Abstract
Chromatin insulators or boundaries are proposed to structure the chromatin fiber into functionally independent domains by promoting the formation of chromatin loops. These elements can block the communication between an enhancer and a gene when placed between them. Interestingly, it has been previously observed that two tandem copies of the Drosophila Su(Hw) insulator abolish this enhancer-blocking activity, presumably through pairing. This bypass effect has not been described with other insulators, however. In this report, we show that the insertion of binding sites for the GAGA factor (GAF) between an enhancer and the Su(Hw) insulator allows bypassing of the insulator. This bypass relies on the activity of both the GAF protein and the Mod(mdg4)-67.2 protein, a factor required for Su(Hw) insulator activity. We show that these two proteins interact in vitro and in vivo, providing molecular evidence of pairing between the GAF sites and the Su(Hw) insulator. Finally, we show that placing the Mcp boundary together with the Su(Hw) insulator between an enhancer and a promoter leads to bypass, again in a GAF- and Mod(mdg4)-dependent manner. Our data provide direct evidence that heterologous insulators can be bypassed by distal enhancers and identify the interaction between GAF and Mod(mdg4) as a possible means to regulate insulator activity.
Insulators and boundaries are chromosomal elements that have been proposed to be involved in the establishment of independent domains within the chromatin fiber. By preventing interactions between regulatory sequences in one domain and promoters in other domains, they participate in the regulation of enhancer/promoter communication. These elements have been experimentally defined by two types of activities, as evidenced by different assays. The first activity can block the communication between an enhancer and a promoter when the insulator is inserted between them (enhancer-blocking activity). The second activity can protect transgenes from position effects and can act as a barrier to the spread of repressive heterochromatin (barrier activity). Numerous insulators have been described in species ranging from yeast to mammals, and some of them contain both enhancer-blocking and barrier activities (reviewed in ref. 1).
Recent data suggest that the establishment of independent loop domains in chromatin can be mediated by interactions between insulators and/or by the tethering of insulators to the nuclear matrix or nuclear envelope. For example, in Drosophila, the scs and scs′ insulators flanking the 15-kb hsp70 region have been shown to be in close proximity to each other in embryonic cells. This pairing seems to be mediated by interactions between the scs-binding protein Zw5 and the scs′-binding protein boundary element-associated factor (BEAF) (2). In yeast, a screen for proteins with boundary activity identified several proteins that interact with the nuclear pore complex (NPC), and tethering of the HML locus to the NPC has been shown to block the spread of repression (3). Finally, the CCCTC-binding factor (CTCF) protein, which binds insulators in the β-globin locus in chickens, mice, and humans, can target an insulator to the nucleolar surface (4).
Strong evidence for the role of insulators in creating loop domains comes from several studies of the Drosophila Su(Hw) insulator. This insulator, originally identified within the gypsy retrotransposon, contains 12 binding sites for the DNA-binding protein Su(Hw) (5). Su(Hw) interacts with an isoform of the Mod(mdg4) protein, a BTB/POZ (broad complex, tramtrack, and brick à brack/poxviruses and zinc finger) domain-containing protein, and both proteins are required for insulator function in vivo (6). Su(Hw) and Mod(mdg4) are associated with >200 loci on polytene chromosomes (7). In diploid cells, however, these proteins form only 20–25 foci per cell nucleus, called insulator bodies, suggesting that several insulators coalesce into higher-order structures (7). Moreover, when two Su(Hw) insulators are inserted in tandem between an enhancer and a promoter, communication between the sequences located outside the domain bordered by the insulators is permitted, allowing the enhancer to bypass the insulators and activate the promoter (8, 9). These results suggest that two insulators positioned in tandem may loop out the sequences between them, bringing the enhancer and promoter into close proximity. Finally, treatment of Drosophila nuclei with 2 M NaCl to unravel chromatin (halo technique) can be used to reveal chromosomal loops attached to the nuclear matrix, and insertion of a gypsy element into one of these loops induces its subdivision into two smaller loops (10). This evidence indicates that the Su(Hw) insulator is involved in the establishment and the anchoring of loops to the nuclear matrix.
Several other boundaries and insulators have been described in Drosophila: the Mcp and Fab-7 boundaries of the Bithorax complex (BX-C) (reviewed in ref. 11), the SF1 boundary (12), the even-skipped (eve) promoter (13), and two regions in the α1-tubulin gene (14). These elements have not been shown to be involved in the creation of loop domains, although long-distance interactions between two Mcp elements and between two Fab-7 elements located in transgenes inserted on different chromosomes have been observed, suggesting that these elements might be capable of structuring chromosomal domains (15, 16). Interestingly all of these insulators and boundaries include binding sites for the GAGA factor (GAF) protein in their sequence, and several of them are regulated and bound by GAF in vivo (13, 17–20). GAF-binding sites are required for the insulator function of both the eve promoter (13) and the SF1 boundary (12). In addition, mutations in the Trithorax-like (Trl) gene, which encodes GAF (21), impair the enhancer-blocking activity of these insulators (12, 13).
GAF is a DNA-binding protein that is involved in the regulation of numerous genes in Drosophila and can participate in gene activation as well as gene repression (reviewed in ref. 22). In particular, GAF is involved in creating nucleosome-free regions by interacting with complexes involved in nucleosome remodeling, as exemplified by its role on the heat shock genes hsp70 and hsp26 (reviewed in ref. 23). All GAF isoforms produced in vivo contain a BTB domain in their N-terminal part (24). The BTB domain [also called POZ (poxviruses and zinc finger) domain] is found in numerous proteins, including transcription factors and actin-binding proteins, and constitutes a protein–protein interaction motif involved in homo- or heterodimer formation (reviewed in ref. 25). The presence of a BTB domain in GAF and Mod(mdg4) proteins is one of the rare common features that has been found in proteins that bind insulators. Interestingly, a genetic interaction between Trl and mod(mdg4) mutants has been described, and both proteins colocalize partially on polytene chromosomes (7), suggesting that they may interact.
Indeed, we describe here that GAF and Mod(mdg4) proteins interact in vivo and in vitro. Moreover, we show that inserting either an array of GAF-binding sites or the Mcp boundary between an enhancer and the Su(Hw) insulator neutralizes the enhancer-blocking activity of the insulator and promotes insulator bypass. In both cases, this neutralization requires the activity of both Mod(mdg4) and GAF proteins. These results suggest that a loop domain is formed between the element bound by GAF and that bound by Mod(mdg4), and that the interaction between these two elements cancels the enhancer-blocking activity of the Su(Hw) insulator and restores the communication between an upstream enhancer and a downstream reporter gene.
Methods
Transgenic Constructs. The 430-bp Su(Hw) insulator was PCR-amplified from the gypsy retrotransposon and inserted between two loxP sites. An 8-kb fragment containing the yellow gene was provided by P. Geyer (University of Iowa, Iowa City). The white regulatory sequences from positions –1084 to –1465 bp relative to the transcription start site, containing the eye enhancer (Ee), were cloned between two frt sites [frt(Ee)]. The Ee or frt(Ee) fragment was inserted between body and wing enhancers at position –1868 relative to the yellow transcription start site. A 690-bp SalI-PstI fragment containing the Mcp fragment was inserted in direct orientation at position –893 relative to the yellow transcription start site. The eight GAF-binding sites obtained by duplication of the 76-bp DNA fragment containing four binding sites from the hsp70 promoter region (between primers 5′-ATTCGTTATTCTCTC and 5′-GTTTACTGTGTGAC) were cloned between loxP or frt sites. For details of the different subcloning steps, see Supporting Methods, which is published as supporting information on the PNAS web site. Transgenic flies were obtained by standard methods. For details of the crosses for genetic experiments, see Supporting Methods. All frt- and loxP-dependent excisions of specific elements were verified by genomic PCR experiments.
Immunoprecipitations and Nuclear Matrix Preparation. Immunoprecipitation (IP) experiments were performed from Drosophila SL-2 cell nuclear extracts, prepared with the Nuclear Extract Kit (Active Motif). Nuclear extract (100 μg) was incubated overnight at 4°C with the sera or preimmune sera (1–2% volume) in the presence of protein A/G plus agarose (Santa Cruz Biotechnology) in 200 μl of a buffer (25 mM Hepes/100 mM KCl/5 mM MgCl2/10% glycerol/0.1% Triton X-100/1mMDTT/1% BSA). Beads were then washed four times in the same buffer without BSA, containing 100 mM KCl for the experiment of Fig. 1A and 300 mM KCl for the experiment of Fig. 1B. The sera used for IPs and Western blots were as follows. For GAF, we generated a rabbit antiserum directed against the full-length GAF-519, bacterially expressed and purified through a Histidine-tag (26). The polyhomeotic (PH) antibody was provided by F. Maschat (Institute of Human Genetics, CNRS, Montpellier, France). For Mod(mdg4), a serum directed against the BTB domain was provided by R. Dorn (Martin Luther University, Halle, Germany). A monoclonal anti-Ubx antibody was provided by Y. Graba (Laboratoire de Génétique et Biologie du Développement, Institut de Biologie du Développement de Marseille, CNRS, Marseille, France), and a monoclonal anti-lamin antibody was provided by D. Arndt-Jovin (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany). The nuclear matrix was prepared according to Fisher and Blobel (27) from 0- to 24-h-old Drosophila embryos. Western blots were performed following standard protocols.
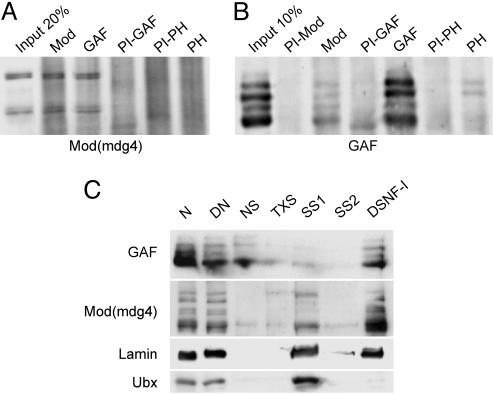
Fig. 1.
GAF and Mod(mdg4) coimmunoprecipitate and are present in the nuclear matrix. (A and B) Coimmunoprecipitation experiments. The serum used for the immunoprecipitation is indicated above each lane. PI indicates that the PreImmune serum was used. Western blots were probed with the anti-Mod(mdg4) serum (A) or the anti-GAF serum (B). (C) Nuclear matrix analysis. The different fractions correspond to the different steps of the nuclear matrix extraction, according to the original protocol (27). N, nuclei; DN, nuclei digested with DNase I and RNase A; NS, nuclease supernatant; TXS, supernatant after Triton X-100 treatment; SS1, first salt supernatant; SS2, second salt supernatant; DSNF-I, Drosophila subnuclear fraction I, which contains nuclear lamina, pore complexes, and the nuclear matrix. Antibodies against GAF, Mod(mdg4), lamin, and Ubx proteins were used to probe the different fractions as indicated.
In Vitro and Two-Hybrid Interactions. GST-Mod(mdg4)-67.2 (28) and His-tagged-GAF (26) were expressed in BL21(DE3) and purified on Glutathione Sepharose 4B (Amersham Pharmacia Biosciences) or nickel-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Valencia, CA), respectively. In vitro translations were performed with TNT Coupled Reticulocyte Lysate Systems (Promega) with 35S-labeled methionine and cysteine (Redivue pro-mix, Amersham Pharmacia Biosciences). To produce the Mod(mdg4)-67.2 protein, a clone in the pING14.1 vector (28) was transcribed with SP6 polymerase. GAF protein and its derivatives, produced from the pKH-GAF clone (26) or from PCR fragments obtained from this clone, were transcribed with T7 polymerase. The binding between the bacterially expressed protein (1–2 μg) retained on beads (glutathione Sepharose 4B or Ni-NTA agarose), and the in vitro translated protein was performed in HB buffer (50 mM Hepes/0.2 mM EDTA/1 mM DTT/0.5% Nonidet P-40) containing 100 mM NaCl and 5 mg/ml BSA, for 1–2 h at room temperature. The beads were washed five times in HB containing 300 mM NaCl and were loaded on an SDS/PAGE. Gels were fixed and dried before exposure. Two-hybrid assays were carried out by using yeast strain pJ694A, plasmids, and protocols obtained from Clontech. For growth assays, yeasts were plated on nonselective media lacking tryptophan and leucine. After growth, plates were replicated on selective media lacking tryptophan, leucine, histidine, and adenine, and growth was compared. Mod(mdg4)-67.2 and its derivatives were cloned in frame in the bait vector pGBT9. For details of the different subcloning steps, see Supporting Methods.
Results
GAF and Mod(mdg4) Interact in Vivo and in Vitro. We first asked whether GAF and Mod(mdg4) proteins interact in vivo by performing coimmunoprecipitations with nuclear extracts prepared from Drosophila SL-2 cells. Fig. 1 A shows that Mod(mdg4) isoforms are immunoprecipitated by an anti-Mod(mdg4) serum and coimmunoprecipitated by an anti-GAF serum, but not by an anti-polyhomeotic (PH) serum or preimmune sera. This result suggests that GAF and Mod(mdg4) can be present in the same complexes in vivo and was confirmed by the reverse experiment: the anti-Mod(mdg4) serum coimmunoprecipitates all GAF isoforms (Fig. 1B). This coimmunoprecipitation is reproducible and robust, because greater amounts of GAF are coimmunoprecipitated with Mod(mdg4) than with PH, a protein previously reported to be present in a complex with GAF in embryos (29). Nevertheless, less GAF was coimmunoprecipitated by Mod(mdg4) antibodies than in the reverse experiment. This result might be due to a different relative abundance of the two proteins in nuclear extracts. However, a quantification of absolute amounts done by comparing the intensities of the bands with fixed amounts of bacterially purified protein isoforms revealed similar levels for the two proteins (data not shown). We therefore decided to analyze their subnuclear distribution.
Recently, the Mod(mdg4)-67.2 isoform has been shown to be present in the nuclear matrix fraction, which is defined experimentally by its resistance to extraction with 2 M NaCl (10). We asked whether GAF is present in the same fraction as Mod(mdg4) by analyzing nuclear matrix preparations from 0- to 24-h Drosophila embryos. Fig. 1C shows that a large fraction of GAF resists the 2-M NaCl extraction and is present in the final fraction, similarly to the positive control lamin (10). This finding is even more pronounced for Mod(mdg4) isoforms, which are strongly concentrated in the lamin fraction. In contrast, the Ubx transcription factor is released from chromatin by this procedure. Together, these results indicate that GAF and Mod(mdg4) interact with each other and with nuclear lamin components in vivo. They might also partially explain the relatively weak coimmunoprecipitation of GAF by anti-Mod(mdg4) antibodies, because matrix-attached protein complexes containing Mod(mdg4) isoforms might be incompletely extracted under the conditions used in coimmunoprecipitation experiments.
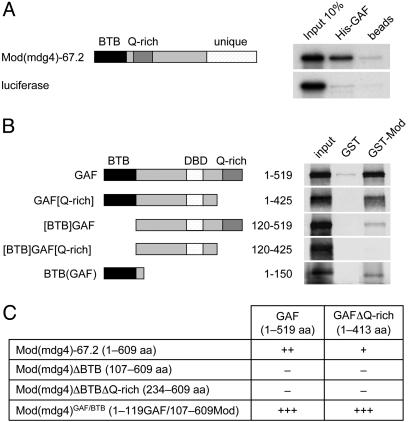
To determine whether the interaction between GAF and Mod(mdg4) is direct, we investigated their interaction in vitro. For this analysis, we used the GAF-519 isoform, which is the predominant form of GAF (30), and the Mod(mdg4)-67.2 isoform, which is the predominant form of Mod(mdg4) and which interacts with Su(Hw) and is required for the enhancer-blocking activity of the Su(Hw) insulator (28, 31). We observed that bacterially expressed GAF is able to interact directly with in vitro-translated Mod(mdg4) (Fig. 2A). To characterize the domains involved in this interaction, we expressed full-length Mod(mdg4) as a GST fusion protein and used it as bait for different in vitro-translated GAF domains (Fig. 2B). The full-length GST-Mod(mdg4) interacts strongly with GAF. This interaction is slightly reduced by removing the C-terminal glutamine-rich (Q-rich) domain from GAF and strongly reduced by removing its BTB domain. Removing both the BTB and the Q-rich domains abolishes the interaction. The GAF BTB domain alone is able to interact with Mod(mdg4). These results show that both the BTB domain and the Q-rich domain of GAF contribute to the interaction with Mod(mdg4). We confirmed this interaction in yeast two-hybrid experiments (Fig. 2C). GAF and Mod(mdg4) interact strongly in this assay, and the interaction is reduced upon removal of the Q-rich domain of GAF, confirming the GST pull-down data. Moreover, this interaction is abolished by deletion of the BTB domain of Mod(mdg4), suggesting that both the GAF as well as the Mod(mdg4) BTB domains are important for the heterologous interaction between the two proteins. Replacing the BTB domain of Mod(mdg4) with that of GAF results in a better interaction of the hybrid protein with full-length GAF or with the GAF variant without Q-rich domain. This result suggests that homodimerization of the GAF BTB domain is preferred to heterodimerization with Mod(mdg4).
Fig. 2.
GAF and Mod(mdg4) interact directly in vitro and in a two-hybrid assay. The relevant protein domains of the proteins are indicated. (A) Interaction between His-tagged GAF and in vitro-translated Mod(mdg4)-67.2 and the luciferase negative control. (B) GST pull-down by using GST-fused Mod(mdg4)-67.2 and the Mod(mdg4) BTB domain or GST alone as the negative control. The various GAF derivatives were produced by in vitro translation. (C) Yeast two-hybrid assay. –, +, ++, and +++ represent no, weak, middle, and strong interaction, respectively.
GAF-Binding Sites Promote Insulator Bypass. We and others have previously observed that the insertion of a second Su(Hw) insulator between an enhancer and a promoter restores enhancer/promoter communication (8, 9). We were interested in identifying other sequences that are capable of bypassing the Su(Hw) enhancer-blocking activity. In view of the interaction between GAF-519 and Mod(mdg4)-67.2, we asked whether GAF-binding sites might represent such a sequence.
For this analysis, we used the miniwhite gene, involved in the pigmentation of the eye, as a reporter gene, as well as its eye-specific enhancer (Ee). It had previously been shown that one copy of the Su(Hw) insulator efficiently blocks communication between Ee and miniwhite (32). We confirmed this result by using the (Ee)Y(S)W transgene, which contains the Ee enhancer separated from miniwhite (W) by the yellow marker gene (Y) and a Su(Hw) insulator (S) (Fig. 3A). The enhancer is flanked by frt sites, allowing its excision by crossing the transgenic flies with flies expressing the FLP recombinase. Similarly, the Su(Hw) insulator is flanked by loxP sites and can be excised by the action of the CRE recombinase. Eleven independent transgenic (Ee)Y(S)W lines were obtained. Their eye color varies from pale-yellow to brown. Excision of the Su(Hw) insulator increased miniwhite expression in all 11 lines [compare (Ee)Y(S)W and (Ee)YW], indicating that the insulator blocks the Ee enhancer. This block is very efficient, because the excision of Ee from the transgene containing the Su(Hw) insulator did not lighten the eye pigmentation [compare (Ee)Y(S)W and Y(S)W], whereas it did in all lines in which the insulator had already been excised [compare (Ee)YW and YW].
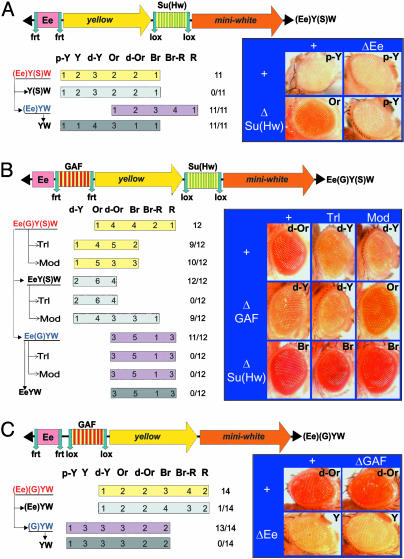
Fig. 3.
GAF-binding sites neutralize the enhancer-blocking activity of the Su(Hw) insulator. Transgenes are schematically represented above each panel. The transgenic lines are sorted according to their eye colors. Wild-type mini-white expression results in bright red eye color (R), whereas the absence of expression results in white eyes (W). Intermediate levels of pigmentation are defined by eye color ranging from pale yellow (p-Y) to yellow (Y), dark-yellow (d-Y), orange (Or), dark orange (d-Or), brown (Br), or brown-red (Br-R), reflecting increasing levels of miniwhite expression. The number of lines in each category is indicated. miniwhite expression levels were determined without excision as well as after excision of the functional elements flanked by loxP and frt sites. Analysis of miniwhite expression in a wild-type, TrlR85 heterozygous mutant and in a Mod(mdg4) homozygous mutant background (mod(mdg4)u1 and mod(mdg4)T6) is indicated (+, Trl, and Mod, respectively). The number of lines in which the eye pigmentation changes after excision of an element or in a mutant background, compared with the original lines, is indicated over the total number of lines. (A) Structure of the (Ee)Y(S)W transgene. Arrows indicate the excision of an element to produce the derivatives Y(S)W, (Ee)YW, and YW. Representative eye pictures are shown. (B) Structure of the Ee(G)Y(S)W transgene and eye colors of the Ee(G)Y(S)W lines and their derivatives. (C) Structure of the (Ee)(G)YW transgene and eye colors of the (Ee)(G)YW lines and their derivatives.
We next tested the consequence of inserting an array of GAF-binding sites between the Ee and the Su(Hw) insulator. This array was obtained by duplication of a fragment from the hsp70 promoter region that contains four GAF-binding sites that are bound by GAF in vivo (33, 34). We constructed the Ee(G)Y(S)W transgene containing the Ee enhancer separated from miniwhite by the yellow gene, which is bordered at its 5′ end by the GAF array (G) and at its 3′ end by the Su(Hw) insulator (S) (Fig. 3B). The cluster of GAF-binding sites is flanked by frt sites, and the Su(Hw) insulator is flanked by loxP sites. Twelve independent transgenic Ee(G)Y(S)W lines were obtained, and derivatives lacking either the GAF-binding sites [EeY(S)W], the Su(Hw) insulator [Ee(G)YW], or both (EeYW), were generated by crossing with flies expressing the appropriate recombinases. For each line, miniwhite expression was studied in a wild-type background as well as in TrlR85/+ and mod(mdg4)u1 or mod(mdg4)T6 mutant backgrounds. TrlR85 is a partial deletion of the Trl gene and is a putative null allele (21). mod(mdg4)u1 and mod(mdg4)T6 are two mutations that disrupt the C-terminal part of the Mod(mdg4)-67.2 isoform only (6, 35). Because the same results were obtained with either alleles in all experiments, we will refer to mod(mdg4) mutant background for simplicity. Ee(G)Y(S)W lines show an eye color ranging from orange to red, which is significantly darker than the pigmentation observed in the previous (Ee)Y(S)W lines. This result suggests that the GAF-binding sites neutralize the enhancer-blocking activity of the Su(Hw) insulator. Indeed, excision of the GAF-binding sites induces a strong reduction of miniwhite expression in all lines [compare Ee(G)Y(S)W and EeY(S)W], indicating that the excision restores the ability of the Su(Hw) insulator to block communication between the Ee enhancer and miniwhite. In contrast, yellow expression was strong in all tissues and all lines tested. In particular, no variation was observed upon excision of GAF-binding sites (data not shown). Thus, the enhancer-blocking activity, as well as the bypass activity, is seen only when the Su(Hw) insulator is placed between enhancer and promoter.
In a mod(mdg4) mutant background, expression of miniwhite increased in 9 of 12 of the EeY(S)W lines, demonstrating that the Su(Hw) insulator blocks communication between the Ee enhancer and the miniwhite promoter through the action of the Mod(mdg4) protein. Interestingly, placing the Ee(G)Y(S)W lines in a mod(mdg4)or TrlR85/+ mutant background resulted in a decrease in eye pigmentation in the majority of the lines (10/12 and 9/12, respectively). This result indicates that both GAF and Mod(mdg4) proteins are involved in the bypass of the Su(Hw) insulator. Excision of the insulator from the construct renders the lines insensitive to Trl and mod(mdg4) mutations as well as to GAF-binding site excision [Fig. 3B, Ee(G)W and EeYW constructs]. It should be noted here that excision of the insulator induced full miniwhite activation [compare the difference between EeY(S)W and EeYW, inducing full loss of enhancer blocking], whereas the Ee is still partially blocked upon mutation of mod(mdg4) [compare EeY(S)W in wild-type or mod(mdg4) mutant backgrounds]. This result suggests that the absence of the Mod(mdg4)-67.2 protein isoform might not completely abolish the function of the Su(Hw) insulator as such, although it is essential for the interaction with GAF sites that is responsible for insulator bypass.
One might postulate that the presence of GAF-binding sites stimulates miniwhite transcription in a manner that is not totally blocked by the Su(Hw) insulator. To test this possibility, we constructed the transgene (Ee)(G)YW, which contains the Ee enhancer flanked by frt sites, the cluster of GAF-binding sites flanked by loxP sites, and the yellow and miniwhite genes (Fig. 3C). We analyzed the eye color of 14 independent insertions. We observed that the excision of the GAF-binding sites did not significantly reduce the eye pigmentation of the transgenic flies, regardless of whether or not the Ee enhancer is present in the construct [Fig. 3C, compare (Ee)(G)YW with (Ee)YW and (G)YW with YW]. This result demonstrates that the mere presence of GAF-binding sites in the transgene does not stimulate miniwhite expression (Fig. 3 B and C). Moreover, GAF-binding sites do not have enhancer-blocking activity per se, because their excision does not induce an increase in miniwhite expression. Taken together, our results show that the interposition of GAF-binding sites between the Ee enhancer and the Su(Hw) insulator allows the insulator's enhancer-blocking activity to be bypassed, restoring communication between Ee and miniwhite.
The Mcp Element Allows Bypass of the Su(Hw) Insulator. We next tested whether a single-copy natural element can neutralize the enhancer-blocking activity of the Su(Hw) insulator. We used a 690-bp Mcp sequence from the BX-C. Genetic analyses indicate that this element behaves as a boundary separating the iab-4 and iab-5 regulatory domains in the BX-C (36). This 690-bp sequence contains the MCP138 element, which is sufficient to maintain silencing of a transgene in embryos and larvae (20). The MCP138 element contains two GAF-binding sites, and both the GAF protein and the GAF-binding sites are required for its silencing activity (20).
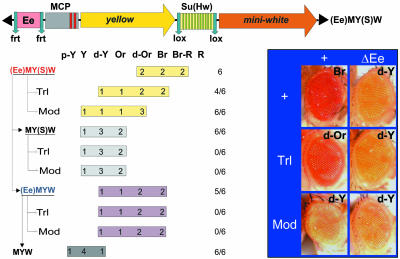
We inserted the 690-bp Mcp element between the Ee enhancer and the yellow gene in (Ee)Y(S)W to produce the (Ee)MY(S)W transgene (Fig. 4). In all six transgenic (Ee)MY(S)W lines, the pigmentation observed in bristle, wing, and body is variegated and independent on excision of functional elements within the construct (data not shown). This finding suggests that Mcp represses the yellow promoter. At the same time, expression of miniwhite is significantly stronger than in the (Ee)Y(S)W lines, suggesting that the presence of Mcp allows the Su(Hw) insulator to be bypassed by the eye enhancer. The eye enhancer activates the miniwhite promoter in the (Ee)MY(S)W transgene, as evidenced by the strong reduction in eye pigmentation seen upon excision of the Ee element [Fig. 4, compare (Ee)MY(S)W and MY(S)W]. Thus, Mcp partially neutralizes the enhancer-blocking activity of the Su(Hw) insulator. Similar results were obtained when the Mcp element was placed in a reverse orientation (data not shown). As observed with the Ee(G)Y(S)W construct, placing the (Ee)MY(S)W transgenes in a mod(mdg4) mutant background decreases miniwhite expression in all of the lines. A similar, although weaker, effect was observed in the TrlR85 mutant background in 4/6 lines. These results indicate that both Mod(mdg4) and GAF proteins are involved in the neutralization of the Su(Hw) insulator by Mcp.
Fig. 4.
GAF is involved in partial neutralization of the Su(Hw) enhancer-blocking activity by Mcp. The structure of the Ee(M)Y(S)W transgene is depicted. The Mcp element contains two GAF-binding sites (indicated as red bars). miniwhite expression was analyzed as described in Fig. 3.
We observed that excision of the insulator in the (Ee)MY(S)W derivatives decreases the eye pigmentation of the flies, both when the enhancer is present [(Ee)MYW lines] and when it has been excised (MYW lines), indicating that Mcp silences miniwhite in the absence of the Su(Hw) insulator. In other words, the silencing activity of Mcp is blocked when the Su(Hw) insulator is present in the transgene. These results suggest that the interaction between Mcp and the Su(Hw) insulator allows the enhancer-blocking activity of the insulator to be bypassed in a manner compatible with blocking of the silencing activity of Mcp. The lack of insulator bypass by the Mcp silencer might depend on the fact that the silencer overlaps the chromatin boundary in Mcp. It is conceivable that the interaction of GAF with Mod(mdg4) might trap the Mcp silencer close to the insulator and thus prevent silencing to cross the Su(Hw) barrier.
Discussion
In this study, we show that inserting either a cluster of GAF-binding sites or the Mcp boundary between an enhancer and the Su(Hw) insulator neutralizes the enhancer-blocking activity of the insulator and restores communication between the upstream enhancer and the downstream promoter. These results are reminiscent of what is observed when two copies of the Su(Hw) insulator are placed in tandem between an enhancer and a reporter gene: both insulators are bypassed. We propose that the GAF-binding sites and the Mcp element can pair with the Su(Hw) insulator and neutralize its enhancer-blocking activity. Given the direct interaction between GAF and Mod(mdg4) proteins reported here, this pairing could be mediated by interactions between the GAF protein bound to the GAF-binding sites and the Mod(mdg4) protein bound to the insulator by means of its interaction with the Su(Hw) protein. Such pairing would loop out the sequences between the GAF-binding sites and the insulator, bringing the upstream enhancer and downstream promoter into close proximity to each other.
Several BTB domain-containing proteins have been implicated in enhancer/promoter communication and loop formation. The GAF protein itself can form oligomers through its BTB domain (37, 38) and can link separate DNA fragments containing GAGA sites in vitro (39). These interactions have been proposed as being involved in enhancer/promoter communication by bringing these elements into close proximity to each other. Our results provide evidence that interactions between different BTB-containing proteins can promote loop formation, and that such interactions can be involved in the regulation of insulator function and/or the regulation of enhancer/promoter communication.
Our results also suggest that a GAF- and Mod(mdg4)-dependent interaction between the Mcp boundary and the Su(Hw) insulator leads to the bypass of the Su(Hw) insulator. Recently, two independent studies (40, 41) examined possible interactions between homologous and heterologous Drosophila insulator pairs in transgenic assays. Except for the pair of Su(Hw) insulators, none of the combinations tested led to insulator bypass. However, there is evidence for insulator bypass in at least two cases in Drosophila. A first set of data strongly suggests that a gypsy insulator inserted within an intron of the yellow gene can be bypassed by downstream enhancers, due to the presence of a second endogenous insulator located between the 3′ end of the gene and the enhancers (42). This endogenous insulator was shown to be capable of enhancer blocking and is bound by the Su(Hw) protein in vivo. Its insulator function, however, was shown to be only moderately dependent on Su(Hw), suggesting that other proteins may participate in its enhancer-blocking activity and in pairing with the gypsy insulator.
A second case of insulator bypass has been described in the BX-C. At this locus, Abd-B expression is regulated by a series of regulatory regions, called iab-5, iab-6, iab-7, and iab-8,9, which are located up to 50 kb downstream of the Abd-B transcription unit and which are believed to be separated from each other by boundaries (reviewed in ref. 11). The Fab-7 and Fab-8 boundaries flank the iab-7 domain, which is involved in Abd-B activation in parasegment 12. In this parasegment, iab-7 has to overcome the Fab-8 boundary to activate Abd-B. Zhou and Levine (43) identified a sequence, called the promoter-targeting sequence (PTS), that is located near the Fab-8 boundary within the iab-7 domain and that promotes bypass of Fab-8. In transgenesis, the PTS is able to neutralize the enhancer-blocking activity of both the Fab-8 boundary and the Su(Hw) insulator when it is inserted between the enhancer and the promoter (43, 44). In addition, mutations in the PTS within the BX-C produce homeotic transformations that reveal a lack of Abd-B activation by iab-7, suggesting that the enhancer is blocked by the Fab-8 boundary in the absence of PTS.
Our data are compatible with GAF sites or the Mcp element behaving in a manner similar to the PTS element of the BX-C, although the PTS is molecularly poorly characterized and thus it is not possible to conclude on how it acts. This work provides direct evidence for a bypass of the Su(Hw) insulator that is induced by heterologous Drosophila DNA sequences and identifies the protein pair GAF/Mod(mdg4) as responsible for the bypass. Neither a cluster of GAF-binding sites nor the Mcp boundary have been previously tested in combination with the Su(Hw) insulator in transgenic assays. No Su(Hw) insulator bypass was found, however, with two other elements that also bind GAF, the SF1 and the Fab-7 boundaries (41). This result may reflect an interference with the GAF/Mod(mdg4) interaction by other proteins bound to the SF1 or Fab-7 boundaries. For instance, although our data demonstrate a role for GAF in insulator neutralization, they do not rule out the possible involvement of other GAGA-motif-binding proteins, such as Pipsqueak (Psq) (45, 46), which might play a role at a subset of the GAF-binding motifs. Moreover, both the SF1 and the Fab-7 insulators are complex elements that may be regulated by specific factors. This is probably the reason why two copies of each of these two insulators are also not bypassed, and it will be interesting to isolate DNA sequences and proteins that promote or counteract the bypass phenomenon. Finally, it has been shown that Su(Hw) insulator activity depends both on insulator strength and on the strength of the elements it counteracts (47, 48). Such a precise balance may be necessary to achieve bypass between two heterologous insulators. Therefore, the bypass of insulators is not a universal phenomenon but is instead involved in the regulation of a subset of genomic loci, depending upon the intrinsic features of the different regulatory DNA sequences and associated factors involved. In this way, at least some characterized insulators may not behave as immutable barriers, but rather participate in the dynamic regulation of gene expression during development and differentiation.
Supplementary Material
Acknowledgments
We thank Donna Arndt-Jovin, Rainer Dorn, Yacine Graba, and Florence Maschat for their generous gift of antibodies. We thank Dale Dorsett (Saint Louis University School of Medicine, St. Louis, MO) for expression clones of Mod(mdg4) derivatives, and Peter Becker (University of Munich, Munich) and Pamela Geyer for DNA clones. F.J. was supported by a postdoctoral fellowship of the Fondation Schlumberger pour L'Éducation et la Recherche (FSER). G.C. was supported by grants from the Centre National de la Recherche Scientifique, the Human Frontier Science Program Organization, and the Association pour la Recherche sur le Cancer. P.G. was supported by the Molecular and Cellular Biology program of the Russian Academy of Science, by the Volkswagen-Stiftung Foundation (Federal Republic of Germany), and by an International Research Scholar award from the Howard Hughes Medical Institute.
Author contributions: G.C. and P.G. designed research; L.M., F.J., N.G., and A.M. performed research; L.M., F.J., G.C., and P.G. analyzed data; and F.J. and G.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PTS, promoter-targeting sequence; BX-C, Bithorax complex; GAF, GAGA factor; Trl, Trithorax-like; Ee, eye enhancer; Y, yellow marker gene; S, Su(Hw) insulator; W, miniwhite; G, GAF array; BTB, broad complex, tramtrack, and brick à brack.
References
- 1.West, A. G., Gaszner, M. & Felsenfeld, G. (2002) Genes Dev. 16, 271–288. [DOI] [PubMed] [Google Scholar]
- 2.Blanton, J., Gaszner, M. & Schedl, P. (2003) Genes Dev. 17, 664–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishii, K., Arib, G., Lin, C., Van Houwe, G. & Laemmli, U. K. (2002) Cell 109, 551–562. [DOI] [PubMed] [Google Scholar]
- 4.Yusufzai, T. M., Tagami, H., Nakatani, Y. & Felsenfeld, G. (2004) Mol. Cell 13, 291–298. [DOI] [PubMed] [Google Scholar]
- 5.Spana, C., Harrison, D. A. & Corces, V. G. (1988) Genes Dev. 2, 1414–1423. [DOI] [PubMed] [Google Scholar]
- 6.Gerasimova, T. I., Gdula, D. A., Gerasimov, D. V., Simonova, O. & Corces, V. G. (1995) Cell 82, 587–597. [DOI] [PubMed] [Google Scholar]
- 7.Gerasimova, T. I. & Corces, V. G. (1998) Cell 92, 511–521. [DOI] [PubMed] [Google Scholar]
- 8.Muravyova, E., Golovnin, A., Gracheva, E., Parshikov, A., Belenkaya, T., Pirrotta, V. & Georgiev, P. (2001) Science 291, 495–498. [DOI] [PubMed] [Google Scholar]
- 9.Cai, H. N. & Shen, P. (2001) Science 291, 493–495. [DOI] [PubMed] [Google Scholar]
- 10.Byrd, K. & Corces, V. G. (2003) J. Cell Biol. 162, 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mihaly, J., Hogga, I., Barges, S., Galloni, M., Mishra, R. K., Hagstrom, K., Muller, M., Schedl, P., Sipos, L., Gausz, J., et al. (1998) Cell Mol. Life Sci. 54, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belozerov, V. E., Majumder, P., Shen, P. & Cai, H. N. (2003) EMBO J. 22, 3113–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohtsuki, S. & Levine, M. (1998) Genes Dev. 12, 3325–3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Donnell, K. H., Chen, C. T. & Wensink, P. C. (1994) Mol. Cell. Biol. 14, 6398–6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bantignies, F., Grimaud, C., Lavrov, S., Gabut, M. & Cavalli, G. (2003) Genes Dev. 17, 2406–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muller, M., Hagstrom, K., Gyurkovics, H., Pirrotta, V. & Schedl, P. (1999) Genetics 153, 1333–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strutt, H., Cavalli, G. & Paro, R. (1997) EMBO J. 16, 3621–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Donnell, K. H. & Wensink, P. C. (1994) Nucleic Acids Res. 22, 4712–4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mishra, R. K., Mihaly, J., Barges, S., Spierer, A., Karch, F., Hagstrom, K., Schweinsberg, S. E. & Schedl, P. (2001) Mol. Cell. Biol. 21, 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Busturia, A., Lloyd, A., Bejarano, F., Zavortink, M., Xin, H. & Sakonju, S. (2001) Development (Cambridge, U.K.) 128, 2163–2173. [DOI] [PubMed] [Google Scholar]
- 21.Farkas, G., Gausz, J., Galloni, M., Reuter, G., Gyurkovics, H. & Karch, F. (1994) Nature 371, 806–808. [DOI] [PubMed] [Google Scholar]
- 22.Lehmann, M. (2004) Trends Genet. 20, 15–22. [DOI] [PubMed] [Google Scholar]
- 23.Wilkins, R. C. & Lis, J. T. (1997) Nucleic Acids Res. 25, 3963–3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benyajati, C., Mueller, L., Xu, N., Pappano, M., Gao, J., Mosammaparast, M., Conklin, D., Granok, H., Craig, C. & Elgin, S. (1997) Nucleic Acids Res. 25, 3345–3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albagli, O., Dhordain, P., Deweindt, C., Lecocq, G. & Leprince, D. (1995) Cell Growth Differ. 6, 1193–1198. [PubMed] [Google Scholar]
- 26.Agianian, B., Leonard, K., Bonte, E., Van der Zandt, H., Becker, P. B. & Tucker, P. A. (1999) J. Mol. Biol. 285, 527–544. [DOI] [PubMed] [Google Scholar]
- 27.Fisher, P. A. & Blobel, G. (1983) Methods Enzymol. 96, 589–596. [DOI] [PubMed] [Google Scholar]
- 28.Gause, M., Morcillo, P. & Dorsett, D. (2001) Mol. Cell. Biol. 21, 4807–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poux, S., Melfi, R. & Pirrotta, V. (2001) Genes Dev. 15, 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soeller, W. C., Oh, C. E. & Kornberg, T. B. (1993) Mol. Cell. Biol. 13, 7961–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh, D., Gerasimova, T. I. & Corces, V. G. (2001) EMBO J. 20, 2518–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roseman, R. R., Pirrotta, V. & Geyer, P. K. (1993) EMBO J. 12, 435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien, T., Wilkins, R. C., Giardina, C. & Lis, J. T. (1995) Genes Dev. 9, 1098–1110. [DOI] [PubMed] [Google Scholar]
- 34.Tsukiyama, T., Becker, P. B. & Wu, C. (1994) Nature 367, 525–532. [DOI] [PubMed] [Google Scholar]
- 35.Mongelard, F., Labrador, M., Baxter, E. M., Gerasimova, T. I. & Corces, V. G. (2002) Genetics 160, 1481–1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karch, F., Galloni, M., Sipos, L., Gausz, J., Gyurkovics, H. & Schedl, P. (1994) Nucleic Acids Res. 22, 3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Espinas, M. L., Jimenez-Garcia, E., Vaquero, A., Canudas, S., Bernues, J. & Azorin, F. (1999) J. Biol. Chem. 274, 16461–16469. [DOI] [PubMed] [Google Scholar]
- 38.Katsani, K. R., Hajibagheri, M. A. & Verrijzer, C. P. (1999) EMBO J. 18, 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmoudi, T., Katsani, K. R. & Verrijzer, C. P. (2002) EMBO J. 21, 1775–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhn, E. J., Viering, M. M., Rhodes, K. M. & Geyer, P. K. (2003) EMBO J. 22, 2463–2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Majumder, P. & Cai, H. N. (2003) Proc. Natl. Acad. Sci. USA 100, 5223–5228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parnell, T. J., Viering, M. M., Skjesol, A., Helou, C., Kuhn, E. J. & Geyer, P. K. (2003) Proc. Natl. Acad. Sci. USA 100, 13436–13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou, J. & Levine, M. (1999) Cell 99, 567–575. [DOI] [PubMed] [Google Scholar]
- 44.Lin, Q., Wu, D. & Zhou, J. (2003) Development (Cambridge, U.K.) 130, 519–526. [DOI] [PubMed] [Google Scholar]
- 45.Huang, D. H., Chang, Y. L., Yang, C. C., Pan, I. C. & King, B. (2002) Mol. Cell. Biol. 22, 6261–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwendemann, A. & Lehmann, M. (2002) Proc. Natl. Acad. Sci. USA 99, 12883–12888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scott, K. C., Taubman, A. D. & Geyer, P. K. (1999) Genetics 153, 787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai, H. N., Zhang, Z., Adams, J. R. & Shen, P. (2001) Development (Cambridge, U.K.) 128, 4339–4347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.