Abstract
We have established that the gene AF4, which had long been recognized as disrupted in childhood leukemia, also plays a role in the CNS. Af4 is mutated in the robotic mouse that is characterized by ataxia and Purkinje cell loss. To determine the molecular basis of this mutation, we carried out a yeast two-hybrid screen and show that Af4 binds the E3 ubiquitin ligases Drosophila seven in absentia (sina) homologues (Siah)-1a and Siah-2 in the brain. Siah-1a and Af4 are expressed in Purkinje cells and colocalize in the nucleus of human embryonic kidney 293T and P19 cells. In vitro binding assays and coimmunoprecipitation reveal a significant reduction in affinity between Siah-1a and robotic mutant Af4 compared with wild-type, which correlates with the almost complete abolition of mutant Af4 degradation by Siah-1a. These data strongly suggest that an accumulation of mutant Af4 occurs in the robotic mouse due to a reduction in its normal turnover by the proteasome. A significant increase in the transcriptional activity of mutant Af4 relative to wild-type was obtained in mammalian cells, suggesting that the activity of Af4 is controlled through Siah-mediated degradation. Another member of the Af4 family, Fmr2, which is involved in mental handicap in humans, binds Siah proteins in a similar manner. These results provide evidence that a common regulatory mechanism exists that controls levels of the Af4/Fmr2 protein family. The robotic mouse thus provides a unique opportunity to understand how these proteins play a role in disorders as diverse as leukemia, mental retardation, and neurodegenerative disease.
Neurodegeneration of the cerebellum is characteristic of numerous human disorders and cerebellar mouse mutants have provided a rich source of model systems in which to study perturbations of the CNS as a whole (1). The distinctive trilaminar structure and small number of cell types of the cerebellum have facilitated mutant mouse studies of ion-channel defect (2), neurotransmitter release (3), and triplet-repeat disorders (4). We have identified a mouse model of autosomal dominant cerebellar ataxia, named robotic, that develops adult-onset Purkinje cell loss as well as cataracts (5). Genetic and physical mapping lead to the identification of the causative mutation in a highly conserved region of acute lymphoblastic leukemia 1-fused gene from chromosome 4 (Af4), which was also shown to be specifically expressed in Purkinje cells in the brain. Chromosomal translocations involving AF4 and the mixed-lineage leukemia gene are implicated in childhood acute lymphoblastic leukemia (6). However, despite B and T cell developmental defects in the Af4 knockout mouse suggesting a role in lymphopoiesis (7), the precise function of AF4 remains poorly understood. Consequently, the robotic mutant provides a function for Af4 in the brain that would not have been predicted from previous studies. In addition, the distinctive region-specific pattern of Purkinje cell loss that occurs is unusual among known cerebellar mutants and presents an opportunity to study new neurodegenerative pathways.
AF4 is a member of the AF4, LAF4, and FMR2 (ALF) family of proline- and serine-rich proteins that also includes the more recently characterized AF5Q31 (8-11). All proteins share a significant degree of homology over a number of regions, and it is hypothesized that they act as transcription factors; it has been demonstrated that the N-terminal region of the ALF family, and can activate transcription in an in vitro reporter system and that LAF4 has the ability to bind DNA nonspecifically (10, 12, 13). Like AF4, AF5Q31 is implicated in leukemogenesis through chromosomal rearrangements, whereas inactivating mutations in FMR2 are associated with fragile site, chromosome Xq28 mental retardation (11, 14, 15). The ALF domain that contains the robotic mutation is highly conserved among all protein family members, suggesting that this region may be functionally significant, although its precise role in the context of the protein is not known.
To understand the cause of neurodegeneration in the robotic mouse, we investigated potential binding protein partners in the brain and the possibility that Af4-mutant/wild-type (mut/wt) proteins might have different affinities for their targets. We identified members of the ubiquitin (Ub)-proteasome pathway, Drosophila seven in absentia (sina) homologues (Siah)-1a and Siah-2, from a yeast two-hybrid screen and showed that the normal rapid turnover of Af4 by the proteasome is significantly reduced when the robotic mutation is present. These data provide insights into the regulation of the AF4 family of transcription factors that are important in the normal function of the CNS.
Methods
Yeast Two-Hybrid Screening. The region of Af4 corresponding to the first 360 aa was cloned into the pGBKT7 (bait) vector before transformation into the haploid yeast strain AH109 (BD Biosciences Clontech, Palo Alto, CA). The Matchmaker-pretransformed adult mouse whole-brain library was screened with wild-type and robotic mutant constructs according to the manufacturer's instructions (BD Biosciences Clontech). Diploids were selected on -Ade/-His/-Leu/-Trp plates, and plasmid DNA was purified from restreaked colonies by using the RPM yeast plasmid isolation kit (QBiogene, Carlsbad, CA). Interacting clones were identified by sequencing pGADT7 (prey) library inserts. To confirm interactions, the ORFs of both Siah-1a and Siah-2 were cloned into pGBKT7 and cotransformed into AH109 yeast cells with the Af4 baits.
Plasmid Construction. Full-length Siah-1a-wt was cloned into the mammalian expression vector pCDNA3 (Invitrogen) to create pCDNA3-Siah-1a-wt. The mutant version, pCDNA3-Siah-1amut, was generated by mutagenizing amino acid 44 of the RING domain from cysteine to serine, using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's instructions. The Siah-1a substrate-binding domain (SBD) (amino acids 82-282) was cloned into pET28(a) (Novagen) to create pET28-SBD-His. Full-length Af4-wt and Af4-mut were subcloned into pCDNA3 with a hemagglutinin (HA) tag to generate pCDNA3-HA-Af4-wt and pCDNA3-HA-Af4-mut. Mouse polyubiquitin was cloned into pCDNA3 with a FLAG tag to generate the pCDNA3-FLAG-Ub. For transactivation studies in yeast, Af4-wt and Af4-mut GAL4 DNA-binding domain (BD) fusions were constructed by cloning the cDNA sequence corresponding to amino acids 1-515 into pGBKT7 (BD Biosciences Clontech) to create pGB-Af4-wt and pGB-Af4-mut. Identical regions were cloned into the pM vector (BD Biosciences Clontech) to create pM-Af4-wt and pM-Af4-mut for transactivation experiments in mammalian cells.
In Situ Hybridization. A 309-bp region of Af4 (3194-3502 bp of GenBank entry AF074266) and a 266-bp region of Siah-1a (1668-1934 bp of GenBank entry Z19579) was used for digoxigenin-labeled riboprobe synthesis and hybridization as described (5).
Production of Recombinant Siah-1a SBD and Peptide-Binding Assay. Expression of His-tagged Siah-1a SBD was induced overnight at 22°C in BL21(DE3) cells (Novagen) and purified from sonicated cells by using BD TALON affinity resin (BD Biosciences Clontech). Purified protein was labeled overnight at 4°C with Europium (Eu3+) labeling reagent according to the manufacturer's instructions (PerkinElmer). Dissociation-enhanced lanthanide fluorescence immunoassay was carried out essentially as recommended by PerkinElmer. Wild-type and robotic 23-mer peptides corresponding to residues 268-290 of Af4 and residues 277-299 of Fmr2 were synthesized with an N-terminal biotin residue (ThermoHybaid, Ulm, Germany) and captured onto streptavidin-coated wells (Nunc) (36 pmol per well). Increasing quantities of Eu3+-labeled Siah-1a SBD were added (0-100 pmol), and Eu3+ fluorescence was measured at 613 nm by using a VICTOR Multilabel counter (Wallac, Gaithersburg, MD).
Cell Culture and Transfection. Human embryonic kidney (HEK) 293T and mouse teratocarcinoma P19 cells were grown in DMEM supplemented with 10% FBS (or 7.5% FBS and 2.5% FCS for P19 cells), 2 mM l-glutamine, 1 mM nonessential amino acids, and 1% ampicillin-streptomycin. HEK293T and P19 cells were transiently transfected for 24 or 72 h, respectively, with equal amounts of pCDNA3 constructs by using FuGENE 6 (Roche Diagnostics, Lewes, U.K.). Before transfection, P19 cell differentiation and the induction of a neuronal-like phenotype were carried out for 3 days by the addition of all-trans retinoic acid (Sigma) at 5 × 10-7 M. After transfection, HEK293T and P19 cells were cultured or were not cultured for 7 h in the presence or absence of the reversible proteasome inhibitor MG132 (Calbiochem) at 10 μM. For Af4 degradation time-course analysis, cells were further treated with cycloheximide (Sigma) at 30 μM for up to 48 h, after MG132 had been washed away. Human Negroid cervix epitheloid carcinoma (HeLa) cells were grown in MEME supplemented as HEK293T cells and were transfected by using Lipofectamine (Invitrogen) for 48 h.
Immunofluorescence Microscopy. HEK293T and P19 cells were cotransfected with pCDNA3-HA-Af4-wt or -mut and pCDNA3-Siah-1a-wt or -mut and treated or not treated in the presence or absence of MG132, as described above. Cells were immunostained by using mouse anti-HA (Sigma) and goat anti-Siah-1 (Santa Cruz Biotechnology) primary antibodies and Alexa Fluor 594 anti-mouse and Alexa Fluor 488 anti-goat secondary antibodies (Molecular Probes) following standard procedures.
Immunoprecipitation and Western Blot Analysis. HEK293T cells were transfected with various combinations of pCDNA3 constructs and treated or not treated in the presence or absence of MG132, as described above. Total cell extracts were prepared in lysis buffer [50 mM Tris·HCl (pH 7.4)/150 mM NaCl/1% Triton X-100] supplemented with a mixture of protease inhibitors (Sigma). For coimmunoprecipitation experiments, Triton X-100 was substituted for 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate in the lysis buffer. Lysates were quantified by using the bicinchoninic acid assay kit (Pierce), and equal amounts of protein were immunoprecipitated overnight with either rabbit anti-HA or goat anti-Siah-1 antibodies. Immunocomplexes were bound to protein G-Sepharose 4B fast flow (Sigma), resolved by SDS/PAGE, and analyzed by Western blot using rabbit anti-HA, mouse anti-FLAG, or goat anti-Siah-1 primary antibodies and donkey anti-rabbit, sheep anti-mouse (Amersham Biosciences, Piscataway, NJ), or donkey anti-goat (Jackson ImmunoResearch) horseradish peroxidase-conjugated secondary antibodies, according to standard procedures. Proteins were visualized by enhanced chemiluminescence detection (Amersham Biosciences) following the manufacturer's instructions. All immunoprecipitation experiments were carried out a minimum of three times.
Yeast Transactivation Assay. Yeast cell transformations and β-galactosidase reporter assays were carried out as described (13). Briefly, pGB-Af4-wt, pGB-Af4-mut, and pGBKT7 vectors were transformed into competent cells from the Saccharomyces cerevisiae Y187 reporter strain. Liquid culture β-galactosidase assays were carried out from individual colonies by using o-nitrophenyl-β-d-galactopyranoside as the substrate.
Mammalian Cell Transactivation Assay. HeLa cells were cotransfected with pM-Af4-wt or pM-Af4-mut constructs and pG5CAT reporter plasmid, as described above. Total cell extracts were prepared in chloramphenicol acetyltransferase (CAT) ELISA lysis buffer (Roche Diagnostics) and quantified by using the bicinchoninic acid protein assay kit. CAT assays were performed with normalized extract concentractions by using the CAT ELISA kit as described by the manufacturer (Roche Diagnostics). To determine plasmid expression levels of transfected cells, total RNA was isolated from 104 cells by using the RNeasy miniprep kit and was treated with DNase according to the manufacturer's instructions (Qiagen, Valencia, CA). Primers specific for pM-Af4-wt/mut and pG5CAT were used to amplify cDNA by using 30 cycles of PCR amplification.
Results
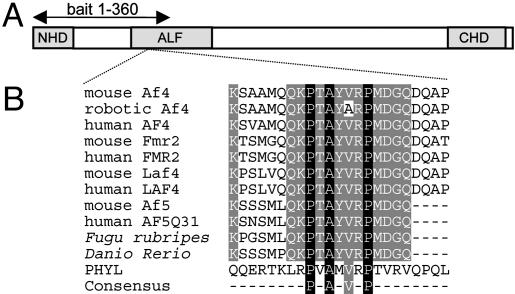
Identification of Siah-1a and Siah-2 as Af4-Binding Partners in Mouse Brain. To identify any differences between protein interactors in the wild-type and robotic cerebellum, we carried out a yeast two-hybrid screen. An Af4 bait construct was designed that contained the robotic mutation but did not include the transactivation domain (Fig. 1A) (8, 13), thus avoiding self-activation of the yeast reporter system. More than 6 × 106 clones were screened independently by using wild-type and mutant baits, and DNA from 75 of the 400 diploid colonies was sequenced. From the wild-type screen, seven clones contained regions of the Siah-1a cDNA sequence and six contained Siah-2, whereas fewer clones (three Siah-1a and two Siah-2) were obtained by using the mutant bait. The interaction of the Af4 bait with full-length Siah-1a/2 was confirmed by cloning the ORFs as GAL4 activation domain fusions and retesting by cotransformation and yeast mating (data not shown).
Fig. 1.
ALF family protein structure. (A) The conserved N- and C-terminal homology (NHD/CHD) and ALF domains are shown and the yeast two-hybrid bait region is marked. (B) Multiple amino acid alignment of the ALF family including the 23-mer Af4 peptides used for the Siah-1a dissociation-enhanced lanthanide fluorescence immunoassay. GenBank accession nos. from top to bottom: NP_598680, AY749168, NP_005926, NP_032058, NP_002016, XP_283603, NP_002276, NP_291043, and NP_055238; Fugu rubripes and Danio rerio sequences were taken from ENSEMBL database-predicted proteins SINFRUP00000169375 and ENSDARP0000009290, respectively, which can be accessed at www.ensembl.org. The Siah-binding sequence of PHYL (NP_725394) is shown, in addition to the consensus motif (18).
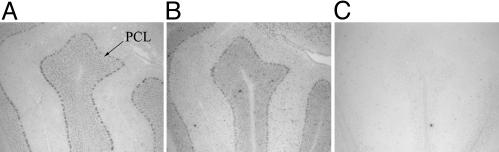
Siah-1a Is Expressed in Purkinje Cells in the Cerebellum. Both Siah-1a and Siah-2 are expressed widely in embryonic and adult mouse tissues (16), and a recent study using a nonspecific Siah-1a/b probe demonstrated that pyramidal neurons of the hippocampus as well as Purkinje cells express high levels of Siah-1 mRNAs (17). To determine whether Siah-1a has an expression profile similar to that of Af4, in situ hybridization was carried out on adult brain sections using a riboprobe designed from the 3′ UTR to avoid crossreactivity with Siah-1b (16). Expression of Siah-1a occurs in the Purkinje cells of the cerebellum and at lower levels in the molecular layer, whereas Af4 expression is restricted to Purkinje cells (Fig. 2). However, no reduction in Siah-1a expression was observed in cerebellar lobe X, as described when using an Af4 riboprobe (5), and identical results were obtained by using both wild-type and robotic samples (data not shown).
Fig. 2.
Coexpression of Af4 and Siah-1a in mouse cerebellum. In situ hybridization was performed from wild-type mouse brain sections by using antisense riboprobes for Af4 (A) and Siah-1a (B). Expression of both genes occurs in the Purkinje cell layer (PCL). (C) A negative control sense probe for Siah-1a is shown. Lobe II of the cerebellum is shown.
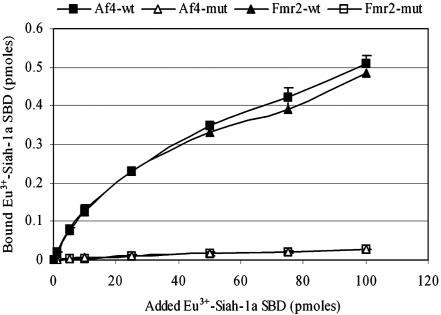
The Robotic Mutation Significantly Reduces the Binding of Af4 and Fmr2 to Siah-1a in Vitro. The robotic V280A mutation in Af4 occurs exactly over the recently identified consensus Siah-binding motif (Fig. 1B) (5, 18). To investigate whether this mutation affects the interaction between Siah-1a and Af4, a quantifiable dissociation-enhanced lanthanide fluorescence immunoassay was performed by using recombinant Eu3+-labeled Siah-1a SBD and immobilized peptides corresponding to wild-type and robotic mutant Af4 (peptide sequences in Fig. 1B). An ≈15-fold decrease in the affinity of mutant peptides for Siah-1a SBD compared with wild-type counterparts was observed (Fig. 3). These data predict defective proteasomal degradation of Af4-mut from decreased interaction with Siah-1a in the robotic mouse. Because Fmr2 has been shown to exhibit an overlapping expression pattern to Af4 in the hippocampus and cerebellum (5, 19), we investigated whether the homologous region of this protein would be sufficient for Siah-1a binding by testing both wild-type and robotic mutant Fmr2 peptides. The results were similar to those obtained by using the Af4 peptides, demonstrating that Fmr2 binds to Siah proteins and suggesting that all members of the ALF family will be regulated by this interaction (Fig. 3).
Fig. 3.
In vitro binding assay of Siah-1a to wild-type and mutant Af4 and Fmr2 peptides. Biotinylated Af4/Fmr2 wild-type and mutant peptides were incubated in the presence of increasing quantities of Eu3+-labeled recombinant Siah-1a SBD. The amount of bound protein was determined by converting the fluorescence count values to that obtained for the Eu3+ standard (0.1 pmol). Data are presented as means + SD for three independent experiments.
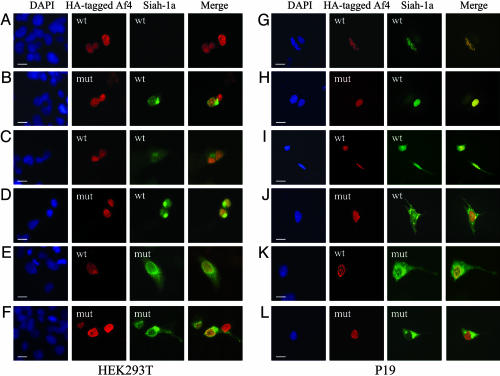
Siah-1a Colocalizes with Af4 in HEK293T and P19 Cells. To verify that Af4 and Siah-1a interact in vivo, we first searched for colocalization in cotransfected HEK293T cells (Fig. 4 A-F). Af4-wt and -mut were detected in the nucleus, whereas Siah-1a was expressed in both the nuclear and cytoplasmic compartments (Fig. 4 A and B). However, Siah-1a was detectable only when coexpressed with Af4-mut, suggesting that the robotic protein is able to stabilize Siah-1a expression. Siah-1 is known to be degraded rapidly through the proteasome pathway (20, 21), and cell treatment with the proteasome inhibitor MG132 indeed efficiently stabilized Siah-1a expression (Fig. 4 C and D), enabling colocalization with Af4-wt to be observed (Fig. 4C). Nuclear colocalization of Af4 and Siah-1a was more evident in cells expressing Siah-1a-mut, a stable nonfunctional mutant Siah-1a (Fig. 4 E and F) (20), with localized accumulation of Siah-1a at the periphery of the nucleus when coexpressed with Af4-mut (Fig. 4F). To confirm colocalization of Af4 and Siah-1a in a neuronal model system, the same study was conducted in differentiated neuron-like P19 cells (Fig. 4 G-L). Higher Siah-1a expression levels in this cell type allowed colocalization with both Af4-wt and Af4-mut to be observed without the use of MG132 or Siah-1a-mut (Fig. 4 G and H), suggesting that the turnover of Siah-1a may be less efficient in neurons than in other cell types. Interestingly, colocalization with Af4-wt occurred with a distinctive punctate appearance in the nucleus (Fig. 4 G, I, and K), whereas a more homogeneous pattern was observed with Af4-mut (Fig. 4 H, J, and L). In addition, a reproducible accumulation of Siah-1a outside of the nucleus occurred in cells coexpressing Af4-mut (Fig. 4 J and L).
Fig. 4.
Siah-1a and Af4 colocalization in mammalian cells. HEK293T cells (A-F) and differentiated P19 cells (G-L) were cotransfected with equal amounts of pCDNA3-HA-Af4-wt (A, C, E, G, I, and K) or pCDNA3-HA-Af4-mut (B, D, F, H, J, and L) and pCDNA3-Siah-1a-wt (A-D and G-J) or pCDNA3-Siah-1a-mut (E, F, K, and L) constructs. Cells were cultured for 7 h in the presence (C, D, I, and J) or absence (A, B, E, F, K, and L) of MG132. Cells were labeled with mouse anti-HA and goat anti-Siah-1 primary antibodies followed by Alexa Fluor 594 anti-mouse and Alexa Fluor 488 anti-goat secondary antibodies. Preparations were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize the nuclei (blue); regions of overlap between Af4 (red) and Siah-1a (green) appear in yellow in the merged images. (Scale bars, 10 μm.)
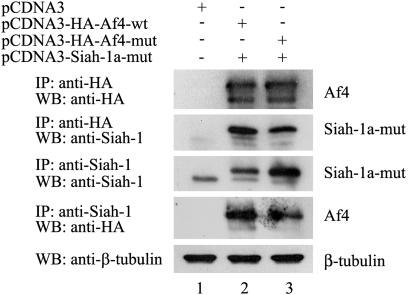
Siah-1a Interacts with Af4 and Mediates Its Degradation by the Ub-Proteasome Pathway in Mammalian Cells. To confirm the interaction between Af4 and Siah-1a, coimmunoprecipitation experiments were performed. Because of the lack of detection of Siah-1a-wt when coexpressed with Af4-wt (Fig. 4A), Siah-1amut was used to enable its detection by Western blot. By using an anti-HA antibody, Af4-wt and Af4-mut were detected as two protein bands with apparent molecular masses of 143 and 183 kDa (Fig. 5, lanes 2 and 3). Because the predicted molecular mass of HA-tagged Af4 is 143 kDa, we discounted the possibility that the larger band might represent the glycosylated form of Af4 (data not shown), although it may be due to other posttranslational modifications or alternative splicing, as is the case for FMR2 (22). It is most likely, however, that the smaller band corresponds to a degradation product of the 183-kDa full-length Af4 protein. The anti-HA antibody efficiently coimmunoprecipitated the 32-kDa Siah-1a-mut protein from cells coexpressing Af4-wt (Fig. 5, lane 2), whereas the levels of Siah-1a coimmunoprecipitated from cells coexpressing Af4-mut were significantly reduced (Fig. 5, lane 3). These data were confirmed by using the reciprocal antibodies, demonstrating a reduction in binding affinity between Siah-1a and Af4-mut.
Fig. 5.
Siah-1a interacts with Af4 in mammalian cells. HEK293T cells were cotransfected with the indicated constructs. Total cell lysates were immunoprecipitated (IP), resolved by SDS/PAGE, and analyzed by Western blot (WB). To verify equal inputs, 20 μg of each total lysate was analyzed by Western blot by using an anti-β-tubulin antibody.
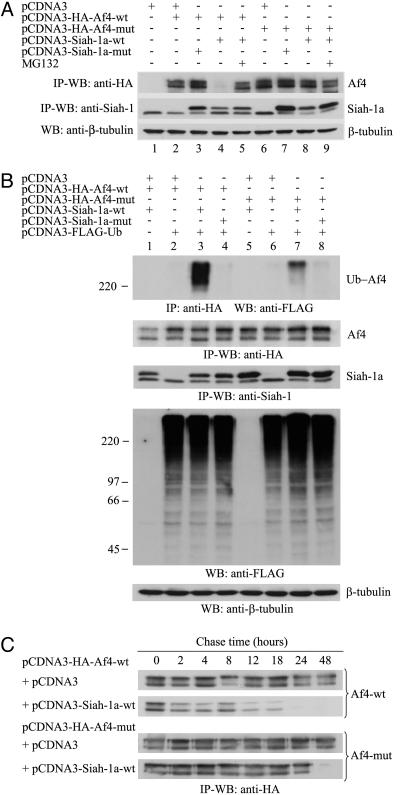
To confirm that this interaction leads to degradation of Af4 by the Ub-proteasome pathway, cotransfected HEK293T cultures were treated in the presence or absence of MG132 (Fig. 6A). An almost complete absence of Af4-wt detection was obtained in cells coexpressing Siah-1a-wt (Fig. 6A, lane 4) whereas, as expected, Siah-1a-mut had no effect (Fig. 6A, lane 3). The levels of Af4-wt were efficiently restored after treatment with MG132 (Fig. 6A, lane 5), confirming Siah-1a-mediated degradation of Af4 by the proteasome. In contrast, there was no detectable reduction in the levels of Af4-mut (Fig. 6A, lane 8). Interestingly, a reproducible stabilization of Siah-1a-wt occurred in the presence of Af4-mut (Fig. 6A, compare lanes 4 and 8). By coexpressing FLAG-tagged Ub (FLAG-Ub), we could also verify that Siah-1a-wt, but not Siah-1a-mut, promotes ubiquitination of Af4 (Fig. 6B, lanes 3 and 4), whereas lower levels of ubiquitinated Af4-mut were observed (Fig. 6B, lane 7).
Fig. 6.
Siah-1a mediates Af4 degradation through the Ub-proteasome pathway. (A-C) HEK293T cells were cotransfected with the indicated pCDNA3 constructs. Unless otherwise stated, cells were cultured for 7 h in the presence of MG132. In C, cells were subsequently washed and chased for the indicated times in the presence of cycloheximide. Total cell lysates were analyzed as in Fig. 5. In B, FLAG-Ub-conjugated proteins were detected by Western blot from 20 μg of each lysate using an anti-FLAG antibody.
Finally, the turnover of both Af4-wt and Af4-mut was assessed in the presence or absence of Siah-1a in a pulse-chase study (Fig. 6C). Results showed that the degradation of Af4-wt is accelerated by coexpression of Siah-1a, with a half-life of ≈12 h. In contrast, Af4-mut showed no detectable degradation up to the final time point at 48 h, illustrating the significance of the mutation on the normal turnover of Af4.
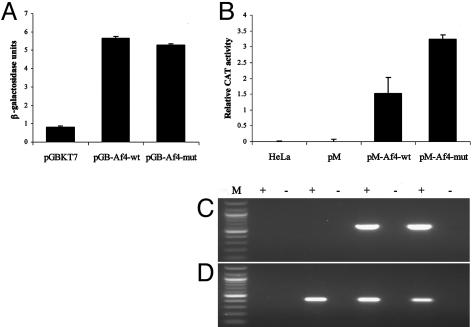
Transcriptional Activation Analysis of Wild-Type and Robotic Af4. To investigate whether the robotic mutation affects the transcriptional properties of Af4, a quantitative reporter assay was carried out by using N-terminal constructs containing the ALF domain and the previously described minimal transactivation domain (8, 13). By using a yeast o-nitrophenyl-β-d-galactopyranoside reporter assay, both Af4 GAL4-BD fusions (pGB-Af4-wt and pGB-Af4-mut) showed an ≈6-fold activation of transcription above the control pGBKT7 vector, although no significant difference was observed between control and robotic mutant constructs (Fig. 7A). However, in HeLa cells using a CAT reporter assay, a significant and reproducible 2-fold increase in activation was obtained with the robotic GAL4-BD construct (pM-Af4-mut) compared with wild-type (pM-Af4-wt) (Fig. 7B). These results were not caused by differences in transfection efficiency or in vivo transcription of the transfected pM-Af4-wt/mut and reporter pG5CAT constructs, as demonstrated by RT-PCR (Fig. 7 C and D).
Fig. 7.
Effect of the robotic mutation on the transactivation potential of Af4. (A) Yeast o-nitrophenyl-β-d-galactopyranoside activation assay. pGBKT7 fusions were transformed into Y187 cells and β-galactosidase activity was assayed. One unit of β-galactosidase is defined as the amount that hydrolyses 1 mmol of o-nitrophenyl-β-d-galactopyranoside to o-nitrophenol and d-galactose per minute. Results are shown + SD. (B) Mammalian cell activation assay. pM fusions were cotransfected into HeLa cells with reporter vector pG5CAT. After transfection, CAT activity was measured by ELISA. Results are shown + SD. (C and D) RT-PCR analysis of mammalian activation assay. Primers specific for pM-Af4-wt/mut (C) or pG5CAT (D) were used to amplify cDNA from the corresponding cotransfections in B. + and -, reactions with and without the presence of reverse transcriptase in the cDNA synthesis step. Reactions were run with a 100-bp ladder (lane M) (New England Biolabs).
Discussion
In this study, we have demonstrated that Af4 interacts with the E3 Ub ligases Siah-1a and Siah-2 and that the point mutation in the robotic mouse would severely impair the normal turnover of this transcription factor due to a reduction in binding affinity. The Ub-proteasome pathway is now recognized as an important mechanism for the regulation of protein stability (23), and an increasing number of unrelated studies have isolated Siah proteins from yeast two-hybrid screens, reflecting the diverse nature of the signaling pathways in which these proteins are involved. Known interactors include the synaptic vesicle protein synaptophysin (24) and a number of transcription factors, including transforming growth factor β-inducible early gene-1 (25) and the protooncogene c-Myb (26). Siah proteins have also been linked to Parkinson's disease by the ubiquitination and degradation of synphillin-1, a major component of Lewy bodies, the characteristic cytoplasmic inclusions found in affected patients (21). Most recently, it has been predicted that the transforming potential of AF4-mixed-lineage leukemia gene fusions is controlled by SIAH-1 and SIAH-2 (27). In HEK293T cells, the normal proteasomal degradation of AF4-mixed-lineage leukemia gene was blocked once it had undergone proteolysis and complex formation, suggesting that SIAH-1/2 may regulate the oncogenic potential of the fusion protein.
Here, we have proved that Af4 is degraded by the Ubproteasome complex and have confirmed and quantified its interaction with Siah-1a by coimmunoprecipitation. Furthermore, immunofluorescence staining of Siah-1a and Af4 in HEK293T and P19 cells showed colocalization in the nucleus, which is consistent with previous overexpression studies of epitope-tagged Siah-1a in HEK293T cells (28) and the known nuclear localization of AF4 (8). In addition, we have demonstrated that the robotic mutation significantly reduces the binding affinity of the Siah-1a SBD with Af4 and Fmr2 peptides, which was not unexpected, due to the 80% reduction in binding affinity of a PHYL peptide with an identical mutation (PHYLV120A) (18). Because of the 100% identity of the PXAX-VXP motif and the presence of conserved nuclear localization signals, we predict that all members of the ALF family will interact with and be degraded by means of Siah-1a-mediated ubiquitination (Fig. 1B).
The evidence presented here demonstrates for the first time, to our knowledge, that the conserved Siah-binding motif links Af4 and Fmr2 regulation. Mutations in FMR2 are known to be responsible for fragile site, chromosome Xq28 mental handicap (15, 16). However, only subtle deficits in long-term potentiation were discovered in the Fmr2 knockout mouse (29); thus, the role of this gene in mental retardation remains elusive. The robotic mouse therefore provides an opportunity to demonstrate that ALF proteins have overlapping functions in the brain. Future work with transgenic mice containing the robotic mutation in Fmr2 would facilitate functional studies of this gene.
We have shown here that both Siah-1a and Af4 show specific expression patterns in Purkinje cells of the cerebellum by in situ hybridization (5). Because we have recently demonstrated that Siah-2, also isolated from our yeast two-hybrid screen, is expressed in Purkinje cells (data not shown), it is likely that Siah-2 is involved in the degradation of ALF proteins. Indeed, it has been shown that, in addition to their high degree of sequence homology and similar expression patterns, Siah proteins possess some overlapping functions. For example, synaptophysin and deleted in colorectal cancer are degraded by the Ub-proteasome pathway by means of an interaction with both Siah-1a and Siah-2 (25, 30). Consequently, we have concentrated our study on Siah-1a.
We have previously postulated that the mutant Af4 allele acts through a gain-of-function mechanism based on expression studies and the lack of CNS abnormalities in the Af4 knockout mice (5). The data presented here suggest that an accumulation of Af4-mut occurs due to a lack of normal Siah-mediated ubiqitination and its subsequent degradation. Both the Af4 null and heterozygous knockout mice show no signs of ataxia or neurodegeneration up to 6 months of age (data not shown), indicating that Af4-mut could be activating downstream targets ultimately leading to the features of the robotic phenotype. Our data also show a consistent accumulation of Siah-1a in cells cotransfected with Af4-mut. This finding suggests either that weak or transient binding between Siah-1a and Af4-mut is sufficient to prevent Siah-1a from self-degradation and therefore from interacting with other proteins (31) or simply that Siah molecules usually bound to Af4-wt are free to target additional proteins for proteasomal degradation in vivo.
Previous studies have defined the N-terminal regions of ALF proteins required for transactivaton in vitro (12, 13). It was concluded that FMR2 was a more potent activator of transcription than AF4 and that the conserved ALF domain may act as a transcriptional repressor; in both yeast and mammalian cells, increased levels of reporter gene expression were described from splice variants in which this region of the gene was deleted (13). We have shown here, by using the same reporter system in yeast, that the robotic mutation had no effect on the transcriptional activation activity of Af4. However, in HeLa cells, the transactivation properties of constructs containing the robotic mutation were greater than wild type. The absence of any known Siah homologue in fungi suggests that the increase in transactivation observed with the Af4-mut construct in HeLa cells is likely to result from defective degradation by endogenous SIAHs. Therefore, we predict that the robotic mutation only indirectly affects the transcriptional properties of Af4 by preventing its normal proteasomal degradation by Siah proteins.
The results presented here provide evidence for a common regulatory mechanism controlling the levels and thereby the activity of the ALF protein family. Therefore, the robotic mouse provides an opportunity to understand more about the normal function of this family of proteins and their contribution to human disorders as diverse as leukemia, mental retardation, and neurodegenerative disease.
Author contributions: P.L.O., E.B., and K.E.D. designed research; P.L.O., E.B., J.C., and E.L.J. performed research; P.L.O., E.B., J.C., E.L.J., and K.E.D. analyzed data; and P.L.O., E.B., and K.E.D. wrote the paper.
Abbreviations: HEK, human embryonic kidney; ALF, AF4, LAF4, and FMR2; SBD, substrate-binding domain; mut, mutant; wt, wild-type; HA, hemagglutinin; Ub, ubiquitin; Siah, Drosophila seven in absentia (sina) homologues; BD, binding domain; CAT, chloramphenicol acetyltransferase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY749168).
References
- 1.Heintz, N. & Zoghbi, H. (2000) Annu. Rev. Physiol. 62, 779-802. [DOI] [PubMed] [Google Scholar]
- 2.Patil, N., Cox, D. R., Bhat, D., Faham, M., Myers, R. M. & Peterson, A. S. (1995) Nat. Genet. 2, 126-129. [DOI] [PubMed] [Google Scholar]
- 3.Zuo, J., De Jager, P. L., Takahashi, K. A., Jiang, W., Linden, D. J. & Heintz, N. (1997) Nature 388, 769-773. [DOI] [PubMed] [Google Scholar]
- 4.Burright, E. N., Orr, H. T. & Clark, H. B. (1997) Brain Pathol. 7, 965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Isaacs, A. M., Oliver, P. L., Jones, E. L., Jeans, A., Potter, A., Hovik, B. H., Nolan, P. M., Vizor, L., Glenister, P., Simon, A. K., et al. (2003) J. Neurosci. 23, 1631-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domer, P. H., Fakharzadeh, S. S., Chen, C. S., Jockel, J., Johansen, L., Silverman, G. A., Kersey, J. H. & Korsmeyer, S. J. (1993) Proc. Natl. Acad. Sci. USA 90, 7884-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Isnard, P., Core, N., Naquet, P. & Djabali, M. (2000) Blood 96, 705-710. [PubMed] [Google Scholar]
- 8.Nilson, I., Reichel, M., Ennas, M. G., Greim, R., Knorr, C., Siegler, G., Greil, J., Fey, G. H. & Marschalek, R. (1997) Br. J. Haematol. 98, 157-169. [DOI] [PubMed] [Google Scholar]
- 9.Gecz, J., Bielby, S., Sutherland, G. R. & Mulley, J. C. (1997) Genomics 44, 201-213. [DOI] [PubMed] [Google Scholar]
- 10.Ma, C. & Staudt, L. M. (1996) Blood 87, 734-745. [PubMed] [Google Scholar]
- 11.Taki, T., Kano, H., Taniwaki, M., Sako, M., Yanagisawa, M. & Hayashi, Y. (1999) Proc. Natl. Acad. Sci. USA 96, 14535-14540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prasad, R., Yano, T., Sorio, C., Nakamura, T., Rallapalli, R., Gu, Y., Leshkowitz, D., Croce, C. M. & Canaani, E. (1995) Proc. Natl. Acad. Sci. USA 92, 12160-12164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillman, M.A. & Gecz, J. (2001) J. Hum. Genet. 46, 251-259. [DOI] [PubMed] [Google Scholar]
- 14.Gecz, J., Gedeon, A. K., Sutherland, G. R. & Mulley, J. C. (1996) Nat. Genet. 13, 105-108. [DOI] [PubMed] [Google Scholar]
- 15.Gu, Y., Shen, Y., Gibbs, R. A. & Nelson, D. L. (1996) Nat. Genet. 13, 109-113. [DOI] [PubMed] [Google Scholar]
- 16.Della, N. G., Senior, P.V. & Bowtell, D. D. (1993) Development (Cambridge, U.K.) 117, 1333-1343. [DOI] [PubMed] [Google Scholar]
- 17.Moriyoshi, K., Iijima, K., Fujii, H., Ito, H., Cho, Y. & Nakanishi, S. (2004) Proc. Natl. Acad. Sci. USA 101, 8614-8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.House, C. M., Frew, I. J., Huang, H. L., Wiche, G., Traficante, N., Nice, E., Catimel, B. & Bowtell, D. D. (2003) Proc. Natl. Acad. Sci. USA 100, 3101-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chakrabarti, L., Bristulf, J., Foss, G. S. & Davies, K. E. (1998) Hum. Mol. Genet. 7, 441-448. [DOI] [PubMed] [Google Scholar]
- 20.Hu, G. & Ferron, E. R. (1999) Mol. Cell. Biol. 19, 724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nagano, Y., Yamashita, H., Takahashi, T., Kishida, S., Nakamura, T., Iseki, E., Hattori, N., Mizuno, Y., Kikuchi, A. & Matsumoto, M. (2003) J. Biol. Chem. 278, 51504-51514. [DOI] [PubMed] [Google Scholar]
- 22.Gecz, J. & Mulley, J. C. (1999) Eur. J. Hum. Genet. 7, 157-162. [DOI] [PubMed] [Google Scholar]
- 23.Weissman, A. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 169-178. [DOI] [PubMed] [Google Scholar]
- 24.Wheeler, T. C., Chin, L. S., Li, Y., Roudabush, F. L. & Li, L. (2002) J. Biol. Chem. 277, 10273-10282. [DOI] [PubMed] [Google Scholar]
- 25.Johnsen, S. A., Subramaniam, M., Monroe, D. G., Janknecht, R. & Spelsberg, T. C. (2002) J. Biol. Chem. 277, 30754-30759. [DOI] [PubMed] [Google Scholar]
- 26.Tanikawa, J., Ichikawa-Iwata, E., Kanei-Ishii, C., Nakai, A., Matsuzawa, S., Reed, J. C. & Ishii, S. (2000) J. Biol. Chem. 275, 15578-15585. [DOI] [PubMed] [Google Scholar]
- 27.Bursen, A., Moritz, S., Gaussmann, A., Moritz, S., Dingermann, T. & Marschalek, R. (2004) Oncogene 23, 6237-6249. [DOI] [PubMed] [Google Scholar]
- 28.Matsuzawa, S., Takayama, S., Froesch, B. A., Zapata, J. M. & Reed, J. C. (1998) EMBO J. 17, 2736-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu, Y., McIlwain, K. L., Weeber, E. J., Yamagata, T., Xu, B., Antalffy, B. A., Reyes, C., Yuva-Paylor, L., Armstrong, D., Zoghbi, H., et al. (2002) J. Neurosci. 22, 2753-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu, G., Zhang, S., Vidal, M., Baer, J. L., Xu, T. & Fearon, E. R. (1997) Genes Dev. 11, 2701-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Polekhina, G., House, C. M., Traficante, N., Mackay, J. P., Relaix, F., Sassoon, D. A., Parker, M. W. & Bowtell, D. D. Nat. Struct. Biol. 9, 68-75. [DOI] [PubMed]