Abstract
The nuclear receptor retinoid X receptor (RXR) is a ligand-activated transcription factor. To create receptors for a new ligand, a structure-based approach was used to generate a library of ≈380,000 mutant RXR genes. To discover functional variants within the library, we used chemical complementation, a method of protein engineering that uses the power of genetic selection. Wild-type RXR has an EC50 of 500 nM for 9-cis retinoic acid (9cRA) and an EC50 of >10 μM for the synthetic retinoid-like compound LG335 in yeast. The library produced ligand–receptor pairs with LG335 that have a variety of EC50 values (40 nM to >2 μM) and activation levels (10–80% of wild-type RXR with 9cRA) in yeast. The variant I268V;A272V;I310L;F313M has an EC50 for LG335 of 40 nM and an EC50 for 9cRA of >10 μM in yeast. This variant has essentially the reverse ligand specificity of wild-type RXR and is transcriptionally active at a 10-fold-lower ligand concentration in yeast. This EC50 is 25-fold lower than the best receptor we have engineered through site-directed mutagenesis, Q275C;I310M;F313I. Furthermore, the variants' EC50 values and activation levels in yeast and mammalian cells correlate. This protein engineering method should be extendable to produce other functional ligand–receptor pairs, which can be selected and characterized from libraries within weeks. Coupling large library construction with chemical complementation could be used to engineer proteins that bind virtually any small molecule for conditional gene expression, applications in metabolic engineering, and biosensors and to engineer enzymes through genetic selection.
The human retinoid X receptor α (RXR) is a ligand-activated transcription factor of the nuclear receptor superfamily. RXR plays an important role in morphogenesis and differentiation and serves as a dimerization partner for other nuclear receptors (1). Like most nuclear receptors, RXR has two structural domains, the DNA-binding domain (DBD) and the ligand-binding domain (LBD), which are connected by a flexible hinge region. The DBD contains two zinc modules, which bind a sequence of six bases (2). The LBD binds and activates transcription in response to multiple ligands including phytanic acid, docosahexaenoic acid and 9-cis retinoic acid (9cRA) (Fig. 5, which is published as supporting information on the PNAS web site) (3–6). RXR is a modular protein; the DBD and LBD function independently. Therefore, the LBD can be fused to other DBDs and retain function. A conformational change is induced in the LBD upon ligand-binding, which initiates recruitment of coactivators and the basal transcription machinery resulting in transcription of the target gene (7).
Nuclear receptors have evolved to bind and activate transcription in response to a variety of small-molecule ligands. The known ligands for nuclear receptors are chemically diverse and include steroid and thyroid hormones, vitamin D, prostaglandins, fatty acids, leukotrienes, retinoids, antibiotics, and other xenobiotics. Evolutionarily closely related receptors (e.g., thyroid hormone receptor and retinoic acid receptor) bind different ligands, whereas some members of distant subfamilies (e.g., RXR and retinoic acid receptor) bind the same ligand (8). This diversity of ligand–receptor interactions demonstrates the versatility of the fold for ligand-binding and suggests that it should be possible to engineer LBDs with a large range of novel specificities.
The crystal structure of RXR bound to 9cRA elucidates important hydrophobic and polar interactions in the LBD binding pocket (9). There are 20 hydrophobic and polar amino acids within 4.4 Å of the bound 9cRA (Fig. 6, which is published as supporting information on the PNAS web site). RXR is a good candidate for creating variants that bind different ligands through site-directed mutagenesis, because side-chain atoms, not main-chain atoms, contribute the majority of the ligand contacts (10–13). A library of RXR LBDs with all 20 amino acids at each of the 20 positions in the ligand-binding pocket screened against multiple compounds could potentially produce many new ligand–receptor pairs. However, the number of possible combinations (2020 ≈ 1026) renders saturation mutagenesis impractical for constructing a complete library.
Codon randomization creates protein libraries with mutations at specific sites (14). We used a modified version of the Lim and Sauer (15) codon randomization method to create a library of binding pocket variants of RXR. This library allowed exploration of a vast quantity of sequence space in a minimal amount of time.
Chemical complementation is a method that links survival of yeast to the presence of a small molecule (12) (B.A., E. I. Chang, L.J.S., and D.F.D., unpublished data). Chemical complementation allows us to test for the activation of protein variants by specific ligands by using genetic selection. Here, we used LG335, a synthetic retinoid-like compound (Fig. 5), to test the viability of chemical complementation for discovery of ligand–receptor pairs from large libraries. LG335 was previously shown to selectively activate an RXR variant and not activate wild-type RXR (11, 16). Combining chemical complementation with a large library of protein variants decreases the time, effort, and resources necessary to find new ligand–receptor pairs. We detail our first attempt at large-scale chemical complementation to engineer ligand–receptor pairs.
Materials and Methods
Ligands. 9cRA (molecular weight of 300.4 g/mol) was purchased from ICN. LG335 (16) was synthesized as described in refs. 17–21 and in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Expression Plasmids. pGAD10BAACTR (22) (B.A., E. I. Chang, L.J.S., and D.F.D., unpublished data), pGBT9Gal4, pGBDRXRα (12), pCMX-hRXR (1), and pCMX-βGAL (10) have been described. Conversion of pGBDRXRα to pGBDRXRαL-SH-ME is described in Supporting Materials and Methods. pCMX-hRXR mutants were cloned from pGBDRXR vectors by using SalI and PstI restriction enzymes and ligated into similarly cut pCMX-hRXR vectors. pLuc_CRBPII_MCS was constructed as described in Supporting Materials and Methods. All plasmids have been confirmed through sequencing.
Genotype Determination. Plasmids were rescued by using either the Powers method (www.fhcrc.org/labs/gottschling/yeast/yplas.html) or the Zymoprep kit (Zymo Research, Orange, CA). The plasmids were then transformed into Z-competent (Zymo Research) XL1-Blue cells (Stratagene). The QIAprep Spin Miniprep kit (Qiagen, Valencia, CA) was used to purify the DNA from the transformants. These plasmids were sequenced.
Quantitation Assays. Solid media. The rescued plasmids were transformed (23) into PJ69-4A containing the pGAD10BAACTR plasmid and plated on synthetic complete (SC) –Trp –Leu medium. These plates were grown for 2 days at 30°C. Colonies were streaked onto the following media: SC, SC –Trp –Leu, SC –Ade –Trp –Leu, and SC –Ade –Trp –Leu plus increasing concentration of LG335 or 9cRA from 1 nM to 10 μM.
Liquid media and mammalian luciferase assay. The method used for quantitation of liquid media in this work was modified from a method developed by Miller (24). Mammalian luciferase assay was performed with HEK 293 cells as previously described (11).
Supporting Materials and Methods. Plasmid and insertion cassette library construction (Fig. 7, which is published as supporting information on the PNAS web site), yeast selection plates and transformation, and molecular modeling are described in Supporting Materials and Methods.
Results
Library Design. The binding pocket of the RXR LBD is composed of primarily hydrophobic side chains plus several positively charged residues that stabilize the negatively charged carboxylate group of 9cRA (9). The target ligand, LG335, contains an analogous carboxylate group, so the positively charged residues were left unchanged. We hypothesized that binding affinity arises from hydrophobic contacts and that specificity arises from binding pocket size, shape, hydrogen bonding, and electrostatics. The randomized amino acids were chosen based on their proximity to the bound 9cRA as observed in the crystal structure and the results of site-directed mutagenesis (Fig. 6) (9, 11). The electrostatic interactions were held constant while the size, shape, and potential hydrogen bonding interactions were varied to find optimum contacts for LG335-binding. A library of RXRs with mutations at six positions was created. At three of the positions (Ile-268, Ala-271, and Ala-272) are four possible amino acids (Leu, Val, Ala, and Pro), and at the other three positions (Ile-310, Phe-313, and Leu-436) are eight possible amino acids (Leu, Ile, Val, Phe, Met, Ser, Ala, and Thr). The combination of six positions and the number of encoded amino acids allowed testing of the library construction while keeping the library size (32,768 amino acid combinations and ≈3 million codon combinations) within reasonable limits. Pro was included in the library as a negative control. Residues 268, 271, and 272 are in the middle of helix 3, which would be disrupted by the inclusion of Pro. Therefore, Pro residues should appear at these positions only in unselected variants and not in the variants that activate in response to ligand. The substitutions at positions 268, 271, and 272 were restricted to small amino acids, allowing access to the positively charged residues at this end of the pocket.
To eliminate contamination of the library with unmutated, wild-type RXR, the gene was modified to create a nonfunctional gene, RXR:3Stop. Forty base pairs were deleted at three separate sites, producing three stop codons in the coding region to create this nonfunctional gene. The deletions correspond to regions in the RXR gene where randomized codons are designed. This plasmid, pGBDRXR:3Stop, was cotransformed into yeast with the library of insertion cassettes containing full-length RXR LBD genes with randomized codons at positions 268, 271, 272, 310, 313, and 436. The insertion cassettes and the plasmid contain homologous regions enabling the yeast to homologously recombine the cassette into the plasmid. Recombination repairs the deletions in the RXR:3Stop gene to make full-length genes with mutations at the six specific sites.
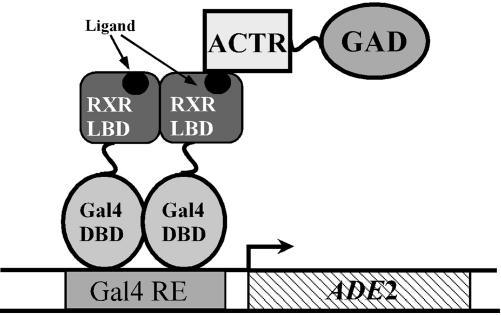
Library Selection. To limit the number of variants to be screened, the library was subjected to chemical complementation (Fig. 1) (B.A., E. I. Chang, L.J.S., and D.F.D., unpublished data). Chemical complementation exploits the power of genetic selection to make the survival of yeast dependent on the presence of a small molecule. The PJ69-4A strain of Saccharomyces cerevisiae has been engineered for use in yeast two-hybrid genetic selection and screening assays. For selection, PJ69-4A contains the ADE2 gene under the control of a Gal4 response element. Plasmids created through homologous recombination in PJ69-4A express the Gal4 DBD fused with a variant RXR LBD (GBD:RXR). A plasmid expressing ACTR, a nuclear receptor coactivator, fused with the Gal4 activation domain (ACTR:GAD), was also transformed into PJ69-4A. When a ligand causes a variant RXR LBD to associate with ACTR, transcription of the ADE2 gene is activated. Expression of ADE2 permits adenine biosynthesis and, therefore, yeast survival on media lacking adenine.
Fig. 1.
Schematic depicting chemical complementation. For selection, yeast strain PJ69-4A has the ADE2 gene under the control of a Gal4 response element (Gal4 RE). This strain is transformed with a plasmid expressing ACTR:GAD (B.A., E. I. Chang, L.J.S., and D.F.D., unpublished data). Plasmids created through homologous recombination in PJ69-4A express a variant, GBD:RXR. In media lacking Ade, yeast will grow only in the presence of a ligand that causes the RXR LBD to associate with ACTR and activate transcription of ADE2. For clarity, only one ACTR:GAD is depicted.
A small amount of the yeast library was plated onto medium (SC –Leu –Trp) selecting only for the presence of the plasmids pGAD10BAACTR (expressing ACTR:GAD and containing a Leu selective marker) and mutant pGBDRXR (expressing variant GBD:RXR and containing a Trp selective marker). The majority of the yeast cells transformed with the RXR library were plated directly onto SC –Leu –Trp –Ade medium containing 10 μM LG335, selecting for Ade production in response to the compound LG335. The transformation efficiency of this library into yeast strain PJ69-4A was 3.8 × 104 colonies per μg of DNA. This number includes both the efficiency of transforming the DNA into the cells and the homologous recombination efficiency. Of the ≈380,000 transformants, ≈300 grew on SC –Ade –Trp –Leu plus 10 μM LG335 selective medium.
Library Characterization. Twenty-one plasmids were rescued from yeast colonies: nine from nonselective plates (SC –Trp –Leu medium) and 12 from selective plates (SC –Ade –Trp –Leu plus 10 μM LG335 medium). The relevant portion of plasmid DNA from these colonies was sequenced to determine the genotype (Table 1). All nine of the plasmid sequences from the nonselective plates contained at least one deletion and were nonfunctional genes. Of the 12 plasmids that grew on the selective media, all contained full-length RXR LBDs with designed mutations. With 95% certainty, we concluded that the unselected library was at least 72% background and the selected library was at least 78% designed sequences (see Supporting Materials and Methods).
Table 1. Genotypes of mutants from unselected and selected libraries.
| Mutant | I268 | A271 | A272 | I310 | F313 | L436 |
|---|---|---|---|---|---|---|
| Unselected library | ||||||
| 1 | Deleted | Deleted | Deleted | Deleted | Deleted | Deleted |
| 2 | Deleted | Deleted | Deleted | Deleted | Deleted | Deleted |
| 3 | GTA(V) | CCT(P) | CCT(P) | TCG(S) | TCG(S) | Deleted |
| 4 | Deleted | Deleted | Deleted | Deleted | Deleted | Deleted |
| 5 | Deleted | Deleted | Deleted | Deleted | Deleted | GCG(A) |
| 6 | Deleted | Deleted | Deleted | Deleted | Deleted | Deleted |
| 7 | Deleted | Deleted | Deleted | Deleted | Deleted | Deleted |
| 8 | Deleted | Deleted | Deleted | Deleted | Deleted | Deleted |
| 9 | Deleted | Deleted | Deleted | Deleted | Deleted | TTC(F) |
| Selected library | ||||||
| 1 | GTG(V) | wtRXR | GTG(V) | TTG(L) | ATG(M) | TTG |
| 2 | GTG(V) | wtRXR | GCA | GTG(V) | TCC(S) | TTG |
| 3 | CTA(L) | GCT | GCA | ATG(M) | GTG(V) | TTG |
| 4 | GCG(A) | wtRXR | GCA | TCC(S) | GTG(V) | TTC(F) |
| 5 | GCT(A) | GCT | GCA | GCC(A) | GCG(A) | TTC(F) |
| 6 | GCT(A) | GCT | GTT(V) | GCC(A) | GCG(A) | TTC(F) |
| 7 | CTT(L) | GCT | GCT | GTC(V) | ATC(I) | TTG |
| 8 | CTG(L) | GTG(V) | GCG | TTG(L) | TTG(L) | TTG |
| 9 | GTG(V) | GTG(V) | GCG | TTG(L) | GTG(V) | TTG |
| 10 | GTA(V) | wtRXR | GTG(V) | ATG(M) | TCC(S) | ATG(M) |
| 11 | GCG(A) | GCG | GCA | ATG(M) | GCG(A) | ACG(T) |
| 12 | GCG(A) | GCT | GCG | TCG(S) | GTC(A) | TTC(F) |
Sequenced codons are followed by the encoded amino acid in parentheses. “wtRXR” indicates that the sequence corresponds to the wild-type RXR codon. “Deleted” indicates the presence of an unmutated 3Stop deletion background cassette.
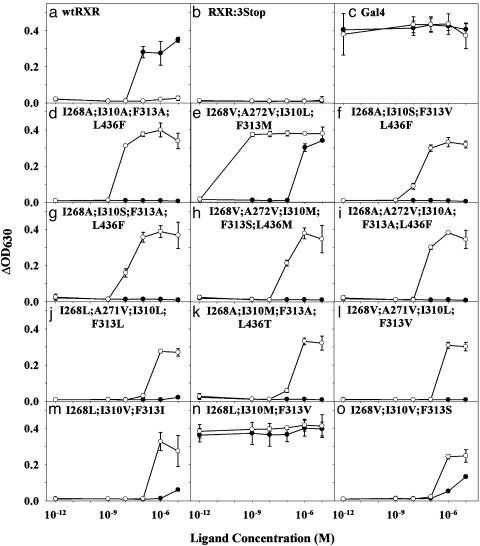
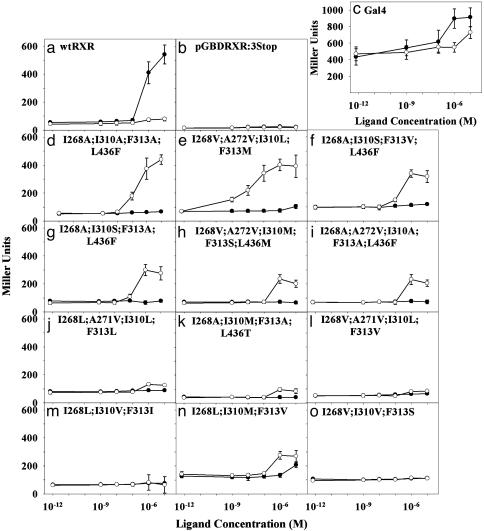
Variant Characterization in Yeast. The 12 plasmids rescued from the selective plates were retransformed (23) into PJ69-4A to confirm that their phenotype is plasmid-linked. The strain PJ69-4A was engineered to contain a Gal4 response element controlling expression of the LacZ gene in addition to the ADE2 gene. Both selection and screening were used to determine the activation level of each variant by 9cRA and LG335. The selection assay quantifies yeast growth occurring through transcriptional activation of the ADE2 gene, whereas the screen quantifies β-galactosidase activity occurring through transcriptional activation of the LacZ gene. Although the selection assay (Fig. 2) is ≈10-fold more sensitive than the screen (Fig. 3), it does not quantify activation level (efficacy) as well as the screen. In the selection assay, there is either growth or no growth, whereas the screen more accurately quantifies different activation levels at various concentrations of ligand (Figs. 2 and 3).
Fig. 2.
Selection assay (SC –Ade –Trp –Leu plus ligand medium) for yeast growth in the presence of 9cRA (•) and LG335 (○) for 43 h. The 10–12 M point is 0 M ligand concentration.
Fig. 3.
Screen assay (SC –Trp –Leu plus ligand medium) for β-galactosidase activity with o-nitrophenyl β-d-galactopyranoside (ONPG) in the presence of 9cRA (•) and LG335 (○). Miller units normalize the change in absorbance at 405 nm for the change in OD at 630 nm, which reflects the number of cells per well. The 10–2 M point is 0 M ligand concentration.
Three plasmids were used as controls in the screen and selection assays. The plasmids pGBDRXRα and pGBT9Gal4 were used as positive controls with which the activation level of the variants can be compared. pGBDRXRα expresses the gene for the “wild-type” GBD:RXR, which grows and is activated by 9cRA but not by LG335. pGBT9Gal4 expresses the gene for the ligand-independent yeast transcription factor Gal4 (25), which is constitutively active in the presence or absence of either ligand. The plasmid pGBDRXR:3Stop serves as a negative control. pGBDRXR:3Stop carries a nonfunctional RXR LBD gene; therefore, yeast transformed with this plasmid does not grow in the selection assay or show activity in the screen. This plasmid provides a measure of background noise in both the selection and screen assays.
Both the selection and screen assays showed that 10 of the 12 variants are selectively activated by LG335. Results of these assays are shown in Figs. 2 and 3. Table 2 summarizes the transcriptional activation profiles of all 12 variants in response to both 9cRA and LG335 compared with wild-type RXR.
Table 2. EC50 and efficacy in yeast and HEK 293 cells for RXR variants.
| 9cRA
|
LG335
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Yeast
|
HEK 293
|
Yeast
|
HEK 293
|
|||||
| Variant | EC50 | Eff | EC50 | Eff | EC50 | Eff | EC50 | Eff |
| WT | 500 | 100 | 220 | 100 | >10,000 | 10 | 300 | 10 |
| I268A;I310A;F313A;L436F | >10,000 | 0 | >10,000 | 0 | 220 | 70 | 30 | 50 |
| I268V;A272V;I310L;F313M | >10,000 | 10 | >1600 | 30 | 40 | 60 | 1 | 30 |
| I268A;I310S;F313V;L436F | >10,000 | 10 | — | — | 470 | 60 | — | — |
| I268A;I310S;F313A;L436F | >10,000 | 0 | >10,000 | 0 | 430 | 50 | 690 | 20 |
| I268V;A272V;I310M;F313S;L436M | >10,000 | 10 | >10,000 | 0 | 680 | 30 | 180 | 30 |
| I268A;A272V;I310A;F313A;L436F | >10,000 | 0 | — | — | 530 | 30 | — | — |
| I268L;A271V;I310L;F313L | >10,000 | 0 | — | — | 530 | 20 | — | — |
| I268A;I310M;F313A;L436T | >10,000 | 0 | >10,000 | 0 | 610 | 10 | 140 | 20 |
| I268V;A271V;I310L;F313V | >10,000 | 0 | — | — | 650 | 10 | — | — |
| I268L;I310V;F313I | >10,000 | 0 | — | — | >2000 | 10 | — | — |
| I268L;I310M;F313V | >10,000 | 20 | — | — | 610 | 20 | — | — |
| I268V;I310V;F313S | >10,000 | 0 | — | — | 440 | 10 | — | — |
EC50 values (given in nm) represent the averages of two screen experiments in quadruplicate for yeast and in triplicate for HEK 293. Efficacy (Eff; given as a percent) is the maximum increase in activation relative to the increase in activation of wild type with 10 μM 9cRA. Values represent the averages of two screen experiments in quadruplicate for yeast and in triplicate in HEK 293.
Mammalian Correlation. Five variants were chosen for testing in mammalian cell culture for comparison of the activation profiles I268A;I310A;F313A;L436F, I268V;A272V;I310L;F313M, I268A;I310S;F313A;L436F, I268V;A272V;I310M;F313S;L436M, and I286A;I310M;F313A;L436T. The genes for these variants were removed from yeast expression plasmids and ligated into mammalian expression plasmids.
Discussion
The chemical complementation system presented here focuses on one small-molecule target ligand and utilizes the power of genetic selection to reveal proteins within the library that bind and activate transcription in response to that small molecule. We were able to isolate functional receptors from a large pool of nonfunctional variants, even from a nonoptimized library. Of the nine plasmids from the unselected library, all contained sequences that were at least partially background. Therefore, the majority of the library contains at least partial sequence from the nonfunctional gene (RXR:3Stop) used in the recombination process. Of the 12 plasmids from the selected library, all contain designed sequences. We conclude that chemical complementation is very efficient at finding functional receptors within a large collection of nonfunctional receptors.
Importantly, we have also demonstrated that we can generate and find ligand–receptor pairs that have various activation properties. Figs. 2 and 3 depict examples of the differing activation profiles, which are summarized in Table 2. Some of the variants approach wild-type and Gal4 levels of activation (>50% efficacy; Fig. 3 a and c–f), whereas some are activated only to low (<25% efficacy; Fig. 3 j–l) or moderate (between 25% and 50% efficacy; Fig. 3 g–i) levels. Most of the ligand–receptor pairs act like switches (i.e., on or off) (Fig. 3h): they are fully activated at one concentration but show no activation at the next lower concentration. However, some act as rheostats (Fig. 3e): transcription levels can be varied by changing ligand concentration over several orders of magnitude. These variants can be used to titrate levels of transcription by varying the concentration of ligand. The ability to titrate transcription levels should be useful in gene therapy to control transcription of therapeutic genes and in agriculture to control transcription of pesticidal or other genes.
Although I268L;I310M;F313V is constitutively active in the selection assay (Fig. 2n) and has high basal activity in the screen assay, both 9cRA and LG335 increase activity at micromolar concentrations (Fig. 3n). This variant may be in an intermediate conformation, with weakly activated transcription that can be improved by ligand-binding. The high basal activation could also be due to a change in the conformation equilibrium with a shift toward the active conformation when ligand is not present.
I268V;I310V;F313S is constitutively active on solid media (data not shown) but shows no activation in the screen (0% efficacy; Table 2 and Fig. 3o) and only grows in the liquid media selection after two days (Fig. 2o). The basal activation level may be below the threshold of detection for the liquid media assays. However, it is also possible that agar, which is not present in the liquid assays, contains some small molecule that activates the receptor.
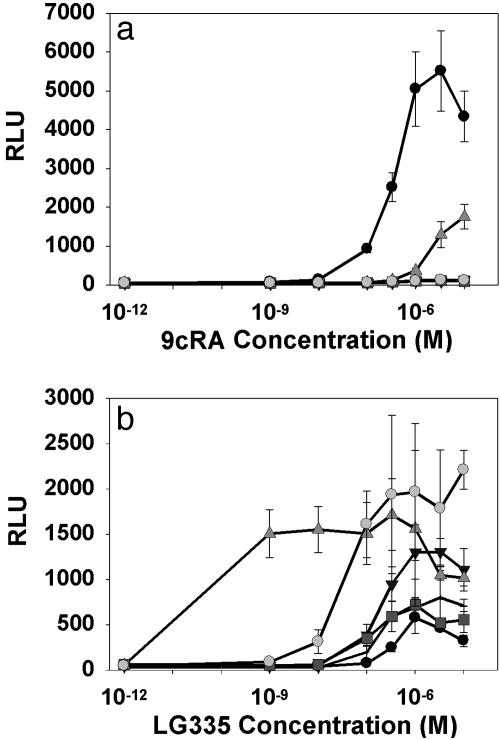
Activation levels and EC50 values correlate in yeast and HEK 293 cells (Fig. 4 and Table 2). For the majority of the variants, 9cRA shows little or no activation in yeast or mammalian cells. Variant I268V;A272V;I310L;F313M is activated slightly by 9cRA in yeast, but in mammalian cells it is activated to the same level by both 9cRA and LG335 (Figs. 2, 3, 4). With one exception, all variants tested have EC50 values within 10-fold in yeast and mammalian cells. However, the EC50 values in mammalian cells are generally lower than in yeast. We speculate that this shift is due to increased penetration of LG335 into mammalian cells vs. yeast.
Fig. 4.
Mammalian cell culture was performed by using a luciferase reporter with wild-type RXR (wtRXR, filled circle), I268A;I310S;F313A;L436F (horizontal line), I268V;A272V;310M;F313S;L436M (downward-pointing triangle), I268A;I310M;F313A;L436T (gray square), I268V;A272V;I310L;F313M (upright-pointing triangle), or I268A;I310A;F313A;L436F (gray circle) in response to 9cRA (a) and LG335 (b). The 10–12 M point is 0 M ligand concentration. RLU, relative light units.
Subtle differences in binding pocket shape can have a drastic effect on specificity. For example, the I268V;A272V;I310L;F313M variant is activated to high levels by LG335 (60% efficacy; Table 2) and is only slightly activated by 10 μM 9cRA in yeast (Fig. 3e), yet the amino acid changes are extremely conservative. The volume difference between Phe and Met side chains is only ≈4 Å3 (26), and their polarity difference is minimal (hydration potentials of the Met and Phe side chains are –0.76 kcal·mol–1 and –1.48 kcal·mol–1, respectively) (27). The other mutations redistribute methyl groups within the binding pocket, with a net difference of one methyl group (≈18 Å3) (26).
The LG335–I268V;A272V;I310L;F313M ligand–receptor pair also represents a 25-fold improvement in EC50 over our previous best LG335 receptor, Q275C;I310M;F313I (40 nM vs. 1 μM in yeast) (11). The Q275C;I310M;F313I variant was created by using site-directed mutagenesis. We would not have predicted that the subtle changes in the I268V;A272V;I310L;F313M variant would have produced a better ligand–receptor pair than the Q275C;I310M;F313I variant. This conclusion is consistent with the observation that nuclear receptors bind ligands through an inducedfit mechanism (28). With current knowledge about protein–ligand interactions, it is not possible to rationally design ligand–receptor pairs with specific activation profiles. Libraries and chemical complementation are a new way to circumvent this problem and obtain functional variants with a variety of activation profiles.
Molecular modeling was used to generate hypotheses about the structural basis of ligand specificity for the variants discovered in the library. First, mutations to smaller or more flexible side chains at positions 310 and 313 are essential to provide space for the propyl group of LG335. All variants activated by LG335 have mutations at these two positions. Second, mutations to amino acids with larger side chains at position 436 sterically clash with the methyl group at position 9 of 9cRA. This interaction may prevent helix 12 from closing properly and therefore prevent activation by 9cRA. The only variant significantly activated by 9cRA, I268V;A272V;I310L;F313M, does not contain a mutation at position 436. Third, we hypothesize that tight packing in the binding pocket may lead to lower EC50 values. The docking results for I268V;A272V;I310L;F313M with LG335 show that the Met and Leu side chains pack tightly against the propyl group of LG335, which may result in tighter binding and, consequently, a lower EC50.
In this study, we were able to take advantage of both structural information and functional data from previous site-directed mutations for our library design. In the absence of functional data, chemical complementation may be used to test more hypotheses about the function of particular residues than would be possible through site-directed mutagenesis. By making a library of changes at a single site, additional information could be obtained about the importance of side-chain size, polarity, and charge compared with the traditional mutation to Ala that is often used to explore single-residue importance. In the absence of structural information, it is possible to make large libraries by using error-prone PCR or gene shuffling. Chemical complementation could also be used to select active variants from these types of libraries.
Chemical complementation allows high-throughput testing of large libraries. Hundreds of thousands of variants can be assayed in one experiment without the spatial resolution necessary for traditional screening methods (i.e., no need for one colony per well). Yeast can be spread on solid media, and, through the power of genetic selection, cells expressing active variants will grow into colonies. Survivors can then be spatially resolved (i.e., transferred to a microplate, one colony per well) for further characterization, decreasing the time and effort required to find new ligand–receptor pairs.
Chemical complementation identifies nuclear receptors with a variety of responses to a specific ligand. Nuclear receptors that activate transcription in response to targeted molecules and not to endogenous compounds have several additional potential applications. The ability to switch a gene on and off in response to any desired compound can be used to build complex metabolic pathways and gene networks and to create conditional knockouts and phenotypes in cell lines and animals. This ability should also be useful in gene therapy and in agriculture to control expression of therapeutic or pesticidal genes. A variety of responses would be useful in engineering biosensor arrays: an array of receptors with differing activation profiles for a specific ligand could provide concentration measurements and increased accuracy of detection. The ability to engineer proteins that activate transcription in response to any desired compound with a variety of activation profiles will provide a general method of engineering enzymes. Receptors that bind the product of a desired enzymatic reaction can be used to select for enzymes that perform this reaction. The stringency of the assay can be adjusted by using ligand–receptor pairs with lower or higher EC50 values. The lack of a general system for genetic selection is currently the limiting step for directed evolution of enzymes (29, 30).
Supplementary Material
Acknowledgments
We thank Peter O'Daniel and the Seley laboratory for assistance with the LG335 synthesis; Dr. Trent Spencer (Emory University, Atlanta) for the gift of HEK 239 cells; Dr. Ron Evans (The Salk Institute for Biological Studies) for the gifts of pCMX-hRXR, pCMX-ACTR and pCMX-βGal; and the members of the Doyle and Bommarius laboratories for insight and critical reading of the manuscript. This work was supported by The Seaver Foundation, the Research Corporation, and National Science Foundation Major Research Instrumentation Program Grant NSF DBI-0320786 to the Center for Fundamental and Applied Molecular Evolution (FAME).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: RXR, human retinoid X receptor α; 9cRA, 9-cis retinoic acid; LBD, ligand-binding domain; DBD, DNA-binding domain; GBD:RXR, Gal4 DBD RXR LBD fusion protein; ACTR:GAD, ACTR Gal4 activation domain fusion protein; SC, synthetic complete.
References
- 1.Mangelsdorf, D. J., Ong, E. S., Dyck, J. A. & Evans, R. M. (1990) Nature 345, 224–229. [DOI] [PubMed] [Google Scholar]
- 2.Zhao, Q., Chasse, S. A., Devarakonda, S., Sierk, M. L., Ahvazi, B. & Rastinejad, F. (2000) J. Mol. Biol. 296, 509–520. [DOI] [PubMed] [Google Scholar]
- 3.de Urquiza, A. M., Liu, S., Sjoberg, M., Zetterstrom, R. H., Griffiths, W., Sjovall, J. & Perlmann, T. (2000) Science 290, 2140–2144. [DOI] [PubMed] [Google Scholar]
- 4.Allegretto, E. A., McClurg, M. R., Lazarchik, S. B., Clemm, D. L., Kerner, S. A., Elgort, M. G., Boehm, M. F., White, S. K., Pike, J. W. & Heyman, R. A. (1993) J. Biol. Chem. 268, 26625–26633. [PubMed] [Google Scholar]
- 5.Leo, C. & Chen, J. D. (2000) Gene 245, 1–11. [DOI] [PubMed] [Google Scholar]
- 6.Torchia, J., Glass, C. & Rosenfeld, M. G. (1998) Curr. Opin. Cell Biol. 10, 373–383. [DOI] [PubMed] [Google Scholar]
- 7.Moras, D. & Gronemeyer, H. (1998) Curr. Opin. Cell Biol. 10, 384–391. [DOI] [PubMed] [Google Scholar]
- 8.Escriva, H., Safi, R., Hanni, C., Langlois, M., Saumitou-Laprade, P., Stehelin, D., Capron, A., Pierce, R. & Laudet, V. (1997) Proc. Natl. Acad. Sci. USA 94, 6803–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egea, P. F., Mitschler, A., Rochel, N., Ruff, M., Chambon, P. & Moras, D. (2000) EMBO J. 19, 2592–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peet, D. J., Doyle, D. F., Corey, D. R. & Mangelsdorf, D. J. (1998) Chem. Biol. 5, 13–21. [DOI] [PubMed] [Google Scholar]
- 11.Doyle, D. F., Braasch, D. A., Jackson, L. K., Weiss, H. E., Boehm, M. F., Mangelsdorf, D. J. & Corey, D. R. (2001) J. Am. Chem. Soc. 123, 11367–11371. [DOI] [PubMed] [Google Scholar]
- 12.Azizi, B., Chang, E. I. & Doyle, D. F. (2003) Biochem. Biophys. Res. Commun. 306, 774–780. [DOI] [PubMed] [Google Scholar]
- 13.Wurtz, J. M., Bourguet, W., Renaud, J. P., Vivat, V., Chambon, P., Moras, D. & Gronemeyer, H. (1996) Nat. Struct. Biol. 3, 87–94, and erratum (1996) 3, 206. [DOI] [PubMed] [Google Scholar]
- 14.Reidhaar-Olson, J. F., Bowie, J. U., Breyer, R. M., Hu, J. C., Knight, K. L., Lim, W. A., Mossing, M. C., Parsell, D. A., Shoemaker, K. R. & Sauer, R. T. (1991) Methods Enzymol. 208, 564–586. [DOI] [PubMed] [Google Scholar]
- 15.Lim, W. A. & Sauer, R. T. (1991) J. Mol. Biol. 219, 359–376. [DOI] [PubMed] [Google Scholar]
- 16.Boehm, M. F., Zhang, L., Badea, B. A., White, S. K., Mais, D. E., Berger, E., Suto, C. M., Goldman, M. E. & Heyman, R. A. (1994) J. Med. Chem. 37, 2930–2941. [DOI] [PubMed] [Google Scholar]
- 17.Held, P., Heck, M. P., Iyer, J., Gronemeyer, H., Lebeau, L. & Mioskowski, C. (1997) J. Labelled Comp. Radiopharm. 39, 501–507. [Google Scholar]
- 18.Kagechika, H., Kawachi, E., Hashimoto, Y., Himi, T. & Shudo, K. (1988) J. Med. Chem. 31, 2182–2192. [DOI] [PubMed] [Google Scholar]
- 19.Chandraratna, R. A. (1994) U.S. Patent 5,344,959.
- 20.Alvi, K. A., Rodriguez, J., Diaz, M. C., Moretti, R., Wilhelm, R. S., Lee, R. H., Slate, D. L. & Crews, P. (1993) J. Org. Chem. 58, 4871–4880. [Google Scholar]
- 21.Faul, M. M., Ratz, A. M., Sullivan, K. A., Trankle, W. G. & Winneroski, L. L. (2001) J. Org. Chem. 66, 5772–2782. [DOI] [PubMed] [Google Scholar]
- 22.Chen, H., Lin, R. J., Schiltz, R. L., Chakravarti, D., Nash, A., Nagy, L., Privalsky, M. L., Nakatani, Y. & Evans, R. M. (1997) Cell 90, 569–580. [DOI] [PubMed] [Google Scholar]
- 23.Geitz, R. D. & Woods, R. A. (2002) Methods Enzymol. 350, 87–96. [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. H. (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 25.Johnston, S. A., Zavortink, M. J., Debouck, C. & Hopper, J. E. (1986) Proc. Natl. Acad. Sci. USA 83, 6553–6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doyle, D. F. (1996) Dissertation (Univ. of North Carolina, Chapel Hill).
- 27.Wolfenden, R., Andersson, L., Cullis, P. M. & Southgate, C. C. (1981) Biochemistry 20, 849–855. [DOI] [PubMed] [Google Scholar]
- 28.Egea, P. F., Mitschler, A., Moras, D. (2002) Mol. Endocrinol. 16, 987–997. [DOI] [PubMed] [Google Scholar]
- 29.Zhao, H. & Arnold, F. H. (1997) Curr. Opin. Struct. Biol. 7, 480–485. [DOI] [PubMed] [Google Scholar]
- 30.Firestine, S. M., Salinas, F., Nixon, A. E., Baker, S. J. & Benkovic, S. J. (2000) Nat. Biotechnol. 18, 544–547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.