Abstract
The F-box proteins were originally identified as the key component of SKP1-Cullin1-F-box E3 ligase complexes that control the stability of their specific downstream substrates essential for cell growth and survival. However, the involvement of these proteins in type I IFN (IFN-I) signaling during innate immunity has not been investigated. In this study we report that the F-box protein FBXO17 negatively regulates IFN-I signaling triggered by double-strand DNA, RNA, or viral infection. We found that FBXO17 specifically interacts with IFN regulatory factor 3 (IRF3) and decreases its dimerization and nuclear translocation. The decrease of IRF3 dimerization and nuclear translocation is due to the recruitment of protein phosphatase 2 (PP2A) mediated by FBXO17, resulting in IRF3 dephosphorylation. Interestingly, PP2A recruitment does not require the F-box domain but instead the F-box associated region of the protein; thus, the recruitment is independent of the canonical function of the SKP1-Cullin1-F-box family of E3 ligase. Together, our studies identify a previously unreported role of FBXO17 in regulating IFN-I signaling and further demonstrate a novel mechanism for IRF3 deactivation by F-box protein-mediated recruitment of PP2A.
Introduction
During studies of cell cycle regulators, the F-box motif was first found in cyclin F and was shown to interact with S-phase kinase-associated protein 1 (SKP1) (1). The family of F-box proteins, which is characterized by this novel motif, was identified through screening proteins that bind SKP1 (2, 3). Traditionally, F-box proteins function as a component of the SKP1-Cullin1-F-box (SCF) E3 ligase complex to initiate ubiquitin-mediated targeted substrate destruction (4, 5). Previous studies have reported that F-box proteins regulate specific substrates in diverse biological processes, including cell growth and division, cell development and differentiation, and cell survival and death (6). Consequently, dysfunction of these proteins is closely linked with cancers and other diseases (6–9). However, the role for F-box proteins in innate immunity remain poorly studied, and thus far there is no report about involvement of F-box proteins in antiviral events through type I IFN (IFN-I) signaling.
In innate immunity, the IFN-Is induced by the activation of IFN-I signaling cascade play a central role in monitoring and defending against viral infection as well as guiding the subsequent adaptive immune responses (10–13). During the processes of viral infection and replication, host pattern recognition receptors sense the nucleic acid and other components from viruses, termed pathogen-associated molecular patterns, to mount a series of signaling cascade resulting in the production of IFN-Is and other cytokines (13–15). These secreted IFNs further induce and amplify the downstream transcriptional activation of a large number of IFN-stimulated genes through the JAK-STAT pathway, which cumulate to an effective restriction and elimination of pathogens by the host (16, 17). In addition, induction of IFN-I signaling pathway is often accompanied by NF-κB activation during microbial infection, which act in concert to induce a number of other proinflammatory molecules required for successful host defense (18). Weak IFN-I signaling results in insufficient IFN-I output needed for antiviral immunity (19). In contrast, chronic activation of cellular antiviral responses might lead to the prolonged and excess production of IFN-Is, which is deleterious and can provoke autoimmune disorders (19–22). Therefore, it is critical to build and integrate multiple positive and negative circuitries for regulating IFN-I signaling to ensure the homeostasis of IFN-I production by the host.
IFN regulatory factor 3 (IRF3) is a member of the IFN regulatory factors family, which plays a central role in IFN-I induction and antiviral response (13, 23, 24). Upon activation of RNA and DNA sensors, the key adaptor molecules TIR domain-containing adapter inducing IFN-β (TRIF), mitochondrial antiviral signaling protein (MAVS), and stimulator of IFN genes (STING) in the downstream pathway recruit and activate the TANK-binding kinase 1 (TBK1) or IκB kinase ε (IKKi) to phosphorylate IRF3 (25–27). The phosphorylated IRF3 undergoes dimerization and then translocates from the cell cytoplasm to the nucleus to initiate the transcription of IFN-Is and other target genes (28–30). Hence, controlling IRF3 activation is critical for the early regulation of IFN-I signaling. Indeed, a body of previous studies have demonstrated that different molecules and mechanisms have been employed for regulating the activation of IRF3. For example, IRF3, through Herc5-mediated ISG15 modification, can promote its self-activation (31). On the other hand, NIMA-interacting 1 (Pin 1), RNA transcriptional activator–associated ubiquitin ligase (RAUL), RBCC protein interacting with PKC1 (RBCK1), and forkhead box protein O1 (FoxO1) have been shown to mediate ubiquitin-dependent IRF3 degradation to impair production of IFN-Is (32–35). Clearly, regulation of IRF3 activation is complex, and many other mechanisms and regulatory circuitries are awaiting to be unraveled.
In this study, to our knowledge, we first identified F-box protein FBXO17 in negative regulation of IFN-I signaling triggered by various innate stimuli, including double-strand RNA, DNA, and viruses. We found that FBXO17 interacted with IRF3 specifically and decreased its dimerization and nuclear translocation. The decrease of IRF3 dimerization and nuclear translocation was rendered by FBXO17-mediated recruitment of protein phosphatase 2 (PP2A) for IRF3 dephosphorylation, which was independent of ubiquitin-mediated IRF3 destruction. Hence, our study provides a new insight into the role of F-box protein FBXO17 in regulation of IFN-I signaling, and further uncovers a novel mechanism of action operated by F-box containing proteins.
Materials and Methods
Cells and reagents
HEK293T, A549, and THP-1 cells were obtained from American Type Culture Collection, and maintained in DMEM or RPMI 1640 medium supplemented with 10% FBS (Thermo Fisher Scientific), 2 mM l-glutamine 100 U/ml penicillin and 100 μg/ml streptomycin at 37°C with 5% CO2 incubation. Polyinosinic-polycytidylic acid [poly(I:C)] and poly(deoxyadenylic-deoxythymidylic) acid [poly(dA:dT)] were obtained from InvivoGen. Vesicular stomatitis virus (VSV) was provided from Dr. Genhong Cheng’s laboratory (University of California, Los Angeles) and Sendai virus (SeV) by Dr. Jingfeng Wang (Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and Peking Union Medical College). HRP-anti-FLAG (M2) was obtained from Sigma-Aldrich. HRP-anti-hemagglutinin (HA) was obtained from Roche. HRP-anti-β-actin was obtained from GenScript. Ab specific to Gluc luciferase was obtained from New England BioLabs. Anti-IRF3 and β-tubulin were obtained from Santa Cruz Biotechnology. Anti-Lamin A/C was obtained from Immunoway. Abs specific to IRF3 phosphorylated at residue Ser396, TBK1 phosphorylated at residue Ser172, and TBK1 without phosphorylation were obtained from Cell Signaling Technology. Ab specific to FBXO17 was obtained from OriGene. Ab to PP2A-Ca was obtained from BD Biosciences. HRP-conjugated donkey anti-rabbit or anti-mouse Abs were obtained from Jackson ImmunoResearch Laboratories.
Constructs and plasmids
The constructs coding for FBXO17, IRF3, and these two truncated mutants were cloned into pcDNA3.1 vector. The PP2A-Ca and RACK1 were cloned from total RNAs isolated from THP-1 cells. The expression plasmids encoding full-length RIG-I, RIG-I (2 CARD), MDA5, MAVS, TBK1, IKKi, IRF3, IRF3-5D, IRF3 S386A, MyD88 were provided by Dr. Jun Cui (Sun Yat-Sen University).
Transfection and reporter assays
HEK293T cells (3 × 104) were seeded in 96-well plates and transfected using Lipofectamine 2000 (Invitrogen) plus an expression plasmid encoding an IFN-β (20 ng) or IFN-sensitive response element (ISRE) (20 ng) luciferase reporter, pRL-TK (Renilla luciferase reporter; 5 ng), 25 ng activating expression plasmids encoding RIG-I (2 CRAD), MAVS, TBK1, IKKi, IRF3, or IRF3-5D, and 50 ng expression plasmid encoding FBXO17. After 24 h, the luciferase activity was measured with the Dual-Luciferase reporter kit (Promega) in a Synergy II luminometer (Biotech) according to the manufacturer’s protocol.
RNA preparation and real-time PCR
Total RNA was extracted from HEK293T, THP-1, and A549 cells using TRIzol (Invitrogen) following the manufacturer’s instructions. cDNAs were transcribed from the extracted total RNA using the PrimeScript RT Reagent Kit with gDNA Eraser (Takara). Real-time PCR was performed using the FastStart Essential DNA Green Master reaction mix (Roche) and a Light-Cycler 96 (Roche). The relative expression of each gene was normalized to the expression of GAPDH as determined by the ΔCt method. IFN-β was amplified with forward primer 5′-CCTACAAAGAAGCAGCAA-3′ and reverse primer 5′-TCCTCAGGGATGTCAAAG-3′; ISG54 with forward primer 5′-GGAGGGAGAAAACTCCTTGGA-3′ and reverse primer 5′-GGCCAGTAGGTTGCACATTGT-3′; CCL5 with forward primer 5′-ATCCTCATTGCTACTGCCCTC-3′ and reverse primer 5′-GCCACTGGTGTAGAAATACTCC-3′; FBXO17 with forward primer 5′-CTGACCCGG TCCTTCAGTG-3′ and reverse primer 5′-CTCCCGTACTGCTCAAAAGATAC-3′; GAPDH with forward primer 5′-GAACGGGAAGCTCACTGG-3′ and reverse primer 5′-GCCTGCTTCACCACCTTCT-3′; and the VSV glycoprotein with forward primer 5′-TGCAAGGAAAGCATTGAACAA-3′ and reverse primer 5′-GAGGAGTCACCTGGACAATCACT-3′.
Immunoprecipitation and immunoblot analysis
For immunoprecipitation, cell lysates were incubated overnight with the appropriate Abs plus A/G beads (Pierce). For immunoprecipitation with anti-FLAG or anti-HA Abs, Ab-coated beads (Sigma) were used. The beads were washed with low-salt lysis buffer, eluted in 2× SDS Loading Buffer and resolved by SDS-PAGE. Proteins were transferred to polyvinylidene difluoride membranes (Millipore) and incubated with the appropriate Abs for detection with the Immobilon Western Chemiluminescent HRP Substrate (Millipore).
Nuclear protein extraction and native PAGE
The cytoplasmic and nuclear fractions of cells were prepared using the cytoplasmic and nuclear protein extraction kit (Beyotime) according to the manufacturer’s instructions. Native PAGE was performed as described in a previous report (36).
RNA interference and CRISPR/Cas9 assay
Small interfering RNAs (siRNAs) were transfected into HEK293T or THP-1 cells with Lipofectamine RNAi Max (Invitrogen) according to the manufacturer’s instructions. The sequence of the siRNA specific for FBXO17 is as follows: 5′-GCCGCAAUCUCAUCUUCAATT-3′. The short guide RNAs (sgRNAs) specific for FBXO17 or PP2A were designed and cloned into an sgRNA expression plasmid. The sequence of the sgRNA specific for FBXO17 is 5′-CCCGTCCACTATGTCGCGCC-3′, and for PP2A-Ca is 5′-GGACGAGAAGGTGTTCACCA-3′. The sgRNA and Cas9 expression plasmids were cotransfected with Lipofectamine 2000 into HEK293T cells. The resulting knockout cells were confirmed by genomic DNA sequencing using the InsTAclone PCR Cloning Kit (Fermentas). For FBXO17, the genomic DNA fragments were amplified with forward primer 5′-TGGGGAAGATTGGGAAAGGC-3′ and reverse primer 5′-TCGCCCATTGGCTACATCTC-3′, and for PP2A-Ca with forward primer 5′-CTGGAGCCTCAGCGAGCG-3′ and reverse primer 5′-CAGCGGCAAGACTCTTACTCAG-3′.
VSV-eGFP infection and ELISA
HEK293T cells were transfected with FBXO17-specific or scrambled siRNAs, and then infected at various multiplicities of infection with VSV. The concentration of IFN-β in the culture medium prepared from HEK293T cells was measured with an ELISA kit (Elabscience) following the manufacturer’s instructions.
Statistical analysis
The two-tailed unpaired t test was used for statistical analysis. All p values <0.05 were considered significant.
Results
Downregulation of IFN-I signaling by FBXO17
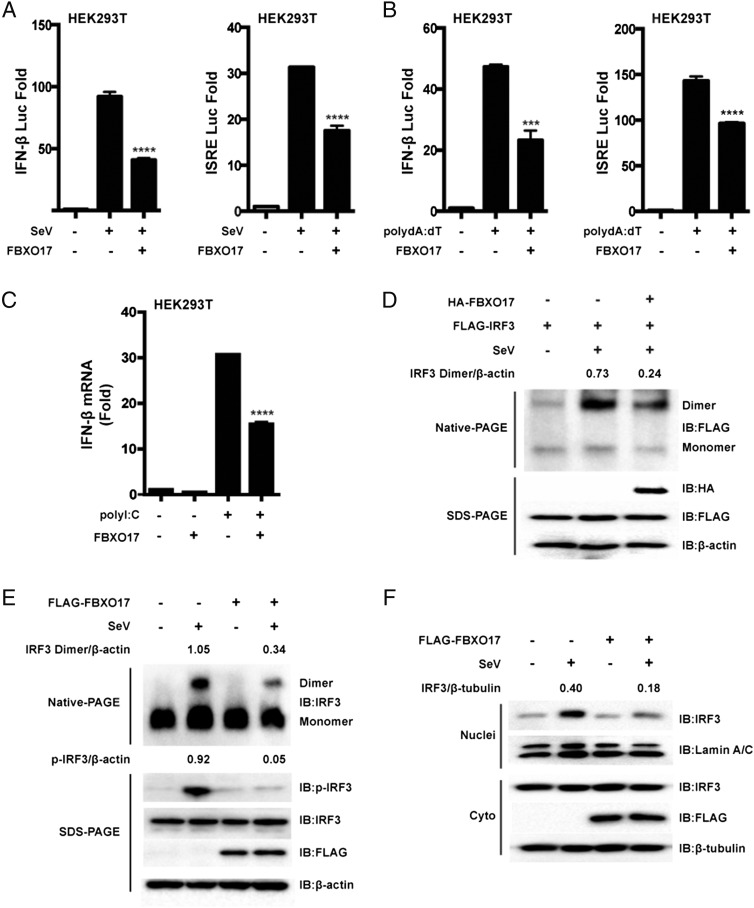
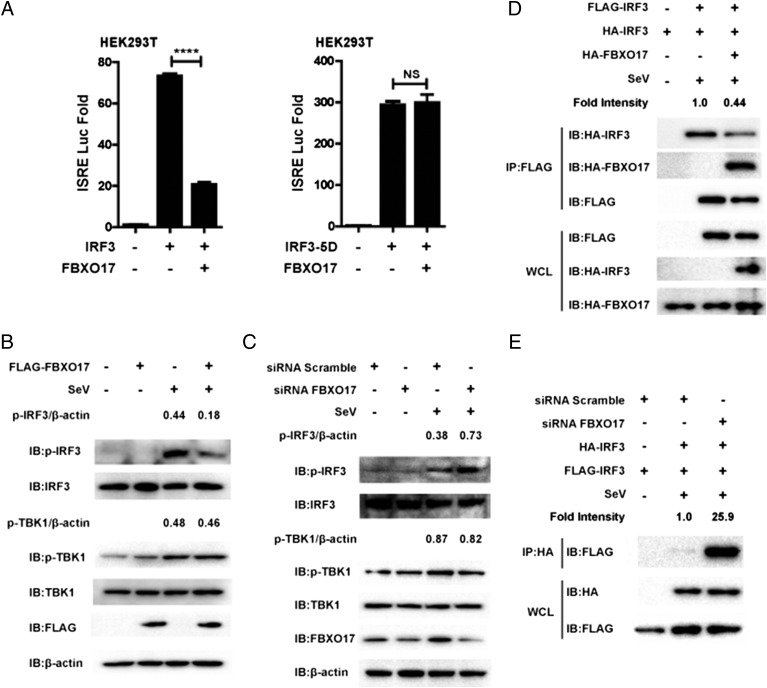
To determine if FBXO17 regulates IFN-I signaling, human embryonic kidney cells (HEK293T cells) were transfected with an IFN-β luciferase reporter and internal control Renilla luciferase reporter, plus the FBXO17 expression plasmid, and then infected with Sendai virus for 12 h. FBXO17 was found to repress SeV-induced IFN-β luciferase activity (Fig. 1A). Because the enhancer motif ISRE is activated by dimerized IRF3 and further induces transcription of the IFN-β gene, we used an ISRE luciferase reporter to confirm the inhibition of IFN-I signaling by FBXO17 and found that ectopically expressed FBXO17 inhibited SeV-triggered ISRE luciferase activity (Fig. 1A). We also obtained similar results from HEK293T cells treated with poly(dA:dT), when the cells were transfected with the FBXO17 expression plasmid (Fig. 1B). Real-time PCR showed that ectopic expression of FBXO17 repressed IFN-β transcription triggered by intracellular poly(I:C) (Fig. 1C). Taken together, these results suggest that FBXO17 acts as a negative regulator in IFN-I signaling.
FIGURE 1.
Downregulation of IFN-I signaling by FBXO17. (A and B) HEK293T cells were transfected with the FBXO17 expression plasmid and internal control Renilla luciferase reporter plus an IFN-β or ISRE luciferase reporter and were then infected with SeV for 12 h or treated with poly(dA:dT) for 18 h before assessing luciferase activity. (C) HEK293T cells were transfected with the FBXO17 expression plasmid and then stimulated with intracellular poly(I:C) for 12 h before real-time PCR assays were performed. (D and E) HEK293T cells were transfected with the indicated plasmids and then infected with SeV for 8 h. The cell lysates were separated by native (top) or SDS (bottom) PAGE and analyzed by immunoblotting with the indicated Abs. Numbers above the lanes indicate the relative accumulation of dimerized and phosphorylated IRF3. (F) Western blot analysis of the level of IRF3 in the cytoplasmic and nuclear fractions of HEK293T cells, which were transfected with the FBXO17 expression vector or an empty vector and then infected with SeV for 8 h. Numbers above the lanes indicate the relative accumulation of IRF3 in the nucleus. All results are representative of three replicate experiments. Student t test. ***p < 0.001, ****p < 0.0001.
Because phosphorylated, dimerized IRF3 is critical for activation of IFN-I signaling, we then considered whether FBXO17 affects the phosphorylation and dimerization of IRF3. As shown in Fig. 1D, FBXO17 repressed IRF3 homodimerization induced by SeV infection. Furthermore, we assessed the effect of FBXO17 on endogenous IRF3 phosphorylation and dimerization and found that FBXO17 decreased the level of phosphorylated and dimerized IRF3 (Fig. 1E). As IRF3 translocation from cytoplasm to nucleus is another hallmark for IRF3 activation, we next examined the translocation of endogenous IRF3 in cells with or without expression of FLAG-FBXO17 and found FBXO17 could decrease the level of IRF3 in the nucleus (Fig. 1F). Accordingly, these results suggest that FBXO17 negatively regulates the activation of IFN-I signaling triggered by double-strand DNA or RNA or by viral infection through decreasing the dimerization and nuclear translocation of IRF3.
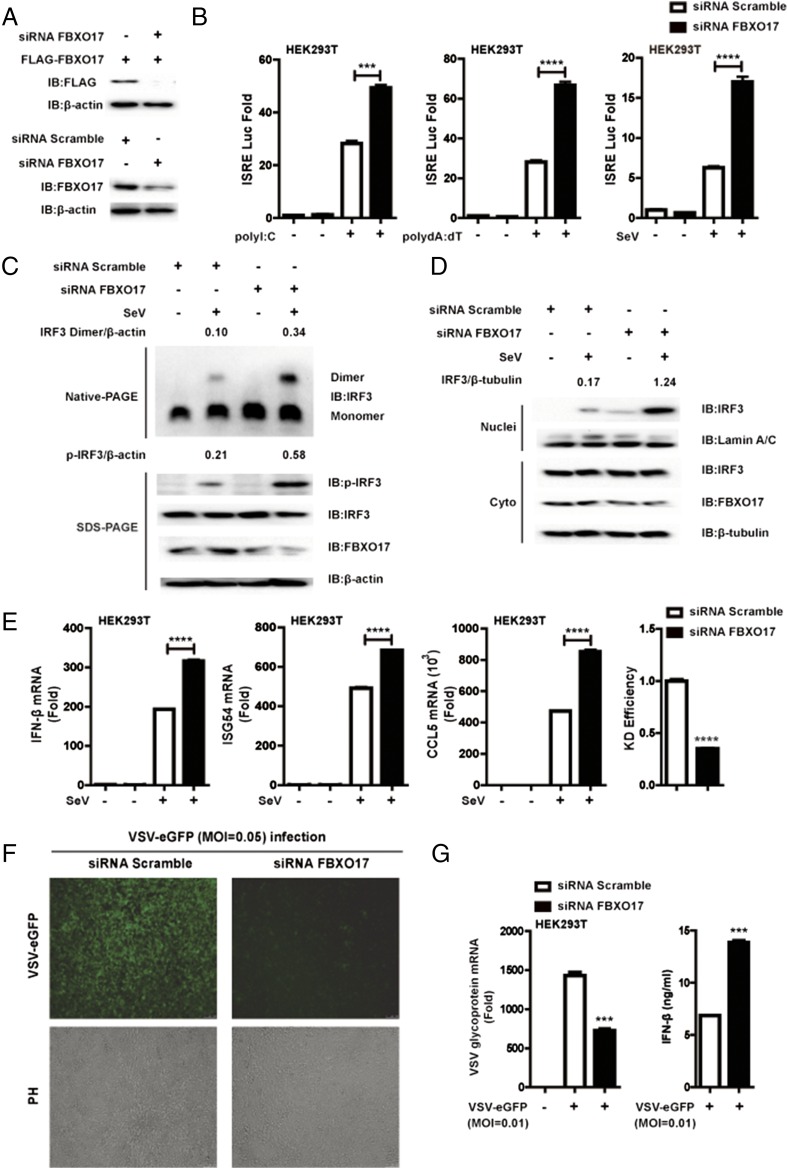
Knockdown of FBXO17 enhances IFN-I signaling and antiviral responses
We next assessed whether knockdown of endogenous FBXO17 would enhance transcription of IFN-β induced by various stimuli. We used an FBXO17-specific siRNA to interfere with FBXO17 expression and found that the siRNA effectively inhibited the expression of exogenous and endogenous FBXO17 (Fig. 2A, Supplemental Fig. 1A, 1B). Next, we observed that knockdown of FBXO17 enhanced ISRE luciferase activity induced by intracellular poly(I:C), poly(dA:dT), or SeV infection (Fig. 2B). Furthermore, we examined the effects of FBXO17 knockdown in IRF3 dimerization and nuclear translocation and found that knockdown of FBXO17 enhanced the formation of phosphorylated and dimerized IRF3 and led to the increase of IRF3 levels in the nucleus (Fig. 2C, 2D). As a complementary approach, we also used IRF3-BiLC reporter system developed in our laboratory to more accurately assess the kinetics of IRF3 dimer formation in A549 cells (37). Indeed we found that FBXO17 knockdown enhanced IRF3-dimer luciferase activities induced by SeV, intracellular poly(I:C), and poly(dA:dT), respectively (Supplemental Fig. 1C). To identify the effects of FBXO17 knockdown on the expression of IFN-β and IFN-stimulated genes, we transfected HEK293T cells with FBXO17-specific or scrambled siRNAs and then infected the cells with SeV. We found that HEK293T cells treated with FBXO17-specific siRNA had higher expression of IFN-β mRNA than those treated with scrambled siRNA (Fig. 2E). We obtained similar results in THP-1 and A549 cells transfected with FBXO17-specific or scrambled siRNAs (Supplemental Fig. 1D). Consistent with these findings, knockdown of FBXO17 also resulted in greater expression of IFN-stimulated gene mRNAs, including ISG54 and CCL5 (Fig. 2E). To assess the relationship between the enhanced production of IFN-β and antiviral responses, HEK293T cells were transfected with FBXO17-specific or scrambled siRNAs, and then infected with VSV-eGFP at a multiplicity of infection (MOI) of 0.05. We observed that the cells treated with scrambled siRNA were more vulnerable to VSV-eGFP infection and resulted in more GFP-positive cells than the cells transfected with FBXO17-specific siRNA (Fig. 2F). Moreover, upon infection with VSV-eGFP (MOI = 0.01), the expression of VSV glycoprotein mRNA in HEK293T cells treated with scrambled siRNA prior to infection was ∼2-fold higher than in HEK293T cells treated with FBXO17-specific siRNA (Fig. 2G). At the same time, the HEK293T cells treated with FBXO17-specific siRNA prior to infection with VSV-eGFP (MOI = 0.01) produced more IFN-β cytokines than those treated with scrambled siRNA (Fig. 2G). These data collectively illustrate that knockdown of FBXO17 enhances the induction of IFN-β gene expression as well as antiviral responses through elevated dimerization and nuclear translocation of IRF3.
FIGURE 2.
Knockdown of FBXO17 enhances IFN-I signaling as well as antiviral responses. (A) HEK293T cells were transfected with the indicated siRNAs and plasmids. The cell lysates were analyzed by immunoblotting with the indicated Abs. (B) HEK293T cells were transfected with FBXO17-specific or scrambled siRNAs, together with an ISRE luciferase reporter and internal control Renilla luciferase reporter, followed by treatment with intracellular poly(I:C) for 18 h, poly(dA:dT) for 18 h, or SeV for 12 h before assessing luciferase activity. (C) HEK293T cells were transfected with FBXO17-specific or scrambled siRNAs and then infected with SeV for 8 h. The cell lysates were separated by native (top) or SDS (bottom) PAGE and analyzed by immunoblotting with the indicated Abs. Numbers above the lanes indicate the relative accumulation of dimerized and phosphorylated IRF3. (D) Western blot analysis of the level of IRF3 in the cytoplasmic and nuclear fractions of HEK293T cells, which were transfected with FBXO17-specific or scrambled siRNAs and then infected with SeV for 8 h. Numbers above the lanes indicate the relative accumulation of IRF3 in the nucleus. (E) Real-time PCR of IFN-β, ISG54, and CCL5 mRNAs in HEK293T cells, which were transfected with FBXO17-specific or scrambled siRNAs and then infected with SeV for 12 h. (F) Fluorescence microscopy assessing VSV-eGFP infection (MOI = 0.05) of HEK293T cells previously transfected with FBXO17-specific or scrambled siRNAs for 12 h. Original magnification (F) ×10. (G) HEK293T cells were transfected with FBXO17-specific or scrambled siRNAs and then infected with VSV-eGFP (MOI = 0.01) for 16 h before real-time PCR and ELISA assays were performed. All results are representative of three replicate experiments. Student t test. ***p < 0.001, ****p < 0.0001.
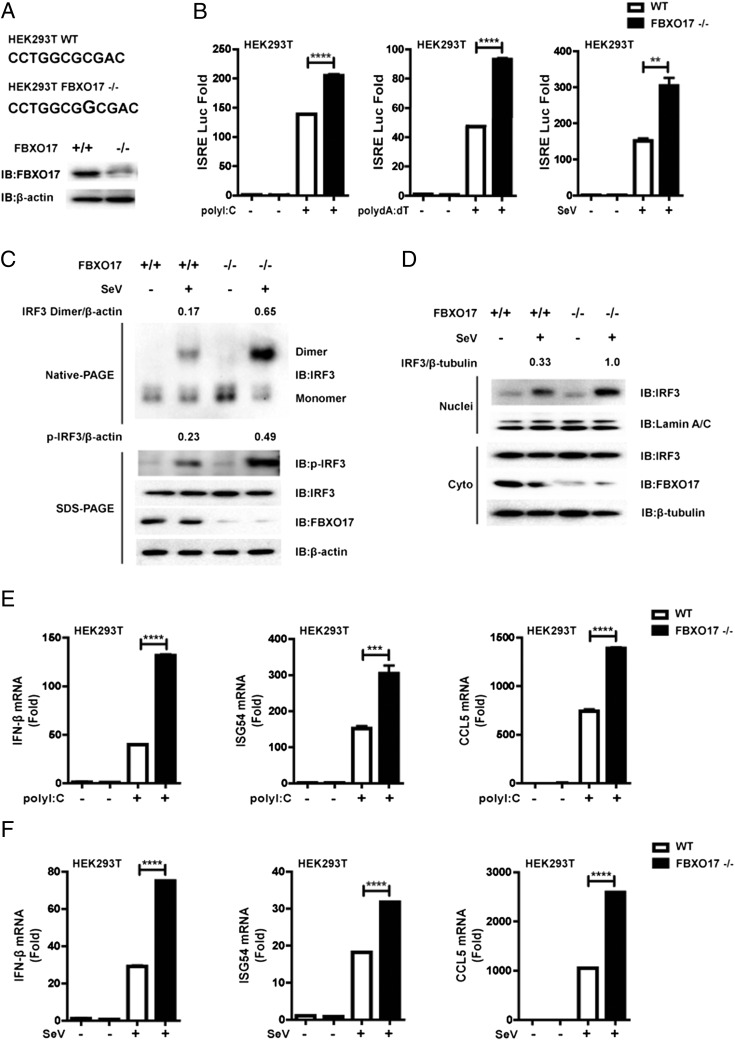
Knockout of FBXO17 potentiates virus-triggered IRF3 activation and IFN-β gene expression
Because the CRISPR/Cas9 assay is an easy and convenient tool for gene editing, we designed a small guide RNA specific for FBXO17 and generated FBXO17 knockout HEK293T cells using the CRISPR/Cas9 system (38, 39). As shown in Fig. 3A and Supplemental Fig. 2A, sequencing showed an insertion of a G nucleotide in the FBXO17 gene of knockout HEK293T cells, and immunoblot analysis confirmed the knockout at the protein level. Next, we determined the effects of FBXO17 knockout on the activation of IFN-I signaling. As we predicted, the ISRE luciferase activity was improved in FBXO17 knockout HEK293T cells compared with wild-type HEK293T cells following stimulation with intracellular poly(I:C), poly(dA:dT), or SeV (Fig. 3B). Furthermore, the results also confirmed that FBXO17 knockout consistently increased the level of phosphorylated and dimerized form of IRF3, and promoted IRF3 nuclear translocation (Fig. 3C, 3D). Real-time PCR further confirmed that FBXO17 knockout HEK293T cells produced more IFN-β mRNA than wild-type HEK293T cells when the cells were treated with intracellular poly(I:C) or SeV (Fig. 3E, 3F). Consistently, knockout of FBXO17 also resulted in enhanced mRNA expression of ISG54 and CCL5 genes (Fig. 3E, 3F). According to these results, we concluded that FBXO17 knockout promotes virus-induced activation of IRF3, and enhances transcription of IFN-β and IFN-stimulated genes.
FIGURE 3.
Knockout (KO) of FBXO17 potentiates virus-triggered activation of IRF3 and IFN-β gene expression. (A) The graphic illustration of the gene modification and Western blot analysis of endogenous FBXO17 in FBXO17 KO HEK293T cells by a CRISPR/Cas9 assay. (B) The luciferase activity in wild-type (WT) and FBXO17 KO HEK293T cells transfected with ISRE luciferase reporter and internal control Renilla luciferase reporter, followed by treatment with intracellular poly(I:C) for 18 h, poly(dA:dT) for 18 h, or SeV for 12 h. (C) The WT and FBXO17 KO HEK293T cells were treated with SeV infection for 8 h. The cell lysates were separated by native (top) or SDS (bottom) PAGE and analyzed by immunoblotting with the indicated Abs. Numbers above the lanes indicate the relative accumulation of dimerized and phosphorylated IRF3. (D) Western blot analysis of IRF3 expression in the cytoplasmic and nuclear fractions of WT and FBXO17 KO HEK293T cells with SeV infection for 8 h. Numbers above the lanes indicate the relative accumulation of IRF3 in the nucleus. (E and F) Real-time PCR of IFN-β, ISG54, and CCL5 mRNAs in WT and FBXO17 KO HEK293T cells treated with intracellular poly(I:C) for 12 h or SeV for 12 h. All results are representative of three replicate experiments. Student t test. **p < 0.01, ***p < 0.001, ****p < 0.0001.
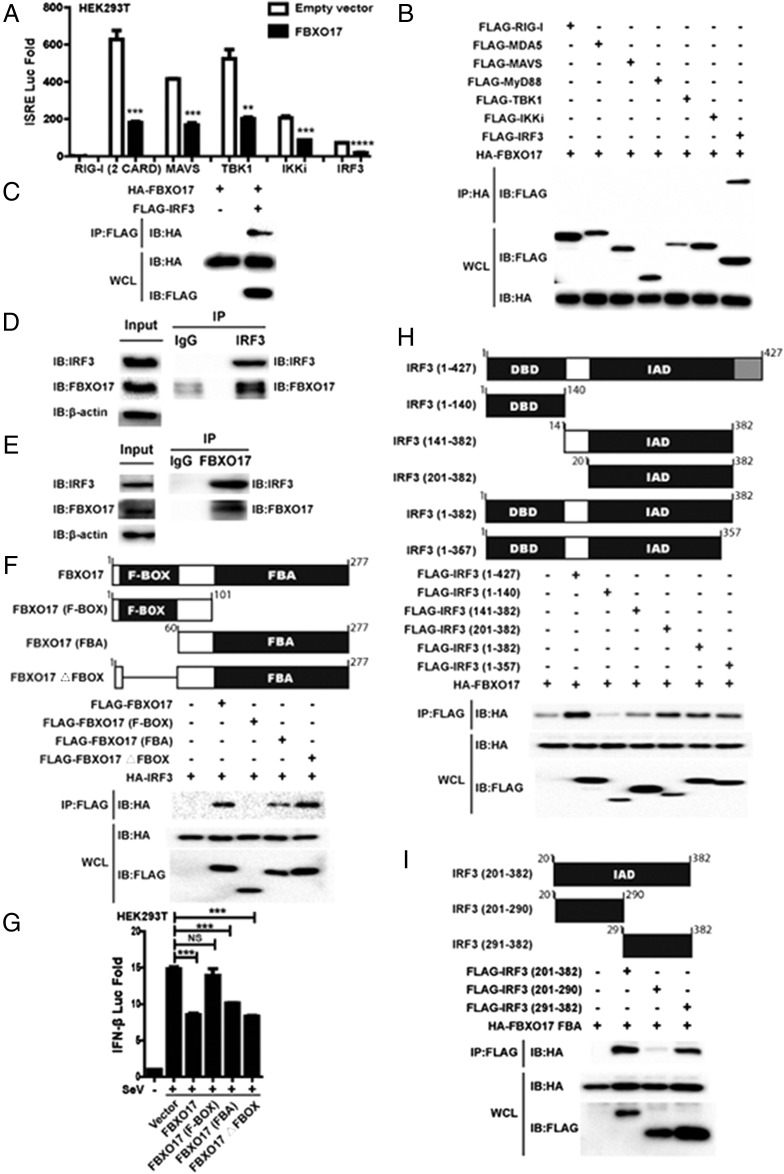
FBXO17 interacts with IRF3 specifically during IFN-I signaling pathway
To clarify the molecular mechanism by which FBXO17 regulates IFN-I signaling, we transfected HEK293T cells with expression plasmids encoding RIG-I (2 CARD), MAVS, TBK1, IKKi, or IRF3, together with the FBXO17 expression plasmid, ISRE luciferase reporter, and internal control Renilla luciferase reporter, and found that FBXO17 inhibited RIG-I (2 CRAD)-, MAVS-, TBK1-, IKKi-, and IRF3-mediated ISRE activation (Fig. 4A). Next, we examined which adaptor protein in IFN-I signaling physically interacts with FBXO17. Fig. 4B illustrated that FBXO17 specifically associated with IRF3 as determined through a coimmunoprecipitation (Co-IP) assay. The association between FBXO17 and IRF3 was again confirmed in Fig. 4C. Furthermore, the endogenous Co-IP indicated that FBXO17 interacted with IRF3 (Fig. 4D, 4E). Accordingly, these results show that FBXO17 interacts specifically with the transcription factor IRF3.
FIGURE 4.
FBXO17 interacts with IRF3 specifically during IFN-I signaling pathway. (A) The luciferase activity of HEK293T cells transfected with the FBXO17 expression plasmid, an ISRE luciferase reporter, and internal control Renilla luciferase reporter, plus RIG-I (2 CRAD), MAVS, TBK1, IKKi, or IRF3. (B and C) HEK293T cells were transfected with the indicated expression plasmids. The cell lysates were immunoprecipitated with anti-HA or anti-FLAG, and immunoprecipitates were analyzed by immunoblotting with the indicated Abs. (D and E) The cell lysates prepared from A549 cells were immunoprecipitated with anti-IRF3 or anti-FBXO17. The immunoprecipitates were analyzed by immunoblotting with the indicated Abs. (F) HA-IRF3 was cotransfected with FLAG-FBXO17 truncated mutants into HEK293T cells. The Co-IP was performed and analyzed by immunoblotting with anti-HA Ab. (G) The luciferase activity of HEK293T cells transfected with FBXO17 truncated mutants, an IFN-β luciferase reporter and internal control Renilla luciferase reporter, followed by SeV infection for 12 h. (H) HA-FBXO17 was transfected with FLAG-IRF3 truncated mutants into HEK293T cells. The Co-IP was performed and analyzed by immunoblotting with anti-HA Ab. (I) HA-FBXO17 FBA domain was transfected with IAD truncated mutants of IRF3 with a FLAG tag into HEK293T cells. The Co-IP was performed and analyzed by immunoblotting with anti-HA Ab. All results are representative of three replicate experiments. Student t test. ***p < 0.001, ****p < 0.0001. DBD, DNA-binding domain; F-BOX, F-box domain.
To further assess the interaction between FBXO17 and IRF3, we constructed truncated mutants of FBXO17 and IRF3, as shown in Fig. 4F and 4H. FBXO17 comprises a conserved F-box domain and an F-box associated (FBA) domain. First, we investigated the interaction between the FBXO17 truncated mutants and IRF3 and found that the FBA domain of FBXO17 interacted with IRF3 (Fig. 4F). We also observed that the FBA region of FBXO17 repressed IFN-β luciferase activity induced by SeV infection (Fig. 4G). IRF3 is composed of an N-terminal DNA-binding domain, an IFN regulatory factor association domain (IAD), and a C-terminal regulatory domain. The Co-IP results showed that the IAD region of IRF3 interacted directly with FBXO17 (Fig. 4H). The interaction between the FBA domain of FBXO17 and the IAD region of IRF3 was confirmed by the Co-IP experiment (Fig. 4I). In addition, we further found that the C-terminal subdomain of the IAD region mediated the association with the FBA domain (Fig. 4I).
FBXO17 attenuates the phosphorylation and homodimer formation of IRF3
Although we confirmed an interaction between FBXO17 and IRF3, the mechanism of how FBXO17 represses IRF3 activation remained unknown. The phosphorylation and homodimerization of IRF3 are required for its activation. To explain whether FBXO17 affects these processes, HEK293T cells were transfected with expression plasmids encoding IRF3 or IRF3-5D (a constitutively active form of IRF3), plus the FBXO17 expression plasmid, ISRE luciferase reporter, and internal control Renilla luciferase reporter. We observed that FBXO17 inhibited IRF3-, but not IRF3-5D–mediated ISRE luciferase activity (Fig. 5A, Supplemental Fig. 2B). Then, we transfected HEK293T cells with the FBXO17 expression plasmid followed by infection with SeV and found that FBXO17 attenuated the level of phosphorylated IRF3 (Fig. 5B). Inversely, the opposite was observed in HEK293T cells treated with FBXO17-specific or scrambled siRNAs followed by SeV infection (Fig. 5C). However, ectopic expression and knockdown of FBXO17 did not affect the level of phosphorylated TBK1 (Fig. 5B, 5C). Together, our results further indicate that FBXO17 targets IRF3 resulting in reduced IRF3 phosphorylation.
FIGURE 5.
FBXO17 attenuates the phosphorylation and homodimer formation of IRF3. (A) The luciferase activity of HEK293T cells transfected with the FBXO17 expression plasmid, an ISRE luciferase reporter, and internal control Renilla luciferase reporter, plus IRF3 or IRF3-5D vector. (B) HEK293T cells were transfected with the FBXO17 expression plasmid and then infected with SeV for 8 h before the immunoblot assay was performed with the indicated Abs. Numbers above the lanes indicate the relative accumulation of phosphorylated IRF3 and TBK1. (C) HEK293T cells were transfected with FBXO17-specific or scrambled siRNAs and then infected with SeV for 8 h before the immunoblot assay was performed with the indicated Abs. Numbers above the lanes indicate the relative accumulation of phosphorylated IRF3 and TBK1. (D) HEK293T cells were transfected with the indicated plasmids and then infected with SeV for 8 h before immunoprecipitation and immunoblot analysis were performed. Numbers above the lanes indicate the fold intensity (ratio of cells transfected with the FBXO17 expression vector to those transfected with an empty vector). (E) HEK293T cells were transfected with the indicated plasmids and siRNAs, and then infected with SeV for 8 h before immunoprecipitation and immunoblot analysis were performed. Numbers above the lanes indicate the fold intensity (ratio of cells transfected with FBXO17 siRNA to those transfected with scrambled siRNA). All results are representative of three replicate experiments. Student t test. ****p < 0.0001.
As FBXO17 interacts with IRF3 and attenuates IRF3 phosphorylation and dimerization, we sought to confirm the role of FBXO17 in the formation of homodimerized IRF3 using a Co-IP assay. HEK293T cells were cotransfected with HA-tagged FBXO17, FLAG-tagged IRF3 and HA-tagged IRF3 followed by infection with SeV and results of the Co-IP assay showed that FBXO17 attenuated homodimerization of IRF3 (Fig. 5D). Consistently, the opposite was observed in FBXO17 knockdown HEK293T cells (Fig. 5E). From our results, we concluded that FBXO17 negatively regulates IFN-I signaling by attenuating the phosphorylation and dimerization of IRF3.
FBXO17 recruits the phosphatase PP2A to promote dephosphorylation of IRF3
Previous studies have demonstrated that the SCF E3 ligase complex uses F-box proteins to recognize and bind targeted substrates for ubiquitin-mediated protein degradation (5, 6). First, we examined whether FBXO17 is involved in degrading the adaptor proteins of IFN-I signaling pathway. HEK293T cells were transfected with expression plasmids encoding TBK1, IKKi, IRF3, or IRF3-5D, together with increasing amounts of the FBXO17 expression plasmid. We observed that FBXO17 did not affect the stability of these proteins, which indicates that the function of FBXO17 does not involve ubiquitin-mediated degradation (Supplemental Fig. 3A). According to another study, the kinase TBK1 is important for IRF3 activation (27). To determine if FBXO17 decreases IRF3 phosphorylation by blocking the interaction of TBK1 and IRF3, we transfected HEK293T cells with FLAG-tagged FBXO17 and FLAG-tagged TBK1, together with HA-tagged IRF3 or HA-tagged IRF3-S386A, and found that FBXO17 did not affect the TBK1-IRF3 interaction or the TBK1-IRF3-S386A interaction using a Co-IP assay (Supplemental Fig. 3B).
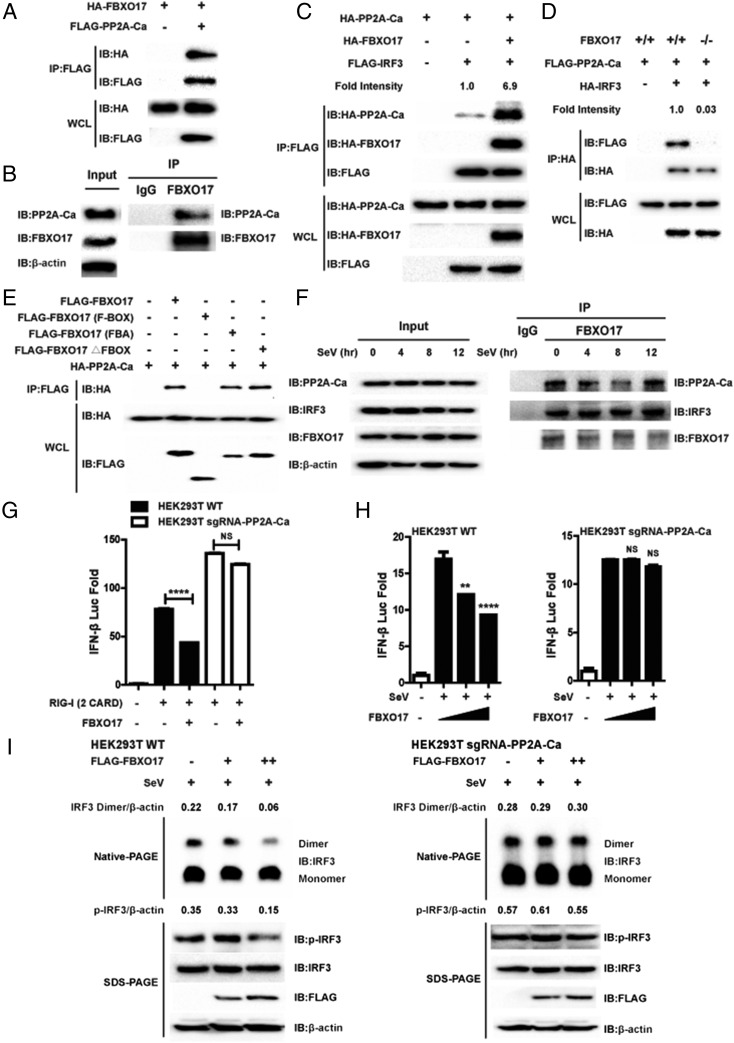
A recent study reported that the phosphatase PP2A is involved in deactivation of IRF3 (40). Therefore, we examined whether FBXO17 would recruit PP2A to promote dephosphorylation of IRF3. In Co-IP experiments, we found that PP2A-Ca interacted with FBXO17 (Fig. 6A), and confirmed this finding for endogenous immunoprecipitation (Fig. 6B). Furthermore, we observed that the interaction between IRF3 and PP2A-Ca in HEK293T cells with ectopically expressed FBXO17 was much higher than that in HEK293T cells transfected with an empty vector, whereas IRF3-PP2A-Ca interacted to a lesser extent when FBXO17 was deleted in FBXO17 knockout HEK293T cells (Fig. 6C, 6D). Then, we found that the FBA domain of FBXO17 interacted with PP2A-Ca (Fig. 6E). Furthermore, endogenous immunoprecipitation showed that PP2A-Ca disassociated from the FBXO17-IRF3 complex upon SeV infection (Fig. 6F). With these results, we concluded that FBXO17 acts as a scaffolding adaptor to bridge the phosphatase PP2A with IRF3.
FIGURE 6.
FBXO17 recruits phosphatase PP2A to promote dephosphorylation of IRF3. (A) The immunoprecipitation and immunoblot analysis of HEK293T cells transfected with HA-FBXO17 and FLAG-PP2A-Ca. (B) The cell lysates prepared from A549 cells were immunoprecipitated with FBXO17 and the immunoprecipitates were analyzed by immunoblot with the indicated Abs. (C) HEK293T cells were transfected with the indicated plasmids before immunoprecipitation and immunoblot analysis were performed. Numbers above the lanes indicate the fold intensity (ratio of cells transfected with FBXO17 expression vector to those transfected with empty vector). (D) Wild-type (WT) and FBXO17 knockout (KO) HEK293T cells were transfected with the indicated plasmids before immunoprecipitation and immunoblot analysis were performed. Numbers above the lanes indicate the fold intensity (ratio of FBXO17 KO cells to WT cells). (E) HA-PP2A-Ca was cotransfected with FLAG-FBXO17 truncated mutants into HEK293T cells. The Co-IP was performed and analyzed by immunoblot with anti-HA Ab. (F) A549 cells were infected with SeV for the indicated times. The cell lysates were immunoprecipitated with FBXO17 Ab and immunoblot analysis was performed with the indicated Abs. (G) The luciferase activity in HEK293T WT and HEK293T sgRNA-PP2A-Ca cells transfected with IFN-β luciferase reporter and internal control Renilla luciferase reporter, plus FBXO17 and RIG-I (2 CARD) expression plasmid. (H) The luciferase activity in HEK293T WT and HEK293T sgRNA-PP2A-Ca cells transfected with IFN-β luciferase reporter and internal control Renilla luciferase reporter, plus FBXO17 expression plasmid, and then infected with SeV for 12 h. (I) HEK293T WT and HEK293T sgRNA-PP2A-Ca were transfected with expression plasmid of FBXO17 with increasing doses and then infected with SeV for 8 h. The cell lysates were separated by native (top) or SDS (bottom) PAGE and analyzed by immunoblotting with the indicated Abs. All results are representative of three replicate experiments. Student t test. **p < 0.01, ****p < 0.0001.
We next sought to examine whether the negative regulation of IFN-I signaling by FBXO17 depends on the phosphatase PP2A. First, we constructed a HEK293T-sgRNA-PP2A-Ca cell line using the CRISPR/Cas9 gene editing technique as described above, and both the immunoblots and genomic DNA sequencing results confirmed the knockout cell line (Supplemental Fig. 2C). In IFN-β luciferase reporter experiments, we found that the inhibition of RIG-I (2 CARD)-mediated IFN-β luciferase activity by FBXO17 was almost completely abrogated in HEK293T-sgRNA-PP2A-Ca cells compared with wild-type HEK293T cells (Fig. 6G). Meanwhile, the repression of SeV-triggered IFN-β activation by FBXO17 was severely impaired in HEK293T-sgRNA-PP2A-Ca cells (Fig. 6H, Supplemental Fig. 2D). Consistent with this finding, we observed that the inhibition of SeV-induced phosphorylation and dimerization of IRF3 by FBXO17 was also impaired in HEK293T-sgRNA-PP2A-Ca cells (Fig. 6I).
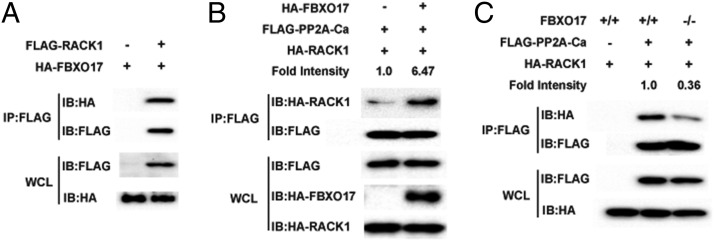
The previous study revealed that RACK1 involves recruiting phosphatase PP2A for IRF3 deactivation (40). Therefore, we next explored the relationship between FBXO17 and RACK1, and found FBXO17 interacts with RACK1 (Fig. 7A). Furthermore, we observed the RACK1-PP2A-Ca interaction in HEK293T cells with ectopic expression of FBXO17 is significantly higher than that in HEK293T cells with empty vector (Fig. 7B). The opposite result was observed in FBXO17 knockout HEK293T cells with the deletion of FBXO17 (Fig. 7C).
FIGURE 7.
FBXO17 interacts with RACK1 to promote RACK1-PP2A-Ca interaction. (A) HEK293T cells were transfected with the indicated plasmids. The Co-IP was performed and analyzed by immunoblotting with anti-HA Ab. (B) HEK293T cells were transfected with the indicated plasmids before immunoprecipitation and immunoblot analysis were performed. Numbers above the lanes indicate the fold intensity (ratio of cells transfected with FBXO17 expression vector to those transfected with empty vector). (C) Wild-type (WT) and FBXO17 knockout (KO) HEK293T cells were transfected with the indicated plasmids before immunoprecipitation and immunoblot analysis were performed. Numbers above the lanes indicate the fold intensity (ratio of FBXO17 KO cells to WT cells). All results are representative of three replicate experiments.
Accordingly, these results suggest that FBXO17 recruits PP2A to promote dephosphorylation of IRF3 for IRF3 deactivation.
Discussion
As earlier studies reported, F-box proteins are involved in the regulation of diverse cellular signaling. Alteration or loss of function of F-box proteins are found to be associated with various pathological conditions and cancers (6). However, our current understanding for roles of F-box proteins in innate immune signaling is very limited. Thus far, no report has described the involvement of the F-box protein family in IFN-I signaling process. In this study, we identified F-box protein FBXO17 as a negative regulator in IFN-I signaling pathway. Our studies demonstrated that FBXO17 inhibited activation of IFN-I signaling induced by various innate stimuli. Moreover, knockdown or knockout of FBXO17 promoted the transcriptional induction of IFN-β and IFN-stimulated genes as well as antiviral activities through increased IRF3 dimerization and nuclear translocation. Collectively, these findings uncovered a previously unappreciated role and function of FBXO17 in negative regulation of IFN-I signaling.
It is generally believed that F-box proteins are a designated component for the formation of SCF E3 ligase complex to mediate ubiquitin-dependent targeted substrate breakdown. In NF-κB signaling pathway, it has been reported that βTrCP1 and FBXW7a mediated the degradation of phosphorylated IκBα and p100, respectively (41, 42). Interestingly, although FBXO17 negatively regulates IFN-I signaling pathway, we found that FBXO17 does not cause the degradation of the key adaptor proteins in signaling pathway, including TBK1, IKKi, IRF3, or a constant active form of IRF3 (IRF3-5D). It has been well established that the F-box motif in F-box family proteins is a critical domain primarily responsible for the interaction with SKP1 to form a SCF complex (1–3). By contrast, our results showed that FBXO17 with the deletion of F-box motif maintained its full activity in repressing SeV-induced IFN-β luciferase reporter expression. Thus, we reasoned that the negative regulatory function of FBXO17 for IFN-I signaling is independent of the conventional mode of action of F-box proteins involving ubiquitin-mediated substrate degradation.
A recent study reported that the phosphatase PP2A is recruited by the adaptor RACK1 to dephosphorylate IRF3 for its deactivation, which exploited a similar regulatory mechanism for IRF3 deactivation as that of FBXO17 (40). To investigate the relationship between RACK1 and FBXO17, we found RACK1 associated physically with FBXO17. Furthermore, we observed FBXO17 enhanced RACK1-PP2A-Ca interaction as well as RACK1-IRF3 interaction (Supplemental Fig. 4), suggesting FBXO17 is likely to serve as a critical adaptor molecule for the formation of a multisubunit complex that brings PP2A and IRF3 together.
FBXO17 represents a subfamily of F-box containing proteins. In this subfamily, FBXO2, FBXO6, FBXO17, FBXO27, and FBXO44 have similar structures comprised of an F-box motif and an FBA domain. Furthermore, the FBA domain in these proteins is related closely to its interaction with glycosylated substrates (43). According to the other reports, FBXO6 interacted with ChK1 and TCR α-chain via the FBA domain (44, 45). FBXO2, also through this domain, bound to preintegrin β1 (46). Consistent with these findings, our results showed this domain in FBXO17 is critical for its association with IRF3 and PP2A-Ca, which further proved the FBA family member recognized and bound target substrates through the FBA domain. More importantly, to our knowledge, this is the first instance of identifying new substrates of FBXO17 aside from its binding selected glycoproteins.
To our knowledge, this study first reported that FBXO17 negatively regulated IFN-I signaling pathway. Thus, this study not only revealed the novel mechanism of FBXO17 controlling IRF3 activation in innate immunity, but also provided a potential therapeutic target for the defense against viral infection and autoimmune disease.
Supplementary Material
Acknowledgments
We thank Dr. Genhong Cheng, Dr. Jingfeng Wang, and Dr. Jun Cui for providing plasmids and reagents as a gift. We also thank Dr. Jun Cui for providing experimental suggestions.
This work was supported by the Chinese Academy of Medical Sciences Initiative for Innovative Medicine (Grant 2016-I2M-1-005), the Ministry of Health (Grant 201302018), the National Major Scientific and Technological Special Project for Significant New Drugs Development (Grant 2015ZX09102023), the National Natural Science Foundation of Key Projects (Grant 81590765), and the Guangdong Innovative Research Team Program (Grant 201001Y0104687244).
The online version of this article contains supplemental material.
- Co-IP
- coimmunoprecipitation
- FBA
- F-box associated
- HA
- HRP-anti-hemagglutinin
- IAD
- IFN regulatory factor association domain
- IFN-I
- type I IFN
- IKKi
- IκB kinase ε
- IRF3
- IFN regulatory factor 3
- ISRE
- IFN-sensitive response element
- MAVS
- mitochondrial antiviral-signaling protein
- MOI
- multiplicity of infection
- poly(dA:dT)
- poly(deoxyadenylic-deoxythymidylic) acid
- poly(I:C)
- polyinosinic-polycytidylic acid
- PP2A
- protein phosphatase 2
- SCF
- SKP1-Cullin1-F-box
- SeV
- Sendai virus
- sgRNA
- short guide RNA
- siRNA
- small interfering RNA
- TBK1
- TANK-binding kinase 1
- VSV
- vesicular stomatitis virus.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Bai C., Sen P., Hofmann K., Ma L., Goebl M., Harper J. W., Elledge S. J. 1996. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell 86: 263–274. [DOI] [PubMed] [Google Scholar]
- 2.Cenciarelli C., Chiaur D. S., Guardavaccaro D., Parks W., Vidal M., Pagano M. 1999. Identification of a family of human F-box proteins. Curr. Biol. 9: 1177–1179. [DOI] [PubMed] [Google Scholar]
- 3.Winston J. T., Koepp D. M., Zhu C., Elledge S. J., Harper J. W. 1999. A family of mammalian F-box proteins. Curr. Biol. 9: 1180–1182. [DOI] [PubMed] [Google Scholar]
- 4.Petroski M. D., Deshaies R. J. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 6: 9–20. [DOI] [PubMed] [Google Scholar]
- 5.Jin J., Cardozo T., Lovering R. C., Elledge S. J., Pagano M., Harper J. W. 2004. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18: 2573–2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skaar J. R., Pagan J. K., Pagano M. 2013. Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol. 14: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duan S., Cermak L., Pagan J. K., Rossi M., Martinengo C., di Celle P. F., Chapuy B., Shipp M., Chiarle R., Pagano M. 2012. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 481: 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frescas D., Pagano M. 2008. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8: 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welcker M., Clurman B. E. 2008. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8: 83–93. [DOI] [PubMed] [Google Scholar]
- 10.Durbin J. E., Fernandez-Sesma A., Lee C. K., Rao T. D., Frey A. B., Moran T. M., Vukmanovic S., Garcia-Sastre A., Levy D. E. 2000. Type I IFN modulates innate and specific antiviral immunity. J. Immunol. 164: 4220–4228. [DOI] [PubMed] [Google Scholar]
- 11.Koyama S., Ishii K. J., Coban C., Akira S. 2008. Innate immune response to viral infection. Cytokine 43: 336–341. [DOI] [PubMed] [Google Scholar]
- 12.Stetson D. B., Medzhitov R. 2006. Type I interferons in host defense. Immunity 25: 373–381. [DOI] [PubMed] [Google Scholar]
- 13.Honda K., Takaoka A., Taniguchi T. 2006. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 25: 349–360. [DOI] [PubMed] [Google Scholar]
- 14.Levy D. E., Garcia-Sastre A. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12: 143–156. [DOI] [PubMed] [Google Scholar]
- 15.Yoneyama M., Suhara W., Fukuhara Y., Fukuda M., Nishida E., Fujita T. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17: 1087–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Platanias L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5: 375–386. [DOI] [PubMed] [Google Scholar]
- 17.Sadler A. J., Williams B. R. 2008. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8: 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiscott J. 2007. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 18: 483–490. [DOI] [PubMed] [Google Scholar]
- 19.Theofilopoulos A. N., Baccala R., Beutler B., Kono D. H. 2005. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 23: 307–336. [DOI] [PubMed] [Google Scholar]
- 20.Banchereau J., Pascual V. 2006. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 25: 383–392. [DOI] [PubMed] [Google Scholar]
- 21.Meyer O. 2009. Interferons and autoimmune disorders. Joint, Bone, Spine 76: 464–473. [DOI] [PubMed] [Google Scholar]
- 22.Gall A., Treuting P., Elkon K. B., Loo Y. M., Gale M., Jr., Barber G. N., Stetson D. B. 2012. Autoimmunity initiates in nonhematopoietic cells and progresses via lymphocytes in an interferon-dependent autoimmune disease. Immunity 36: 120–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akira S., Uematsu S., Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124: 783–801. [DOI] [PubMed] [Google Scholar]
- 24.Honda K., Taniguchi T. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6: 644–658. [DOI] [PubMed] [Google Scholar]
- 25.Wu J., Chen Z. J. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu. Rev. Immunol. 32: 461–488. [DOI] [PubMed] [Google Scholar]
- 26.Liu S., Cai X., Wu J., Cong Q., Chen X., Li T., Du F., Ren J., Wu Y. T., Grishin N. V., Chen Z. J. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347: aaa2630. [DOI] [PubMed] [Google Scholar]
- 27.Fitzgerald K. A., McWhirter S. M., Faia K. L., Rowe D. C., Latz E., Golenbock D. T., Coyle A. J., Liao S. M., Maniatis T. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4: 491–496. [DOI] [PubMed] [Google Scholar]
- 28.Lin R., Mamane Y., Hiscott J. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19: 2465–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori M., Yoneyama M., Ito T., Takahashi K., Inagaki F., Fujita T. 2004. Identification of Ser-386 of interferon regulatory factor 3 as critical target for inducible phosphorylation that determines activation. J. Biol. Chem. 279: 9698–9702. [DOI] [PubMed] [Google Scholar]
- 30.Yoneyama M., Suhara W., Fujita T. 2002. Control of IRF-3 activation by phosphorylation. J. Interferon Cytokine Res. 22: 73–76. [DOI] [PubMed] [Google Scholar]
- 31.Shi H. X., Yang K., Liu X., Liu X. Y., Wei B., Shan Y. F., Zhu L. H., Wang C. 2010. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 30: 2424–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saitoh T., Tun-Kyi A., Ryo A., Yamamoto M., Finn G., Fujita T., Akira S., Yamamoto N., Lu K. P., Yamaoka S. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7: 598–605. [DOI] [PubMed] [Google Scholar]
- 33.Yu Y., Hayward G. S. 2010. The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3. Immunity 33: 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang M., Tian Y., Wang R. P., Gao D., Zhang Y., Diao F. C., Chen D. Y., Zhai Z. H., Shu H. B. 2008. Negative feedback regulation of cellular antiviral signaling by RBCK1-mediated degradation of IRF3. Cell Res. 18: 1096–1104. [DOI] [PubMed] [Google Scholar]
- 35.Lei C. Q., Zhang Y., Xia T., Jiang L. Q., Zhong B., Shu H. B. 2013. FoxO1 negatively regulates cellular antiviral response by promoting degradation of IRF3. J. Biol. Chem. 288: 12596–12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwamura T., Yoneyama M., Yamaguchi K., Suhara W., Mori W., Shiota K., Okabe Y., Namiki H., Fujita T. 2001. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6: 375–388. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z., Ji J., Peng D., Ma F., Cheng G., Qin F. X. 2016. Complex regulation pattern of IRF3 activation revealed by a novel dimerization reporter system. J. Immunol. 196: 4322–4330. [DOI] [PubMed] [Google Scholar]
- 38.Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., Zhang F. 2013. Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mali P., Yang L., Esvelt K. M., Aach J., Guell M., DiCarlo J. E., Norville J. E., Church G. M. 2013. RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long L., Deng Y., Yao F., Guan D., Feng Y., Jiang H., Li X., Hu P., Lu X., Wang H., et al. 2014. Recruitment of phosphatase PP2A by RACK1 adaptor protein deactivates transcription factor IRF3 and limits type I interferon signaling. Immunity 40: 515–529. [DOI] [PubMed] [Google Scholar]
- 41.Winston J. T., Strack P., Beer-Romero P., Chu C. Y., Elledge S. J., Harper J. W. 1999. The SCFbeta-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IkappaBalpha and beta-catenin and stimulates IkappaBalpha ubiquitination in vitro. Genes Dev. 13: 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Busino L., Millman S. E., Scotto L., Kyratsous C. A., Basrur V., O’Connor O., Hoffmann A., Elenitoba-Johnson K. S., Pagano M. 2012. Fbxw7alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat. Cell Biol. 14: 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glenn K. A., Nelson R. F., Wen H. M., Mallinger A. J., Paulson H. L. 2008. Diversity in tissue expression, substrate binding, and SCF complex formation for a lectin family of ubiquitin ligases. J. Biol. Chem. 283: 12717–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y. W., Brognard J., Coughlin C., You Z., Dolled-Filhart M., Aslanian A., Manning G., Abraham R. T., Hunter T. 2009. The F box protein Fbx6 regulates Chk1 stability and cellular sensitivity to replication stress. Mol. Cell 35: 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoshida Y., Tokunaga F., Chiba T., Iwai K., Tanaka K., Tai T. 2003. Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J. Biol. Chem. 278: 43877–43884. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida Y., Chiba T., Tokunaga F., Kawasaki H., Iwai K., Suzuki T., Ito Y., Matsuoka K., Yoshida M., Tanaka K., Tai T. 2002. E3 ubiquitin ligase that recognizes sugar chains. Nature 418: 438–442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.