observations, both seasonal isolation and isolation due to the mating system occurs: self-pollinated Ae. sharonensis blossom 1–1.5 months earlier than cross-pollinated Ae. speltoides. In Fig. 1, photographs of Ae. sharonensis (1c) and Ae. speltoides (1d) were taken on the same day. Although Ae. speltoides was blossoming, Ae. sharonensis was already fully mature. Third, despite the close spatial proximity of the two species, no mixed groups were found that testify to ecological divergence, both temporal and edaphic (26).
Possible Mechanisms of rDNA Repatterning. We propose that the repatterning process of speciation-related rDNA sites starts from a simple “traveling” of rDNA to a new site. The mobility of rDNA clusters has been described in many plant species. It may involve major loci, small numbers of copies of the repeat unit, or fragments of the unit (14, 27). Ectopic recombination and/or activity of mobile elements were proposed to be the mechanism for this “traveling” (13). The latter implicated mobility of rDNA sites in genomes of individual plants of Ae. speltoides from the Kishon population associated with activation of En/Spm transposons in meiosis, which was recently described by us (13). Analysis of the dynamics of rDNA clusters at the stage of male gametogenesis is the key for understanding the possible mechanisms of rDNA repatterning. Fluorescent in situ hybridization experiments revealed the inter- (Fig. 4a) and intrachromosomal (Fig. 4b, chromosome 5) transfer of 5S rDNA together with En/Spm elements. As a result, one of the conjugated homologues of chromosome 5 (Fig. 4b) carries an additional 5S rDNA block, and, importantly, this character is heritable because the progenitor also possesses chromosome 5 with extra-5S rDNA sites at the same position (Fig. 4c). Besides, the progeny preserves maternal distribution of rDNA on chromosome 1, i.e., one homologue inherits an extra 5S rDNA site concurrent with 45S rDNA, whereas the other homologue inherits one more additional intercalary 5S rDNA site (Fig. 4c).
Regardless of the mechanisms of rDNA transfer to a new place on the chromosomes, new rDNA clusters must be stabilized. This stabilization can be achieved by homozygotization in two alternative ways: (i) by selfing (Fig. 2 k and l) or crossing with similar genotypes and (ii) by non-Mendelian epigenetic mechanisms of En/Spm-mediated intragenomic transfer of rDNA (13). The latter explains the occurrence of two homozygous but different genotypes from one spike: genotypes 4-2 vs. 4-1 (Fig. 3), or 1-2 (Fig. 2 f–h) vs. 1 (Fig. 3) when both homologs of chromosome 5 carry or do not carry the additional 45S rDNA block. We propose that such events may occur in the zygote after fertilization (Figs. 2 and 3). In its turn, the emergence of a new rDNA site may induce a number of intragenomic events as a consequence of heterologous synapses and recombination between chromosomes that carry extended rDNA arrays (Fig. 4 d and e). In Fig. 4d, the bridge between 45S rDNA blocks of chromosomes 1 and 6 is a relict of this meiotic heterologous recombination. Moreover, on the other bivalent of the same meiotic plate, we observed a combined 5S plus 45S rDNA block in the telomeric position that was also a consequence of heterologous recombination in meiosis or in a premeiotic cell. Because both chromosomal plates presented in Fig. 4 c and d were obtained from the same anther, it is evident that the third extra-5S rDNA block of chromosome 5 (Fig. 4c) was involved in this particular cell-specific heterologous recombination: chromosome 5 does not carry an additional 5S rDNA block in Fig. 4d and seems to be “revertant” to the norm.
In the other case, the emergence of an additional 45S rDNA site on chromosome 5 provoked heterologous synapses with chromosome 1 (Fig. 4e). It is very possible that chromosome 6 could also be involved in heterologous synapses with chromosomes 1 and/or 5 because the presence of additional rDNA sites and a reduction of satellite on the short arm of chromosome 6 in a number of genotypes (Figs. 2 f and g and 3, genotype 1) and their progeny (Figs. 2 k and l and 4c) were observed.
However, the limitation for the quantity of extra rDNA sites most probably exists due to a deleterious effect of increasing heterologous recombination that may trigger a fatal imbalance of the plant genome.
Conclusions
All of the aforementioned make it possible to conclude that our assumption, most probably, is true, and Ae. sharonensis is indeed a derivative of Ae. speltoides adapted for special edaphic and climatic temporal environments. As is obvious from the geno-type descriptions, in a small peripheral population of Ae. speltoides, a series of permanent chromosomal rearrangements emerged, resulting in the onset of several new genomic forms with modified karyotypes (Figs. 2, 3, 4). This finding provides the basis for the particular adaptive combination of genes when the peripheral population of Ae. speltoides is degraded. A limited number of naturally selected fertile, homozygous, and self-compatible forms (Fig. 2 f and k) with sharonensis-types of rDNA chromosomal patterns may colonize soils unfit for the maternal species (Fig. 1b). The latter is confirmed by the fact that Ae. sharonensis from the Kishon population still possesses some features of rDNA chromosomal distribution of intermediate genotypes, i.e., similarity of chromosome 5 of Ae. sharonensis and of Ae. speltoides (genotype 4-2), as well as chromosome 6 of Ae. sharonensis and of Ae. speltoides (genotype 1) (Fig. 3). Thus, we observe a certain “evolutionary channel” leading from the plastic Ae. speltoides genome to that of Ae. sharonensis.
Mode and Tempo of Speciation. Close proximity of the two populations (Fig. 1b) may point to rapid quantum speciation. If the Kishon population of Ae. sharonensis is a derivative of Ae. speltoides population, which is most probable, then it is possible to presume that rapid quantum speciation took place. It implies a new species originating as a new structural homozygote in a small, ecologically marginal population on the periphery of the parent species (3, 7, 8). One of the main characteristic features of this type of speciation is the absence of a hybrid zone surrounding the area of the occurrence of the two species due to the strong genetic isolation of the neospecies observed in the studied populations. Alternatively, if the current coexistence of Ae. speltoides and Ae. sharonensis populations is due to a secondary contact, it is quite possible that Ae. sharonensis arose in another area by geographical isolation (3, 8, 28).
Causes of Speciation. Another important question is, Why does speciation take place in a certain geographical zone, in a certain time, and in a certain group of species? We can hypothesize that in the Aegilops case the cause may be climatic changes in the region during the Quarternary period. Under the influence of the European glaciation, up to the late Pleistocene, the floral zone of ancient Ae. speltoides extended southward to the present, down to northern Africa, but in the Holocene period a gradual recession to the north took place (29). The impact of the glacial period and the close proximity of a large Afro-Arabian desert domain most probably played a leading role in the formation of the Sitopsis group. Geographical location of the Sitopsis species from Egypt (Ae. bicornis) to Syria (Ae. longissima and Ae. searsii) (11) and their special ecological preference made it possible to propose that all four species originated during the recession in the same way as it was described above for Ae. sharonensis, as a result of Ae. speltoides northward retreat at its southern border. In the Quarternary period, there were at least 6 interchanges of cold–temperate climate (29, 30), and the recession scenario may have happened repeatedly. Thus, these species growing largely on sand soils and adapting to xeric environments could be a “trace” of the ancient Ae. speltoides area “pulsation.” Indirectly, it is confirmed by our previously published data from comparative genomic in situ hybridization, when Ae. bicornis, the most southern and probably first of the derived Sitopsis species, exhibited the farthest divergence from Ae. speltoides in a repetitive noncoding DNA fraction (23, 31). If this scenario is substantiated, then the Sitopsis species represent a speciation trend following Grant's (32) theory, when the phyletic group keeps pace with the Pleistocene/Holocene climatic and edaphic changes by means of a succession trend of species.
Acknowledgments
We thank Verne Grant, Maria Puertas, and Tomas Naranjo for helpful comments. We thank Elena Ivanitskaya, Irina Solovei, and Jan Barber for helpful suggestions. This work was supported by grants from the Research Authority of the University of Haifa and the Ancell–Teicher Research Foundation of Genetics and Molecular Evolution.
Abbreviation: En/Spm, Enhancer/Suppressor-mutator.
References
- 1.Timofeeff-Ressovsky, N. W., Vorontsov, N. N. & Yablokov, A. V. (1977) Evolutionary Theory Essay (Nauka, Moscow).
- 2.White, M. J. D. (1978) Modes of Speciation (Freeman, San Francisco).
- 3.Grant, V. (1981) Plant Speciation (Columbia Univ. Press, New York).
- 4.Nevo, E. (2000) in Nature Encyclopedia of Life Sciences (Nature Publishing Group, London), pp. 1–11.
- 5.Forsdyke, D. R. (2003) J. Biol. Sys. 11, 341–350. [Google Scholar]
- 6.Gottlieb, L. D. (2004) New Phitol. 161, 71–82. [Google Scholar]
- 7.Lewis, H. (1962) Evolution 16, 257–271. [Google Scholar]
- 8.Grant, V. (1963) The Origin of Adaptation (Columbia Univ. Press, New York).
- 9.Zhukovsky, P. M. (1928) Proc. Appl. Bot. Select. Genet. 18, 417–609. [Google Scholar]
- 10.Dvorak, J. & Zhang, H. B. (1992) Theor. Appl. Genet. 84, 419–429. [DOI] [PubMed] [Google Scholar]
- 11.Kimber, G. & Feldman, M. (1987) Wild Wheat: An Introduction (Univ. Missouri Press, Columbia, MO).
- 12.Nevo, E., Kirzhner, V., Beiles, A. & Korol, A. (1997) Philos. Trans. R. Soc. London B. 352, 381–389. [Google Scholar]
- 13.Raskina, O., Belyayev, A. & Nevo, E. (2004) Chromosome Res. 12, 153–161. [DOI] [PubMed] [Google Scholar]
- 14.Dubkovsky, J. & Dvorak, J. (1995) Genetics 140, 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Badaeva, E. D., Friebe, B. & Gill, B. S. (1996) Genome 39, 1150–1158. [DOI] [PubMed] [Google Scholar]
- 16.Maestra, B. & Naranjo, T. (2000) Chromosomes Today 13, 155–167. [Google Scholar]
- 17.Baum, B. R., Bailey, L. G., Belyayev, A., Raskina, O. & Nevo, E. (2004) Genome 47, 519–529. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, J. & Gill, B. S. (1994) Chromosoma 103, 179–185. [DOI] [PubMed] [Google Scholar]
- 19.Zohary, M. (1970) in Atlas of Israel, eds. Amiran, D. H. K., Elster, J., Gilead, M., Rosenan, N., Kadmon, N. & Parah, U. (Elsevier, Amsterdam).
- 20.Taketa, S., Ando, H., Takeda, K., Harrison, G. E. & Heslop-Harrison, J. S. (2000) Theor. Appl. Genet. 100, 169–176. [Google Scholar]
- 21.Baum, B. & Bailey, G. (2001) Genet. Res. Crop Evol. 48, 35–51. [Google Scholar]
- 22.Cox, A. V., Bennet, S. T., Parokonny, A. S., Kenton, A., Callimassia, M. A. & Bennet, M. D. (1993) Ann. Bot. 72, 239–247. [Google Scholar]
- 23.Belyayev, A. & Raskina, O. (1998) Chromosome Res. 6, 559–565. [DOI] [PubMed] [Google Scholar]
- 24.Belyayev, A., Raskina, O. & Nevo, E. (2001) Chromosome Res. 9, 129–136. [DOI] [PubMed] [Google Scholar]
- 25.Sarkar, P. & Stebbins, G. L. (1956) Am. J. Bot. 43, 297–304. [Google Scholar]
- 26.Lewis, H. & Roberts, M. R. (1956) Evolution 10, 126–138. [Google Scholar]
- 27.Heslop-Harrison, J. S. (2000) Chromosomes Today 13, 45–56. [Google Scholar]
- 28.Mayr, E. (1954) in Evolution as a Process, ed. Huxley, J. (Allen and Unwin, London).
- 29.Wulff, E. V. (1936) Historical Geography of Plants (State Agricultural Publishing Company, Moscow).
- 30.Tchernov E. (1988) in The Zoogeography of Israel, eds. Yom-Tov, Y. & Tchernov, E. (Kluwer, Dordrecht, The Netherlands), pp. 159–251.
- 31.Belyayev, A., Raskina, O. & Nevo, E. (2001) Heredity 86, 738–742. [DOI] [PubMed] [Google Scholar]
- 32.Grant, V. (1989) Am. Nat. 133, 604–612. [Google Scholar]