Abstract
TCF3-PBX1 (E2A-PBX1) is a recurrent gene fusion in B-cell precursor acute lymphoblastic leukemia (BCP-ALL), which is caused by the translocation t(1;19)(q23;p13). TCF3-PBX1 BCP-ALL patients typically benefit from chemotherapy; however, many relapse and subsequently develop resistant disease with few effective treatment options. Mechanisms driving disease progression and therapy resistance have not been studied in TCF3-PBX1 BCP-ALL. Here, we aimed to identify novel treatment options for TCF3-PBX1 BCP-ALL by profiling leukemia cells from a relapsed patient, and determine molecular mechanisms underlying disease pathogenesis and progression. By drug-sensitivity testing of leukemic blasts from the index patient, control samples and TCF3-PBX1 positive and negative BCP-ALL cell lines, we identified the phosphatidylinositide 3-kinase delta (p110δ) inhibitor idelalisib as an effective treatment for TCF3-PBX1 BCP-ALL. This was further supported by evidence showing TCF3-PBX1 directly regulates expression of PIK3CD, the gene encoding p110δ. Other somatic mutations to TP53 and MTOR, as well as aberrant expression of CXCR4, may influence additional drug sensitivities specific to the index patient and accompanied progression of the disease. Our results suggest that idelalisib is a promising treatment option for patients with TCF3-PBX1 BCP-ALL, whereas other drugs could be useful depending on the genetic context of individual patients.
Introduction
The t(1;19)(q23;p13) translocation is a recurrent aberration occurring in 3–5% of acute lymphoblastic leukemias (ALLs) and results in the fusion protein TCF3-PBX1, which is an oncogenic transcription factor.1 The majority of human TCF3-PBX1 BCP-ALLs are pre-B-cell receptor positive,2, 3, 4 and the expression of pre-B-cell receptor genes has been shown to be directly upregulated by TCF3-PBX1.4 The pre-B-cell receptor signaling pathway is activated in TCF3-PBX1 BCP-ALLs, and its inhibition has been identified as a promising approach for treating this disease.1, 5
With current treatment regimens, the prognosis in adult TCF3-PBX1 BCP-ALL patients is similar to that of other adult ALLs.6 Most patients with TCF3-PBX1 BCP-ALL benefit from chemotherapy, however, the disease often recurs, at which point there are few effective treatment options. Targeted drugs may offer further opportunities for improving treatment outcome, and may also be associated with lower toxicity. However, few studies have sought to identify effective new drugs to treat TCF3-PBX1 BCP-ALL. Furthermore, mechanisms driving disease progression in TCF3-PBX1 BCP-ALL are unknown.
In this study, we aimed to identify novel treatment options for TCF3-PBX1 BCP-ALL by profiling samples from a 25-year-old relapsed t(1;19)-positive ALL patient. Using a drug-sensitivity assay testing 302 investigational and approved anti-neoplastic drugs, we identified several targeted therapies showing efficacy towards TCF3-PBX1 BCP-ALL. Molecular profiling of the patient cells by exome and RNA sequencing, plus phospho-proteome analysis provided supporting evidence and rationale for the efficacy of specific inhibitors. Validation using cell lines and control samples provided support for the use of idelalisib, an inhibitor of phosphatidylinositide 3-kinase delta (p110δ), for TCF3-PBX1 BCP-ALL.
Materials and methods
Patient samples
Bone marrow (BM) aspirates, peripheral blood and skin biopsy samples were collected after written informed consent was obtained and according to protocols approved by local Institutional Review Boards in accordance with the Declaration of Helsinki. Blast counts for the index patient samples were 76-90%. Cohort samples were taken from patients diagnosed with Ph+ and Ph− ALL, B-cell ALL, T-cell ALL, chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), chronic myeloid leukemia (CML), chronic myelomonocytic leukemia (CMML) and acute myeloid leukemia (AML). In addition, BM aspirates were collected from healthy donors (n=13). The mononuclear cell (MNC) fractions of the BM and blood samples were isolated by gradient centrifugation (Ficoll-Paque PLUS, GE Healthcare, Little Chalfont Bucks, UK). The BCP-ALL patient samples and their characteristics are listed in Supplementary Tables S1 and S2.
Ex vivo drug-sensitivity testing
Ex vivo drug-sensitivity testing was performed as previously described.7 Freshly isolated, patient-derived leukemia cells were cultured in Mononuclear Cell Medium (PromoCell, Heidelberg, Germany) and seeded in pre-drugged 384-well plates that included 302 active agents. The compound collection included the majority of FDA/EMA-approved anticancer drugs and several investigational compounds. The drugs were plated in five different concentrations in 10-fold dilutions using an acoustic liquid handling device (Echo 550; Labcyte Inc., Sunnyvale, CA, USA). After 72 h incubation, cell viability was measured using the CellTiter-Glo luminescence assay (Promega, Madison, WI, USA) with the PHERAstar (BMG LABTECH, Ortenberg, Germany) or SpectraMax Paradigm (Molecular Devices, Sunnyvale, CA, USA) plate readers. Drug sensitivities were quantified using a drug-sensitivity score, which is a modified area under the curve-based metric that has been described previously.8
Drug testing of BCP-ALL cell lines
TCF3-PBX1 positive (697, KASUMI-2, RCH-ACV) and negative cell lines (TOM-1, MHH-CALL-4, MUTZ-5) were purchased from DSMZ (Leibniz Institute, Braunschweig, Germany) and cultured in Gibco RPMI medium (Thermo Scientific, Carlsbad, CA, USA) supplemented with 15% fetal bovine serum, 2 mm l-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. The compounds were pre-plated in 384-well plates at seven different concentrations using the Echo 550 acoustic dispenser. The cells were seeded in 25 μl volume of medium at the following densities: 697 and RCH-ACV, 2500 cells/well; TOM-1, KASUMI-2, 5000 cells/well; and MUTZ-5 and MHH-CALL-4, 7000 cells/well. After 72 h, cell viability was measured using the CellTiter-Glo assay. The data were normalized to the negative control (dimethyl sulfoxide vehicle only) and the positive control wells (100 μmol/l benzethonium chloride).
Exome sequencing and mutation analysis
Diagnosis BM, two relapse BM and skin biopsy samples were exome sequenced. DNA was isolated with the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany). Exome capture was performed using 3 μg DNA and the NimbleGen SeqCap EZ v2 capture kit (Roche NimbleGen, Madison, WI, USA). Sequencing was performed on the HiSeq 2500 instrument (Illumina, San Diego, CA, USA). For the skin biopsy and BM tumor samples, 4 × 107 and 1 × 108 2 × 100-bp reads were sequenced per sample, respectively.
Reads were processed and aligned to the GRCh37 reference-genome as previously described.9 Somatic mutations were called from the exome-capture target regions using the VarScan2 somatic algorithm10 with the following parameters: strand-filter 1, min-coverage-normal 8, min-coverage-tumor 6, somatic-P-value 0.01, min-var-freq 0.05. Mutations were annotated using SnpEff 4.03(ref. 11) with Ensembl v68 database. Mutations were visually validated using the Integrative Genomics Viewer (Broad Institute, Cambridge, MA, USA). Copy number aberrations were identified as described in the Supplementary Information.
RNA sequencing
RNA sequencing was performed on two relapsed BM samples from the index patient and in a cohort of acute leukemias consisting of AMLs and ALLs (N=66). RNA was prepared with Qiagen AllPrep kit then ribosomal RNA was depleted using a Ribo Zero rRNA Removal Kit (Epicenter, Madison, WI, USA). Ribo-depleted RNA was reverse-transcribed to double-stranded cDNA using a SuperScript Double-Stranded cDNA Synthesis Kit (Thermo Scientific). Random hexamers (New England BioLabs, Ipswich, MA, USA) were used to prime the first-strand synthesis reaction. The Epicenter NextEra kit was used to prepare the RNA sequencing libraries. High-molecular-weight buffer and 50 ng of cDNA was used for tagmentation. The fragmented cDNA was purified using Agencourt AMPure XP SPRI beads (Beckman Coulter, Brea, CA, USA). Sequencing libraries were size-selected to 350–700 bp fragments in 2% agarose gel and then purified using the Qiagen QIAquick gel-extraction kit. The Illumina HiSeq2500 platform with the TruSeq SBS Kit v3-HS reagent kit was used to perform paired-end sequencing to 100 bp read length. The NextEra Read Primers 1 and 2 as well as the NextEra Index Read Primer were used for paired-end sequencing and index-read sequencing, respectively. RNA-sequence data were analyzed as described previously.12 Exome and RNA sequence data has been deposited at the European Genome-phenome Archive (EGA, http://www.ebi.ac.uk/ega/), which is hosted by the EBI, under accession number EGAS00001001876.
Results
Case history of the index patient
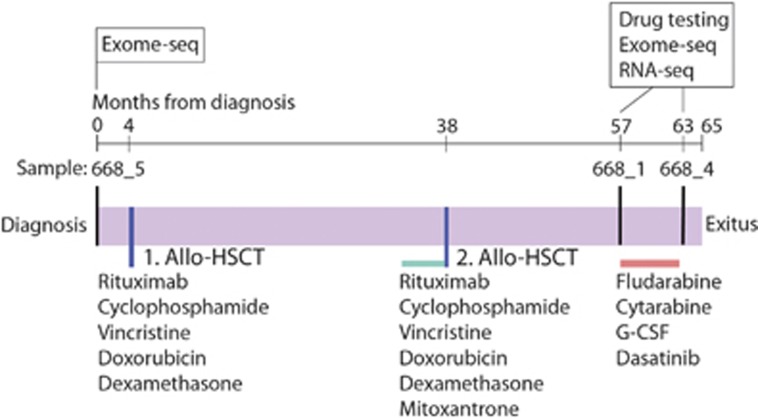
The index patient (668) was a previously healthy 25-year-old male when diagnosed with BCP-ALL. The patient presented with lymphadenopathy and splenomegaly. Leukemic blasts were CD20-positive. Conventional karyotyping and fluorescence in situ hybridization analyses of BM cells revealed the presence of an unbalanced der(19)t(1;19)(q23;p13) translocation and isochromosome i(9q). The patient received rituximab, cyclophosphamide, vincristine, doxorubicin and dexamethasone induction and consolidation therapy, followed by an allogeneic hematopoietic stem cell transplant from a matched unrelated donor while in the first complete remission (Figure 1). The fluorescence in situ hybridization analysis was negative for t(1;19) cells before transplant. The first relapse occurred at 2 years and 4 months after the initial transplantation. Cytogenetic analysis showed a t(1;19), i(9q) karyotype. The patient received rituximab, cyclophosphamide, vincristine, doxorubicin and dexamethasone induction therapy that resulted in a complete morphological remission, but with minimal residual disease detected by fluorescence in situ hybridization. The patient was consolidated with clofarabine, cytarabine and etoposide before a second allogeneic hematopoietic stem cell transplant. After 18 months, the patient again relapsed, with karyotype analyses indicating further clonal evolution. The patient achieved a complete remission after receiving a combination of fludarabine, cytarabine and G-CSF (FLAG) therapy, but minimal residual disease was detected by immunophenotyping. He then received a 14-week course of consolidation therapy with dasatinib (25–50 mg daily) and pegylated interferon α (30 μg/week). Due to cytopenias, the treatment was not administered continuously. The minimal residual disease level remained stable during the treatment until disease progression. FLAG therapy was restarted with no response. The patient died at 5 years and 5 months after the initial diagnosis.
Figure 1.
Clinical timeline of TCF3-PBX1 BCP-ALL index patient 668 with treatment, sampling and analysis history. Allo-HSCT, allogeneic hematopoietic stem cell transplant.
Ex vivo drug-sensitivity testing reveals several active targeted agents for relapsed TCF3-PBX1 BCP-ALL
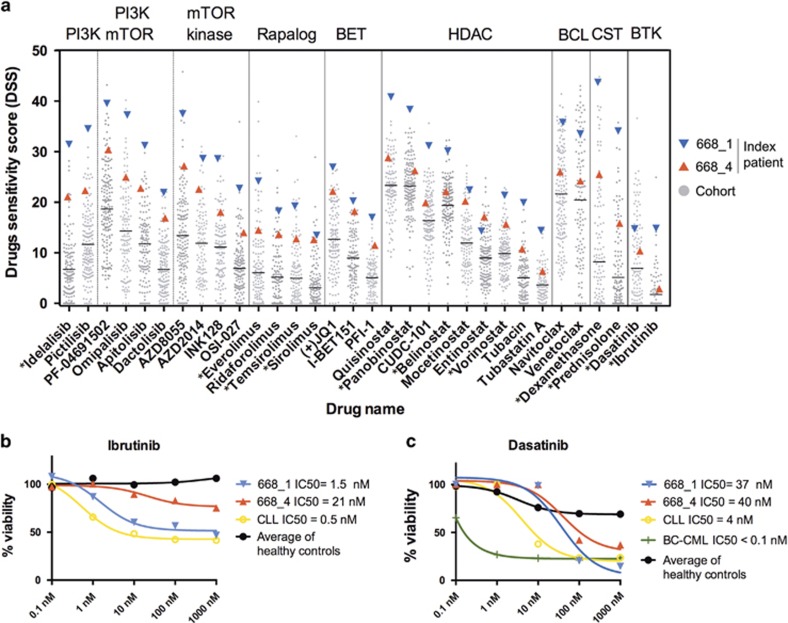
To identify novel treatment options for relapsed TCF3-PBX1 BCP-ALL, we assessed the ex vivo sensitivity of BM mononuclear cells from the index patient against a panel of 302 small molecule inhibitors. The cells were highly sensitive to many targeted agents compared with other samples analyzed under similar conditions. Identified sensitivities included PI3K inhibitors, PI3K/mTOR dual kinase inhibitors, ATP-competitive mTOR kinase inhibitors, rapalogs, bromodomain (BET) inhibitors, histone deacetylase inhibitors, BCL2 inhibitors and corticosteroids (Figure 2a and Supplementary Table S4). Compared with the first profiled relapse sample 668_1, the later sample 668_4 was less sensitive to most drugs, indicating that drug-resistant clones were selected as the disease progressed. Interestingly, although sensitivity was reduced for most drugs, the sensitivity to BET inhibitor I-BET151 or the histone deacetylase inhibitors mocetinostat and entinostat did not change (Figure 2a).
Figure 2.
Drug-sensitivity results for the index patient cells compared with other tested patient samples. (a) Cells from TCF3-PBX1 BCP-ALL patient 668 are compared with a cohort of hematological tumors tested under similar conditions, including ALL, AML and CLL. The values establish the activity range for each compound in the ex vivo assay. Asterisks indicate drugs approved for clinical use. (b) Dose response of cells from TCF3-PBX1 BCP-ALL patient 668 and a CLL patient to ibrutinib. Results are normalized to the viability of cells treated with the vehicle (DMSO). (c) Dose response of cells from TCF3-PBX1 BCP-ALL patient 668, a CLL patient and a BC-CML patient to dasatinib. DMSO, dimethyl sulfoxide.
Ibrutinib and dasatinib have previously been suggested to be effective against TCF3-PBX1 BCP-ALLs.4, 5 The cells from the first relapse sample (668_1) were sensitive to ibrutinib and had sensitivity comparable to cells from a chronic lymphocytic leukemia (CLL) patient (Figure 2b). However, the later relapse sample (668_4) lost sensitivity to ibrutinib (Figure 2b). Compared with other patient samples tested, the cells from both relapse samples of the TCF3-PBX1 BCP-ALL patient exhibited average sensitivity to dasatinib (Figure 2a). This was in contrast to cells from a blast-crisis chronic myeloid leukemia (BC-CML) patient, which were highly sensitive to dasatinib (Figure 2c).
TCF3-PBX1 ALLs are sensitive to the PI3K-delta inhibitor idelalisib
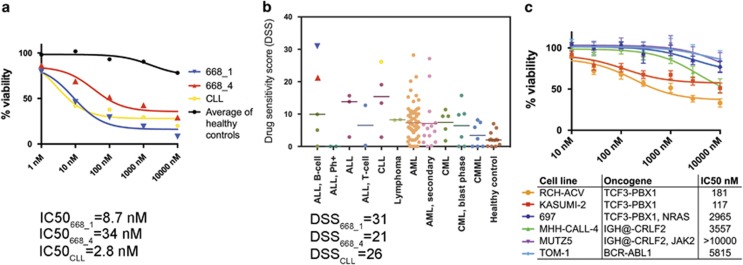
Both relapse samples of the TCF3-PBX1 BCP-ALL index patient were highly sensitive to idelalisib (CAL-101, GS-1101). The samples exhibited similar sensitivity as a CLL sample, which was used as a positive control for idelalisib sensitivity (Figure 3a). Although cells from the later relapse sample were slightly less sensitive to idelalisib compared with the earlier sample, the cells remained highly sensitive compared with other hematological tumors and healthy controls. Notably, the index patient cells were among the most sensitive to idelalisib compared with other hematological tumors (n=112) and healthy bone marrow controls (n=13; Figure 3b).
Figure 3.
Idelalisib sensitivity of TCF3-PBX1 BCP-ALL patient cells and cell lines compared with cells lacking the fusion. (a) Ex vivo drug-sensitivity testing results for leukemia cells obtained from a TCF3-PBX1 BCP-ALL index patient samples (668_1 and 668_4). The values for the patient samples are shown compared with the average values for BM-MNCs obtained from 13 healthy control individuals and to a CLL sample. The data are normalized to the viability of cells treated with the vehicle (DMSO). (b) The distribution of ex vivo drug sensitivities for a cohort of hematological tumors and BM-MNCs obtained from healthy individuals. (c) Viability in TCF3-PBX1 positive and negative BCP-ALL-cell lines in response to idelalisib after a 3-day incubation. The means of three replicate experiments are shown, and the error bars indicate±s.d. DMSO, dimethyl sulfoxide; MNC, mononuclear cell.
To determine whether idelalisib sensitivity is common to all BCP-ALL or specific for TCF3-PBX1 positive BCP-ALL, we assessed the sensitivity of both TCF3-PBX1 positive and negative BCP-ALL cell lines. We found that two of three TCF3-PBX1 positive cell lines tested, Kasumi-2 and RCH-ACV, were sensitive to idelalisib; whereas the three TCF3-PBX1 negative cell lines tested were insensitive. The TCF3-PBX1 positive line 697, which lacked sensitivity to idelalisib, harbors an oncogenic NRAS p.G12D mutation,13 potentially affecting idelalisib response.
We also tested sensitivity to BET, histone deacetylase and BCL2 inhibitors as well as to temsirolimus and ruxolitinib in the same cell lines. However, we did not observe a consistent difference in sensitivity to these compounds between the TCF3-PBX1 positive and negative cell lines (Supplementary Figure S1).
Clonal evolution of TCF3-PBX1 BCP-ALL
To determine the mechanisms mediating drug sensitivity and resistance as well as disease progression in TCF3-PBX1 BCP-ALL, we performed exome sequencing on BM mononuclear cells collected at diagnosis and two relapses from the index patient (Figure 1). We identified an MTOR p.M1724_Q1725insV in-frame insertion mutation, a PHF6 loss-of-function mutation, plus chromosomal aberrations t(1;19) and i(9q) in all the samples (Table 1). The formation of isochromosome i(9q) causes the heterozygous deletion of 9p, which contains the tumor-suppressor gene CDKN2A as well as B-cell transcription factor PAX5.
Table 1. Somatic mutations and chromosomal aberrations in known cancer genes in leukemia cells from the TCF3-PBX1 BCP-ALL index patient.
| Gene | Mutation type |
Mutant variant allele frequency |
||
|---|---|---|---|---|
| Diagnosis | Relapse (668_1) | Relapse (668_4) | ||
| PHF6 | p.Q121* | 86% | 92% | 84% |
| MTOR | p.M1724_Q1725insV | 47% | 46% | 42% |
| TP53 | p.R273_V274insAGP | 0% | 47% | 60% |
| TP53 | p.R283_T284insR | 0% | 0% | 16% |
| Translocation and CNAs | ||||
| TCF3-PBX1 | t(1;19)(q23;p13) | x | x | x |
| CDKN2A | i(9q) Heterozygous deletion | x | x | x |
| CDKN2A | Homozygous deletion | x | ||
| PAX5 | i(9q) Heterozygous deletion | x | x | x |
| TBL1XR1 | Heterozygous deletion | x | x | |
| TP53 | Subclonal deletion | x | ||
Abbreviations: BCP-ALL, B-cell precursor acute lymphoblastic leukemia; CNA, copy number aberration.
Relapse-specific mutations included TP53 loss-of-function mutations (Table 1). Following the second allogeneic hematopoietic stem cell transplant, the leukemia cells had acquired TP53 p.R273_V274insAGP. In the subsequent relapse, the cells had additionally acquired TP53 p.R283_T284insR and a subclonal del(17p). To determine at which point during the disease evolution the observed TP53 insertion mutations occurred, we validated the mutations using high-sensitivity amplicon sequencing of the diagnosis and relapse samples. The TP53 mutations were not detected in the diagnosis sample, indicating that these mutations were acquired during therapy. An additional focal deletion of CDKN2A was present in the earlier 668_1 relapse sample but was not detected in the later 668_4 sample, indicating that the dominant clone changed between these time points. Non-synonymous mutations and chromosomal aberrations identified in the index patient's samples are listed in Supplementary Table S5, and genes with aberrant somatic copy number are listed in Supplementary Table S6.
TCF3-PBX1 regulates PIK3CD expression by binding to leukocyte-specific promoters
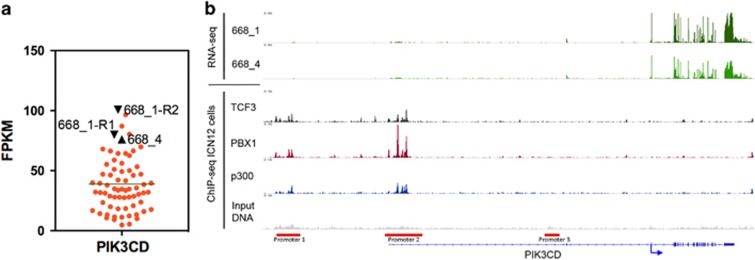
To further define mechanisms mediating drug responses in TCF3-PBX1 BCP-ALL, we profiled the patient cells by RNA sequencing. The PIK3CD gene encodes the idelalisib target protein p110δ. PIK3CD expression has been shown to be upregulated in the majority of TCF3-PBX1 BCP-ALLs and to be a regulatory target of TCF3-PBX1.4 We found that compared with data from 66 leukemia samples with other aberrations, both relapsed samples from the index patient had higher expression of PIK3CD (Figure 4a).
Figure 4.
PIK3CD gene expression in TCF3-PBX1 BCP-ALL. (a) PIK3CD expression was measured using RNA sequencing in 66 leukemias. The samples from TCF3-PBX1 patient 668 (668_1 and 668_4) are shown in black. Sample 668_1 was sequenced in replicates (668_1-R1, 668_1-R2). Gene expression levels are shown as FPKM values (fragments per kilobase of exon per million fragments mapped). (b) PIK3CD RNA-sequence expression signal from TCF3-PBX1 BCP-ALL index patient's relapse samples (668_1 and 668_4). TCF3, PBX1 and p300 ChIP-sequence binding signal from TCF3-PBX1 ALL patient ICN12. Input DNA is the background signal for ChIP-sequencing. The locations of PIK3CD promoters are indicated by red bars. ChIP, chromatin immunoprecipitation.
To better understand the mechanism that underlies the regulation of PIK3CD in TCF3-PBX1 BCP-ALL, we analyzed publicly available chromatin immunoprecipitation sequence data of TCF3, PBX1 and p300 from TCF3-PBX1 ALL ICN12.4 P300 is a transcriptional co-activator that is known to bind to the TCF3-PBX1 protein. Our analysis showed that the TCF3-PBX1 protein binds to promoters 1 and 2 of PIK3CD (Figure 4b). These promoters have been shown to be used by leukocytes to drive high level of PIK3CD expression.14, 15
High CXCR4 expression in relapse samples from the index TCF3-PBX1 BCP-ALL patient
By RNA sequencing, we also observed that the chemokine receptor gene CXCR4 was among the most highly expressed protein-coding genes in both relapse samples of the index patient. The patient cells had high CXCR4 expression compared with 66 acute leukemia samples with other aberrations (Figure 5a).
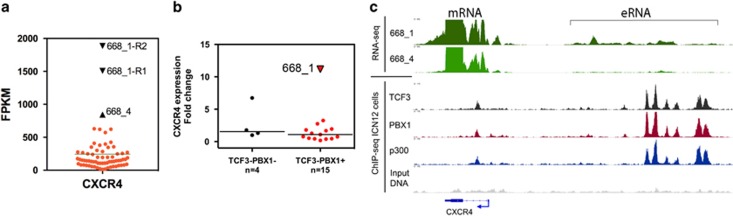
Figure 5.
CXCR4 gene expression in TCF3-PBX1 BCP-ALL. (a) CXCR4 gene expression was measured using RNA sequencing in 66 acute leukemias. Expression levels are indicated as FPKM (fragments per kilobase of exon per million fragments mapped) values. Sample 668_1 was sequenced twice (668_1-R1, 668_1-R2). (b) CXCR4 mRNA expression measured using qRT-PCR in the TCF3-PBX1 BCP-ALLs and ALLs lacking TCF3-PBX1. (c) RNA-sequence expression signal from the CXCR4 locus for the index patient's relapse samples (668_1 and 668_4). TCF3, PBX1 and p300 ChIP-sequence binding signal is from TCF3-PBX1 ALL patient ICN12. ChIP, chromatin immunoprecipitation.
To determine whether CXCR4 expression is common in B-ALL or a consequence of the t(1;19), we analyzed CXCR4 expression in leukemia cells from other B-ALL patients. By qRT-PCR analysis, CXCR4 expression was exceptionally high in a relapse sample from the index patient (668_1), whereas diagnosis phase TCF3-PBX1 BCP-ALL samples (n=14) showed lower CXCR4 expression levels. The remaining B-ALL cases (n=4) had other aberrations. The results showed that the index patient sample obtained at relapse was the only TCF3-PBX1 BCP-ALL sample with high CXCR4 expression (Figure 5b).
Analysis of chromatin immunoprecipitation sequence data showed that TCF3-PBX1 binds to the CXCR4 promoter, intron and upstream enhancer region indicating that TCF3-PBX1 participates in the regulation of CXCR4 gene expression in TCF3-PBX1 positive cells. RNA-sequence data from the index patient cells showed expression of non-coding enhancer RNA (eRNA) from the CXCR4 enhancer region (Figure 5c), indicating the presence of open chromatin that enables binding of transcription factors.
Novel MTOR p.M1724_Q1725insV somatic mutation associated with sensitivity to mTOR inhibitors
The drug profiling results showed that the index patient cells were highly sensitive to mTOR inhibitors including rapalogs and mTOR kinase inhibitors (Figure 2a). Exome sequence showed the cells harbored a heterozygous MTOR somatic mutation p.M1724_Q1725insV, which was present in all samples from diagnosis to relapse (Table 1). To assess mTOR activity in the patient cells, we analyzed the phosphorylation of downstream targets of mTOR kinase: S6K1 and Akt by phospho-proteome array (Supplementary Table S7) and immunoblotting. Both assays showed higher levels of phosphorylation in the index patient cells compared with healthy controls (Supplementary Figure S2), indicating that mTOR kinase signaling was highly active in the patient cells.
To determine whether lymphoid tumors and TCF3-PBX1 BCP-ALLs are typically sensitive to mTOR inhibitors we compared the drug sensitivities of the index patient cells to cells from patients with other lymphoid malignancies lacking the fusion and to previously published mTOR inhibitor sensitivity data from TCF3-PBX1 ALLs. The results showed that sensitivity to mTOR inhibitors is variable in lymphoid tumors (Supplementary Figure S3). Previously published ex vivo data from five TCF3-PBX1 BCP-ALLs show limited sensitivity to mTOR inhibitors.16 These results indicate that the index patient leukemia cells were atypically sensitive to mTOR inhibitors possibly due to mutation to MTOR.
Discussion
Using an unbiased, drug-sensitivity profiling assay, we found that leukemic blasts from a TCF3-PBX1 BCP-ALL index patient obtained at relapse were highly sensitive to the p110δ inhibitor idelalisib. The cells from the index patient were the most sensitive to idelalisib out of a cohort of hematological tumors, although they were obtained from a heavily treated relapsed patient. At the time of the index patient's relapse idelalisib was not yet approved for clinical use. By testing of BCP-ALL cell lines, we found that two of three TCF3-PBX1 positive cell lines were sensitive to idelalisib, whereas TCF3-PBX1 negative cell lines were insensitive. The idelalisib insensitive TCF3-PBX1 BCP-ALL cell line 697 harbors an activating NRAS mutation, which may cause resistance to p110δ inhibition. An earlier study showed pre-BCR-positive ALLs to exhibit sensitivity to idelalisib in vitro.4 Our results with patient samples and cell lines suggest that most TCF3-PBX1 BCP-ALLs are highly sensitive to idelalisib, and provide additional data to support a mechanistic rationale for the use of idelalisib in this group of patients.
The target of idelalisib is p110δ encoded by the PIK3CD gene. Previously, PIK3CD was identified as a regulatory target of TCF3-PBX1 along with pre-B-cell receptor genes.4 In agreement with these results, we observed high levels of PIK3CD expression in cells from the TCF3-PBX1 BCP-ALL index patient. We also found that the TCF3-PBX1 protein binds to two experimentally characterized PIK3CD promoter regions that have been shown to drive high PIK3CD expression in leukocytes.14, 15 The ability of TCF3-PBX1 to directly regulate expression of the idelalisib target p110δ provides important supportive evidence for the use of this drug in TCF3-PBX1 BCP-ALL patients.
The BTK inhibitor ibrutinib and the multi-kinase inhibitor dasatinib, which inhibits both BTK and SRC kinases in addition to other kinases, have been suggested to be effective against TCF3-PBX1 BCP-ALLs.1, 4, 5 Dasatinib sensitivity of cells from the index patient was similar to what has been reported previously for TCF3-PBX1 BCP-ALL.5, 16 Notably, cells from the index patient exhibited substantially lower sensitivity to dasatinib compared with the sensitivity of cells from a BC-CML patient. The patient received low-dose dasatinib (25–50 mg daily) with interferon for 14 weeks, but this combination could not be administered continuously due to the development of cytopenias and eventually failed to prevent relapse. As the affinity of dasatinib is lower for targets in the pre-BCR pathway than for BCR-ABL, the dose required for effective inhibition of pre-BCR signaling is likely to be higher than doses used in CML. However, the high dasatinib doses required may be challenging to administer in the relapse setting due to toxicity. The cells from the first relapse sample of the TCF3-PBX1 BCP-ALL patient were sensitive to ibrutinib, but had lost sensitivity in the final relapse indicating that pre-B-cell receptor positive BCP-ALLs can overcome BTK inhibition.
CXCR4 is the cell surface receptor for CXCL12, a chemokine that is produced by bone marrow stromal cells. Adhesion of leukemia cells to stromal cells through CXCR4/CXCL12 causes adhesion-mediated drug resistance.17 Leukemia cells from the index patient obtained at relapse expressed very high levels of CXCR4 in contrast to diagnostic phase TCF3-PBX1 BCP-ALLs, which did not show similar overexpression. However, chromatin-immunoprecipitation-sequencing results revealed that TCF3-PBX1 binds to enhancer elements upstream of the CXCR4 gene, indicating that TCF3-PBX1 does participate in the regulation of CXCR4 expression in cells with this fusion. Overexpression of CXCR4 was observed at relapse, thus, TCF3-PBX1 alone may not be sufficient to cause aberrant CXCR4 expression, and that in addition to TCF3-PBX1, other alterations may be required. In breast cancer cells, p53 represses CXCR4 expression and TP53 mutations cause upregulation of CXCR4 expression.18 Therefore, the TP53 mutations acquired at relapse in the index patient cells could have contributed to the upregulation of CXCR4. However, we were unable to obtain RNA from the diagnostic phase sample that lacked TP53 mutations to test whether CXCR4 expression was associated with the acquisition of these mutations, and additional studies are needed to investigate this hypothesis. Nevertheless, our results suggest that upregulation of CXCR4 is a mechanism of therapy resistance in relapsed TCF3-PBX1 BCP-ALL. Similar observations have been made in BCP-ALL19 as well as pediatric AML,20 and provide a rationale for targeting CXCR4 either by direct antagonists such as plerixafor21 or by downstream inhibitors of CXCR4 signaling such as dasatinib.22
In CLL, idelalisib inhibits chemokine receptor signaling in leukemia cells causing mobilization of leukemia cells into the bloodstream, which manifests clinically as transient lymphocytosis.23, 24 In vitro, idelalisib has been shown to reduce chemotaxis of CLL cells towards CXCL12.25 Furthermore, PI3K inhibitors have been shown to inhibit CXCR4 signaling in CLL cells.26 These observations suggest that in CLL, the efficacy of idelalisib is in part owing to inhibition of CXCR4 signaling.27 CXCR4/CXCL12 interactions retain B-cell precursors in the bone marrow niche created by CXCL12 expressing stromal cells.28 In analogy to what has been observed in CLL, idelalisib is likely to block CXCR4 signaling via p110δ in TCF3-PBX1 BCP-ALL causing disruption of stromal cell contacts and mobilization of leukemia cells into the blood. Lymphocytosis resulting from mobilization of leukemia cells should be evaluated in future in vivo studies. Mobilized leukemia cells are more sensitive to chemotherapy, and idelalisib may, therefore, be an effective drug for overcoming the therapy resistance caused by CXCR4.
Drug-sensitivity profiling also showed good efficacy of mTOR inhibitors towards cells from the index patient. This is in contrast to previous in vitro drug-testing study on TCF3-PBX1 BCP-ALLs that did not show mTOR inhibitor sensitivity,16 suggesting our observations were specific for the index patient. MTOR mutations have been shown to cause constitutive activation of mTOR kinase and to induce sensitivity to mTOR inhibitors in cell lines and in patient samples.29, 30 Our results suggest that the MTOR p.M1724_Q1725insV mutation present in the index patient cells may have contributed to the high sensitivity to mTOR inhibitors.
In conclusion, on the basis of our results, idelalisib shows promise as a treatment for TCF3-PBX1 BCP-ALL. Idelalisib (Zydelig, Gilead Sciences, Foster City, CA, USA) has been approved as a second-line treatment for CLL, follicular lymphoma and small lymphocytic lymphoma.31 Similar to what has been observed in these diseases, the majority of human TCF3-PBX1 BCP-ALLs depend on pre-B-cell receptor signaling. Idelalisib is a highly specific inhibitor of the p110δ, which is the primary PI3K isoform in B cells, and is expressed mainly by hematopoietic cells. As a result of this specificity, idelalisib has a relatively benign toxicity profile,32 which makes it a promising option for treating patients with TCF3-PBX1 BCP-ALL.
Acknowledgments
We thank the patients who participated in the study. We acknowledge the personnel of the High Throughput Biomedicine Unit and Maija Lepistö, Minna Suvela, Pekka Ellonen, Aino Palva, Pirkko Mattila, Jesus Lopez Martí and Henrikki Almusa from the Institute for Molecular Medicine Finland (FIMM) Technology Centre University of Helsinki for their expert technical assistance. We thank Dr. Brian Grabiner for his assistance and discussions. The Finnish Funding Agency for Technology and Innovation, the European Regional Development Fund, The Finnish Cancer Societies, the Blood Disease Foundation, the Finnish Association of Hematology, the Orion-Farmos Research Foundation and the Academy of Finland supported this work.
Author contributions
SE designed the study, analyzed the data and wrote the manuscript. HK performed the cell line and western analysis experiments. MK collected the sample material and clinical data from the index patient. MK and KP treated the index patient and obtained institutional review board approval for the study. MMM performed drug-sensitivity testing and phospho-proteome analysis. TP performed drug-sensitivity testing of patient cells. AP performed the qRT-PCR experiments and analysis. HE participated in the sequence data analysis. TB and NG provided patient specimens and clinical data. KP, OK, KW and CAH conceived and designed the study, analyzed the data and supervised the work. All the authors contributed to and approved the final manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Müschen M. Rationale for targeting the pre–B-cell receptor signaling pathway in acute lymphoblastic leukemia. Blood 2015; 125: 3688–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunger SP. Chromosomal translocations involving the E2A gene in acute lymphoblastic leukemia: clinical features and molecular pathogenesis. Blood 1996; 87: 1211–1224. [PubMed] [Google Scholar]
- Williams DL, Look AT, Melvin SL, Roberson PK, Dahl G. New chromosomal translocations correlate with specific immunophenotypes of childhood acute lymphoblastic leukemia. Cell 1984; 36: 101–109. [DOI] [PubMed] [Google Scholar]
- Geng H, Hurtz C, Lenz KB, Chen Z, Baumjohann D, Thompson S et al. Self-enforcing feedback activation between BCL6 and Pre-B cell receptor signaling defines a distinct subtype of acute lymphoblastic leukemia. Cancer Cell 2015; 27: 409–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicocca VT, Chang BH, Masouleh BK, Müschen M, Loriaux MM, Druker BJ et al. Crosstalk between ROR1 and the pre-B cell receptor promotes survival of t(1;19) acute lymphoblastic leukemia. Cancer Cell 2012; 22: 656–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister T, Gökbuget N, Schwartz S, Fischer L, Hubert D, Sindram A et al. Clinical features and prognostic implications of TCF3-PBX1 and ETV6-RUNX1 in adult acute lymphoblastic leukemia. Haematologica 2010; 95: 241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemovska T, Kontro M, Yadav B, Edgren H, Eldfors S, Szwajda A et al. Individualized systems medicine strategy to tailor treatments for patients with chemorefractory acute myeloid leukemia. Cancer Discov 2013; 3: 1416–1429. [DOI] [PubMed] [Google Scholar]
- Yadav B, Pemovska T, Szwajda A, Kulesskiy E, Kontro M, Karjalainen R et al. Quantitative scoring of differential drug sensitivity for individually optimized anticancer therapies. Sci Rep 2014; 4: 5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskela HLM, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmäki H, Andersson EI et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med 2012; 366: 1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L et al. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012; 22: 568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly 2014; 6: 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgren H, Murumägi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K et al. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol 2011; 12: R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H et al. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res 2015; 43: D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead MA, Bombardieri M, Pitzalis C, Vanhaesebroeck B. Isoform-selective induction of human p110δ PI3K expression by TNFα: identification of a new and inducible PIK3CD promoter. Biochem J 2012; 443: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok K, Nock GE, Verrall EAG, Mitchell MP, Hommes DW, Peppelenbosch MP et al. Regulation of p110δ PI 3-kinase gene expression. PLoS One 2009; 4: e5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U, Forster M, Rinaldi A, Risch T, Sungalee S, Warnatz H-J et al. Genomics and drug profiling of fatal TCF3-HLF−positive acute lymphoblastic leukemia identifies recurrent mutation patterns and therapeutic options. Nat Genet 2015; 47: 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer 2009; 9: 665–674. [DOI] [PubMed] [Google Scholar]
- Mehta SA, Christopherson KW, Bhat-Nakshatri P, Goulet RJ, Broxmeyer HE, Kopelovich L et al. Negative regulation of chemokine receptor CXCR4 by tumor suppressor p53 in breast cancer cells: implications of p53 mutation or isoform expression on breast cancer cell invasion. Oncogene 2006; 26: 3329–3337. [DOI] [PubMed] [Google Scholar]
- van den Berk LCJ, van der Veer A, Willemse ME, Theeuwes MJGA, Luijendijk MW, Tong WH et al. Disturbed CXCR4/CXCL12 axis in paediatric precursor B-cell acute lymphoblastic leukaemia. Br J Haematol 2014; 166: 240–249. [DOI] [PubMed] [Google Scholar]
- Sison EAR, McIntyre E, Magoon D, Brown P. Dynamic chemotherapy-induced upregulation of CXCR4 expression: a mechanism of therapeutic resistance in pediatric AML. Mol Cancer Res 2013; 11: 1004–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uy GL, Rettig MP, Motabi IH, McFarland K, Trinkaus KM, Hladnik LM et al. A phase 1/2 study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood 2012; 119: 3917–3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaig AM, Cosimo E, Leach MT, Michie AM. Dasatinib inhibits CXCR4 signaling in chronic lymphocytic leukaemia cells and impairs migration towards CXCL12. PLoS One 2012; 7: e48929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furman RR, Sharman JP, Coutre SE, Cheson BD, Pagel JM, Hillmen P et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med 2014; 370: 997–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Byrd JC, Coutre SE, Benson DM, Flinn IW, Wagner-Johnston ND et al. Idelalisib, an inhibitor of phosphatidylinositol 3-kinase p110δ, for relapsed/refractory chronic lymphocytic leukemia. Blood 2014; 123: 3390–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoellenriegel J, Meadows SA, Sivina M, Wierda WG, Kantarjian H, Keating MJ et al. The phosphoinositide 3'-kinase delta inhibitor, CAL-101, inhibits B-cell receptor signaling and chemokine networks in chronic lymphocytic leukemia. Blood 2011; 118: 3603–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedermeier M, Hennessy BT, Knight ZA, Henneberg M, Hu J, Kurtova AV et al. Isoform-selective phosphoinositide 3'-kinase inhibitors inhibit CXCR4 signaling and overcome stromal cell-mediated drug resistance in chronic lymphocytic leukemia: a novel therapeutic approach. Blood 2009; 113: 5549–5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger JA, Okkenhaug K. Idelalisib—targeting PI3K. Nat Rev Clin Oncol 2014; 11: 184–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasawa T. Microenvironmental niches in the bone marrow required for B-cell development. Nat Rev Immunol 2006; 6: 107–116. [DOI] [PubMed] [Google Scholar]
- Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov 2014; 4: 554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Grabiner BC, Van Allen EM, Hodis E, Jacobus S, Supko JG et al. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer Discov 2014; 4: 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BW, Przepiorka D, de Claro RA, Lee K, Nie L, Simpson N et al. FDA approval: idelalisib monotherapy for the treatment of patients with follicular lymphoma and small lymphocytic lymphoma. Clin Cancer Res 2015; 21: 1525–1529. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Cantley LC. Idelalisib—a PI3Kδ inhibitor for B-cell cancers. N Engl J Med 2014; 370: 1061–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.