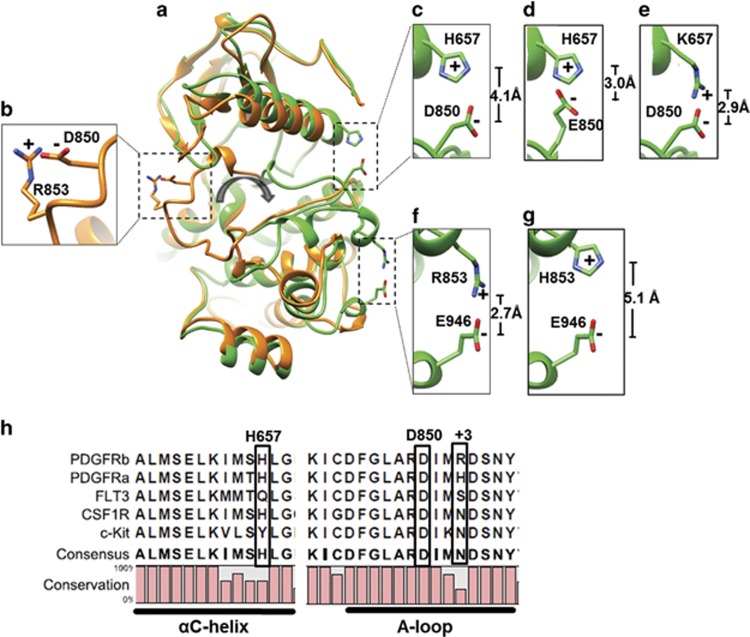
We have identified a novel fusion gene NDEL1-PDGFRB in an 18-month-old child with a myeloid neoplasm and eosinophilia.1, 2 In contrast to earlier data on fusion genes involving PDGRFB in myeloid malignancies, which were generally responsive to treatment with imatinib,3 the patient presented became refractory to both imatinib and nilotinib. Sequencing of a pertinent gene panel including NRAS, KRAS, NF1, PTPN11, CBL, FLT3, c-KIT, PDGFRA, CSF1R, CSF3R, SF3B1, SRSF2, ZRSR2, SH2B3, RUNX1, EZH2, ASXL1, SETBP1, DNMT3A, TET2 and PTEN at the time of diagnosis and both relapses revealed no mutations. However, sequence analysis of the entire tyrosine kinase domain (TKD) of PDGFRB revealed the D850E mutation in the activation loop (A-loop). This mutation was identified in the peripheral blood and bone marrow specimens from both relapses in virtually all cells belonging to the leukemic clone, but was undetectable in the diagnostic peripheral blood or bone marrow. In order to elucidate the structural effects mediated by the D850E mutation in the PDGFRβ TKD, we have generated structure models of the kinase domain both in active (DFG-in) and inactive (DFG-out) conformations, which interact preferentially with type-I and type-II tyrosine kinase inhibitors (TKIs), respectively (Figure 1).4 As the structure of PDGFRβ TKD at the level of atomic resolution is not yet available, we have modelled the kinase domain on the basis of crystallographic structures of closely related homologous proteins including c-KIT, CSF1R and VEGFR2. All structural models indicated that the observed type-II TKI resistance of cells expressing the D850E mutation in NDEL1-PDGFRβ was conceivably related to stabilization of the A-loop in the active conformation. In this conformation, the DFG triad serving as a hypomochlion for the A-loop adopts the so-called DFG-in position.5 The modelled structure of the inactive DFG-out conformation (Figure 1a; orange) revealed the typical auto-inhibitory interaction between D850 and the amino acid at the +3 position, R853, which is commonly observed in inactive TKDs of other receptor tyrosine kinases (RTKs) from the PDGFR family, and is believed to stabilize the A-loop in the inactive conformation (Figure 1b).6, 7, 8 However, modelling of the mutant PDGFRβ TKD in the inactive conformation could not explain the resistance to type-II TKIs and the enhanced kinase activity addressed below, because the negatively charged E850 is also able to form a salt bridge with the positively charged side chain of R853. By contrast, the DFG-in model suggested the occurrence of two intriguing amino acid interactions upon transition of the A-loop from inactive to the active state (Figure 1a; green). One interaction implicated the negatively charged D850 and the positively charged, conserved H657 in the αC-helix (Figure 1c), which is expected to stabilize the A-loop in the active conformation.9 This interaction can be further enhanced by the D850E mutation, because the longer side chain of glutamate in comparison with aspartate brings the negatively charged carboxylic group 1.1 Å closer to the positively charged histidine. This increases the stability of the A-loop in the active conformation (Figure 1d), because the forces of electrostatic interaction between opposite charges increase with the second power of decreasing distance, and become largely ineffective at distances exceeding 4.5 Å.10 The structural model also suggested that the mutation H657K would have an effect similar to the mutation D850E in terms of stabilizing the active conformation (Figure 1e). The other interaction involved R853 and E946 in the C-lobe of the TKD (Figure 1f). The +3 position to D850 is one of the least conserved positions in the A-loop of RTKs from the PDGFR family (Figure 1h), and the arginine at this position in PDGFRβ (R853) has the longest side chain among all members. The DFG-in model suggested that the positively charged side chain of R853 can reach a distance of ~2.7 Å to the negatively charged carboxyl group of E946, which may facilitate electrostatic bonds and provide additional stabilization of the DFG-in conformation of the PDGFRβ TKD. The structural model therefore suggested resistance of NDEL1-PDGFRB with the D850E mutation to type-II TKIs, which can only bind to the inactive conformation of the PDGFRβ TKD, but indicated sensitivity to type-I TKIs binding to the active conformation. To address the predictions provided by the protein model, we have introduced several mutations affecting the aforementioned interactions, and tested the sensitivity of generated constructs against a panel of TKIs.
Figure 1.
Protein models of the PDGFRβ TKD structure. (a) The modelled DFG-out (orange) and DFG-in (green) conformations of PDGFRβ TKD are displayed. The zoom-in windows show the relevant stabilizing electrostatic interactions with the corresponding distances between charges: (b) D850-R853 in the DFG-out model; (c) H657-D850, (d) H657-E850, (e) K657-D850, (f) R853-E946 and (g) H853-E946 in the DFG-in model. Oxygen atoms carrying negative charge are marked in red, nitrogen atoms carrying positive charge in blue. The gray arrow in the center indicates rotation of the A-loop upon transition from inactive to active state. (h) Sequence alignment of RTKs from the PDGFR family depicting the region covering αC-helices and A-loops of PDGFRα, PDGFRβ, FLT3, CSF-1 R and c-Kit.
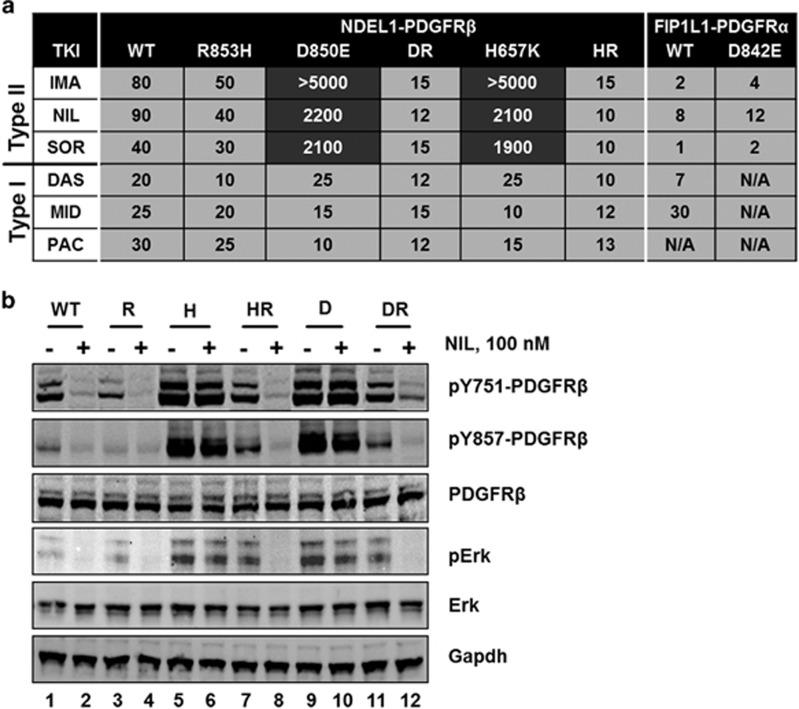
To assess the oncogenic potential of the newly identified fusion gene, the murine cell line Ba/F3 was stably transduced with wild-type or mutant NDEL1-PDGFRB constructs by employing a transposon-based system.11 In addition to the D850E mutation observed in the patient, a construct carrying the H657K mutation was generated. This mutation was expected to strengthen the electrostatic interaction between D850 and the αC-helix, thus stabilizing the DFG-in conformation of the PDGFRβ TKD (Figure 1e). In order to determine the influence of R853 in the PDGRFβ TKD on the kinase activity and TKI-sensitivity, constructs carrying R853H were generated (Figure 1g). Ba/F3 cells expressing the H657K and D850E mutant versions of NDEL1-PDGFRB displayed an elevated in vitro kinase activity, whereas constructs with the R853H mutation showed a kinase activity identical to cells carrying wild-type NDEL1-PDGFRB. The phosphorylation level of PDGFRβ and one of its downstream targets, Erk, which is activated via the Ras-pathway,12 was higher in Ba/F3-NDEL1-PDGFRB cells carrying the H657K or D850E mutations in comparison with wildtype or R853H-carrying fusion gene constructs (Figure 2b).
Figure 2.
TKI responsiveness of wild-type and mutant NDEL1-PDGFRB genes. (a) Displayed are IC50 values of different TKIs against Ba/F3 cells expressing wild-type (wt) or mutant NDEL1-PDGFRβ fusion proteins. The corresponding IC50 values for Ba/F3 expressing FIP1L1-PDGFRα wt and D842E are given for comparison. (b) Western blot analysis of Ba/F3 cells transduced with wild-type or mutant (R=R853H, H=H657K, HR=H657K/R853H, D=D850E and DR=D850E/R853H) NDEL1-PDGFRB genes. The phosphorylation levels of NDEL1-PDGFRβ at Y751 and Y857, and Erk are displayed. Shown are also the total expression levels of NDEL1-PDGFRβ, Erk and the control gene Gapdh upon mock treatment for 4 h with DMSO (indicated by ‘−‘) or with 100 nm nilotinib (indicated by ‘+').
The in vitro responsiveness of Ba/F3-NDEL1-PDGFRB cells to different TKIs of type-I (dasatinib, midostaurin and pacritinib) and type-II (imatinib, nilotinib and sorafenib) was determined by MTT assays. Cells expressing wild-type NDEL1-PDGFRB were sensitive to all TKIs tested (Figure 2a). By contrast, proliferation of Ba/F3 cells carrying the D850E mutation in the NDEL1-PDGFRB gene could only be inhibited by the indicated type-I TKIs at sub-micromolar concentrations (Figure 2a). The observation of TKI resistance apparently induced by the D850E mutation in the kinase domain of PDGFRβ was in contrast to the same amino acid exchange at the corresponding site in PDGFRα (D842E).13 This finding raised questions regarding important structural differences between the two highly homologous RTKs. While PDGFRβ displays an arginine in the +3 position to the mutation site (R853), PDGFRα has the much shorter and less basic histidine in the corresponding position (H845) (Figure 1h). It appeared conceivable, therefore, that the interaction between the side chains of R853 and E946 in the mutant PDGFRβ TKD could stabilize the active conformation in the presence of H657K or D850E mutations, thus mediating resistance to type-II TKIs. To address this notion, the mutation R853H was introduced into NDEL1-PDGFRB constructs, thus mimicking the sequence of the A-loop in the PDGFRα TKD (Figure 1h). In line with the properties of the D842E mutation in the FIP1L1-PDGFRα fusion, this change restored the TKI type-II sensitivity of cells carrying one of the activating mutations, H657K or D850E, in NDEL1-PDGFRβ (Figure 2).
The position of the newly identified mutation D850E in PDGFRβ corresponds to well-known mutation sites within the activation loop of other members of the PDGFR-RTK family including c-KIT (D816),14 PDGFRα (D842)13, 15 and FLT3 (D835)5 (Figure 1h). Mutations converting the aspartate residue at the indicated positions into a bulky hydrophobic residue were shown to mediate resistance to type-II TKIs.5, 15, 16 Structural studies revealed that the interaction of the residues D816 in c-KIT and D835 in FLT3 with the residue at the +3 position (N819 in c-KIT and S838 in FLT3) may maintain the auto-inhibitory inactive conformation of the A-loop.6 Notably, clinically relevant activating mutations D816V in c-KIT and D835V/Y in FLT3 converting the aspartate into a residue with bulky hydrophobic or aromatic side chain would disrupt the hydrophilic interactions with the corresponding residue at +3 position. However, a conversion of aspartate to glutamate, representing an exchange between two hydrophilic and negatively charged amino acids at this position, has never been associated with resistance to type-II TKIs.5, 13, 16 In marked contrast to the clinical and in vitro data presented, the identical mutation at the corresponding site in the TKD of PDGFRα (D842E) in the fusion gene FIP1L1-PDGFRA associated with chronic eosinophilic leukemia did not cause any significant increase in IC50 values of the type-II TKIs imatinib, nilotinib and sorafenib13 (Figure 2a). Similarly, the same mutation at the corresponding site of FLT3 associated with acute myeloid leukemia, D835E, did not significantly increase the IC50 to the type-II TKIs sunitinib and sorafenib5, 16 but enhanced the kinase activity of FLT3, and mediated growth factor-independent proliferation of the murine hematopoietic cell line Ba/F3.17 However, although these analogies are in line with the enhanced kinase activity of the D850E PDGFRβ mutant, they do not explain the observed resistance to type-II TKIs. In this regard, it is important to examine the protein model in the context of intramolecular interactions specific for PDGFRβ TKD. Protein modelling of the PDGFRβ TKD suggested that the residue in +3 position, R853, which differs from corresponding sites in all other RTKs of the PDGFR family, mediates a critical amino acid interaction stabilizing the A-loop in the active conformation, thereby preventing the interaction of type-II TKIs with their target sites in the kinase domain. This notion was confirmed by mutating R853 to histidine, the amino acid present at the corresponding site in PDGFRα, which decreased the kinase activity and restored sensitivity of NDEL1-PDGFRβ with D850E or H657K to type-II TKIs. The findings not only confirm the important role of R853 in establishing the resistant phenotype of the mutant NDEL1-PDGFRB, but also underline the potential of protein modelling for prediction of sensitivity and resistance to TKI treatment. The data presented provide new insights into specific amino acid interactions in mutant RTKs that are of clinical relevance for improved selection of appropriate TKI treatment.
Acknowledgments
This work was supported by the Austrian Science Fund (FWF), SFB Grants F4705-B20 (TL), F4704-B20 (PV) and F4711-B20 (GS-F).
The authors declare no conflict of interest.
References
- Valent P, Klion AD, Horny HP, Roufosse F, Gotlib J, Weller PF et al. Contemporary consensus proposal on criteria and classification of eosinophilic disorders and related syndromes. J Allergy Clin Immunol 2012; 130: 607–612 e609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016; 127: 2391–2405. [DOI] [PubMed] [Google Scholar]
- Cheah CY, Burbury K, Apperley JF, Huguet F, Pitini V, Gardembas M et al. Patients with myeloid malignancies bearing PDGFRB fusion genes achieve durable long-term remissions with imatinib. Blood 2014; 123: 3574–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garuti L, Roberti M, Bottegoni G. Non-ATP competitive protein kinase inhibitors. Curr Med Chem 2010; 17: 2804–2821. [DOI] [PubMed] [Google Scholar]
- Smith CC, Lin K, Stecula A, Sali A, Shah NP. FLT3 D835 mutations confer differential resistance to type II FLT3 inhibitors. Leukemia 2015; 29: 2390–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Dougan DR, Schneider TR, Skene RJ, Kraus ML, Scheibe DN et al. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J Biol Chem 2004; 279: 31655–31663. [DOI] [PubMed] [Google Scholar]
- McTigue M, Murray BW, Chen JH, Deng YL, Solowiej J, Kania RS. Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors. Proc Natl Acad Sci USA 2012; 109: 18281–18289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illig CR, Manthey CL, Wall MJ, Meegalla SK, Chen J, Wilson KJ et al. Optimization of a potent class of arylamide colony-stimulating factor-1 receptor inhibitors leading to anti-inflammatory clinical candidate 4-cyano-N-[2-(1-cyclohexen-1-yl)-4-[1-[(dimethylamino)acetyl]-4-piperidinyl]phenyl]-1H-imidazole-2-carboxamide (JNJ-28312141). J Med Chem 2011; 54: 7860–7883. [DOI] [PubMed] [Google Scholar]
- Elling C, Erben P, Walz C, Frickenhaus M, Schemionek M, Stehling M et al. Novel imatinib-sensitive PDGFRA-activating point mutations in hypereosinophilic syndrome induce growth factor independence and leukemia-like disease. Blood 2011; 117: 2935–2943. [DOI] [PubMed] [Google Scholar]
- Donald JE, Kulp DW, DeGrado WF. Salt bridges: geometrically specific, designable interactions. Proteins 2011; 79: 898–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter K, Kowarz E, Marschalek R. Functional characterisation of different MLL fusion proteins by using inducible sleeping beauty vectors. Cancer Lett 2014; 352: 196–202. [DOI] [PubMed] [Google Scholar]
- Demoulin JB, Essaghir A. PDGF receptor signaling networks in normal and cancer cells. Cytokine Growth Factor Rev 2014; 25: 273–283. [DOI] [PubMed] [Google Scholar]
- von Bubnoff N, Gorantla SP, Engh RA, Oliveira TM, Thone S, Aberg E et al. The low frequency of clinical resistance to PDGFR inhibitors in myeloid neoplasms with abnormalities of PDGFRA might be related to the limited repertoire of possible PDGFRA kinase domain mutations in vitro. Oncogene 2011; 30: 933–943. [DOI] [PubMed] [Google Scholar]
- Sotlar K, Fridrich C, Mall A, Jaussi R, Bultmann B, Valent P et al. Detection of c-kit point mutation Asp-816 –> Val in microdissected pooled single mast cells and leukemic cells in a patient with systemic mastocytosis and concomitant chronic myelomonocytic leukemia. Leuk Res 2002; 26: 979–984. [DOI] [PubMed] [Google Scholar]
- Weisberg E, Wright RD, Jiang J, Ray A, Moreno D, Manley PW et al. Effects of PKC412, nilotinib, and imatinib against GIST-associated PDGFRA mutants with differential imatinib sensitivity. Gastroenterology 2006; 131: 1734–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bubnoff N, Engh RA, Aberg E, Sanger J, Peschel C, Duyster J. FMS-like tyrosine kinase 3-internal tandem duplication tyrosine kinase inhibitors display a nonoverlapping profile of resistance mutations in vitro. Cancer Res 2009; 69: 3032–3041. [DOI] [PubMed] [Google Scholar]
- Clark JJ, Cools J, Curley DP, Yu JC, Lokker NA, Giese NA et al. Variable sensitivity of FLT3 activation loop mutations to the small molecule tyrosine kinase inhibitor MLN518. Blood 2004; 104: 2867–2872. [DOI] [PubMed] [Google Scholar]