Abstract
We present the complete genome sequence of Luteibacter rhizovicinus type strain LJ96T, a yellow-pigmented gammaproteobacterium isolated from the rhizosphere of barley (Hordeum vulgare) Johansen et al. (2005) , a species with numerous potential applications. The genome sequence was deposited to NCBI GenBank with the accession number CP017480.
Keywords: Rhizosphere bacteria, Barley, Genome sequencing
| Specifications | |
| Organism | Luteibacter rhizovicinus |
| Strain | LJ96T |
| Sequencer or array type | PacBio RS II |
| Data format | Analyzed |
| Experimental factors | Bacterial type strain |
| Experimental features | Whole genome analysis and gene annotation of LJ96T |
| Sample source location | Rhizosphere soil of spring barley (H. vulgare) in an organic field at Højbakkegaard, Taastrup, Denmark. |
1. Direct link to deposited data
2. Experimental design, material and methods
The genus Luteibacter belongs to the family Xanthomonadaceae in the class Gammaproteobacteria [1]. Since it was first isolated from rhizosphere of barley [1], Luteibacter sp. has typically been associated with soil or rhizosphere [2], [3], [4], [5]. Luteibacter sp. has shown the ability to degrade PCB [3], produce lipases [5], metabolize cephalomannine [4], chelate ferric ions and solubilize monocalcium phosphate in vitro, as well as to promote plant growth [6]. Genomic DNA from L. rhizovicinus type strain LJ96T [1] was extracted using Genomic-tip 500/G kit (Qiagen GmbH, Hilden, Germany). A library was created using PacBio (Pacific Biosciences, California, USA) 20 kb library preparation protocol and whole genome sequencing was performed using PacBio RS II. The library was sequenced using P6-C4 chemistry with 360 min movie time on one single-molecule real-time (SMRT) cell. The reads were assembled using HGAP v3 (Pacific Biosciences, SMRT Analysis Software v2.3.0). The Minimus2 software of the Amos package was used to circularize the contig, which was confirmed by a dot plot to contain the same sequence at the beginning and end of the contig. RS_Resequencing.1 software (SMRT Analysis version v2.3.0) was used to map reads back to the assembled and circularized sequence in order to correct the sequence after circularization. The sequencing service was provided by the Norwegian Sequencing Centre (www.sequencing.uio.no), a national technology platform hosted by the University of Oslo and supported by the “Functional Genomics” and “Infrastructure” programs of the Research Council of Norway and the Southeastern Regional Health Authorities.
2.1. Data description
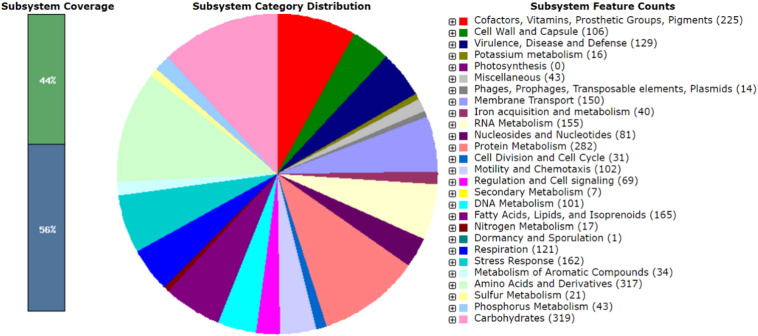
The genome of L. rhizovicinus type strain LJ96T was annotated using the NCBI Prokaryotic Genome Annotation Pipeline [7], GeneMarkS + v 3.3 and the Rapid Annotation System Technology (RAST) server [8]. Fig. 1 presents an overview of the count of each subsystem feature and the subsystem coverage. The genome had a GC content of 64.7%, consisted of 4,765,486 bp and contained 4247 coding sequences, 6 rRNA genes, 51 tRNAs, and 4 noncoding RNA genes.
Fig. 1.
Subsystem category distribution of major protein coding genes of Luteibacter rhizovicinus type strain LJ96T as annotated by the RAST annotation server. The bar chart shows the subsystem coverage in percentage (blue bar corresponds to percentage of proteins included). The pie chart shows percentage distribution of the 27 most abundant subsystem categories.
2.2. Nucleotide accession number
This whole genome project has been deposited at NCBI GenBank under the accession number CP017480.
Acknowledgements
This work would not have been possible without the courtesy of Professor Niels Kroer (the Head of Department of Biology at University of Copenhagen, Denmark) that shared the bacterial strain with us. We are grateful to Hege Særvold Steen for technical assistance. This work was financed by The Foundation for Research Levy on Agricultural Products/Agricultural Agreement Research Fund (233993/E50)/Research Council of Norway (research grant 233993/E50) with support from the industry partners Graminor, Orkla Foods Norge, Bayer Crop Science, Yara Norge, Norske Felleskjøp, Strand Unikorn, Felleskjøpet Agri, Lantmännen Cerealia, Norgesmøllene, Bakers.
References
- 1.Johansen J.E., Binnerup S.J., Kroer N., Mølbak L. Luteibacter rhizovicinus gen. nov., sp. nov., a yellow-pigmented gammaproteobacterium isolated from the rhizosphere of barley. Hordeum vulgare L.Int. J. Syst. Evol. Microbiol. 2005;55:2285–2291. doi: 10.1099/ijs.0.63497-0. [DOI] [PubMed] [Google Scholar]
- 2.Wang X., Song M., Gao C., Dong B., Zhang Q., Fang H., Yu Y. Carbendazim induces a temporary change in soil bacterial community structure. J. Environ. Sci. 2009;21:1679–1683. doi: 10.1016/s1001-0742(08)62473-0. [DOI] [PubMed] [Google Scholar]
- 3.Leigh M.B., Prouzová P., Macková M., Macek T., Nagle D.P., Fletcher J.S. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl. Environ. Microbiol. 2006;72:2331–2342. doi: 10.1128/AEM.72.4.2331-2342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J., Dai J., Chen X., Zhu P. Microbial transformation of cephalomannine by Luteibacter sp. J. Nat. Prod. 2007;70:1846–1849. doi: 10.1021/np0701531. [DOI] [PubMed] [Google Scholar]
- 5.Bresciani F.R., Santi L., Macedo A.J., Abraham W.-R., Vainstein M.H., Beys-da-Silva W.O. Production and activity of extracellular lipase from Luteibacter sp. Ann. Microbiol. 2014;64:251–258. [Google Scholar]
- 6.Guglielmetti S., Basilico R., Taverniti V., Arioli S., Piagnani C., Bernacchi A. Luteibacter rhizovicinus MIMR1 promotes root development in barley (Hordeum vulgare L.) under laboratory conditions. World J. Microbiol. Biotechnol. 2013;29:2025–2032. doi: 10.1007/s11274-013-1365-6. [DOI] [PubMed] [Google Scholar]
- 7.Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Ciufo S., Li W. second ed. National Center for Biotechnology Information; Bethesda, MD: 2013. Prokaryotic Genome Annotation Pipeline, the NCBI Handbook. [Google Scholar]
- 8.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., Meyer F., Olsen G.J., Olson R., Osterman A.L., Overbeek R.A., McNeil L.K., Paarmann D., Paczian T., Parrello B., Pusch G.D., Reich C., Stevens R., Vassieva O., Vonstein V., Wilke A., Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:1. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]