Abstract
Botulinum neurotoxins (BoNTs) act as zinc-dependent endopeptidases that cleave proteins required for neurotransmitter release. To detect toxin activity, fragments of the toxin substrate proteins, synaptobrevin (Syb) or synaptosome-associated protein of 25 kDa (SNAP-25), were used to link cyan fluorescent protein (CFP) to yellow fluorescent protein (YFP). Cleavage of these fusion proteins by BoNTs abolished fluorescence resonance energy transfer between the CFP and YFP, providing a sensitive means to detect toxin activity in real-time in vitro. Furthermore, using full-length SNAP-25 and Syb as the linkers, we report two fluorescent biosensors that can detect toxin activity within living cells. Cleavage of the SNAP-25 fusion protein abolished fluorescence resonance energy transfer between CFP and YFP, and cleavage of Syb resulted in spatial redistribution of YFP fluorescence in cells. This approach provides a means to carry out cell-based screening of toxin inhibitors and to study toxin activity in situ. By using these biosensors, we found that the subcellular localizations of SNAP-25 and Syb are critical for efficient cleavage by BoNT/A and B, respectively.
Botulinum neurotoxins (BoNT) are the most lethal substances known and thus pose a bioterrorism threat (1). BoNTs have also emerged as important medical tools to treat muscle dysfunction, inflammation, and chronic pain (2–5). There are seven related toxins (BoNT/A–G). Each toxin is composed of a heavy chain, which mediates the entry of toxin into neurons, and a light chain, which functions as a zinc-dependent endoprotease inside cells (6). BoNT/A, -E, and -C cleave the peripheral plasma membrane protein SNAP-25 (synaptosome-associated protein of 25 kDa); BoNT/C also cleaves the integral plasma membrane protein syntaxin; and BoNT/B, -D, -F, and -G cleave the secretory vesicle membrane protein synaptobrevin (Syb) (see ref. 6 for a review). Cleavage of these substrates inhibits neuronal exocytosis (6).
Characterizing BoNT activity traditionally relies on functional assays to monitor the effect of the toxin (e.g., toxicity in mice, blockade of exocytosis) or biochemical assays (e.g., immunoblot/immunoassays, chromatography) to detect cleaved substrate molecules (7). Several methods, based on intramolecular quenching of fluorigenic amino acid derivatives, have been recently explored and used to screen toxin inhibitors in vitro (8–10). However, this approach is limited to a subset of toxins and cannot be used to monitor the action of the toxins in live cells. At present, there are no available methods that are amenable to cell-based high-throughput screening of BoNT inhibitors.
Here we report a recently developed method based on monitoring fluorescence resonance energy transfer (FRET) changes between cyan (CFP) and yellow (YFP) fluorescent protein pairs, to detect toxin activity in real time in vitro. Furthermore, using CFP/YFP pairs, we are able to detect BoNT activity in living cells, which enabled us to study toxin substrate cleavage in situ.
Materials and Methods
cDNA Constructs and Recombinant Protein Purification. YFP cDNA (Clontech) was inserted into pECFP-C1 vector (Clontech) by using EcoRI/BamHI, resulting in the pECFP-YFP construct. cDNA encoding amino acids 33–94 and 33–116 of rat Syb II were inserted into the pECFP-YFP vector by using XhoI/EcoRI sites, resulting in the CFP-Syb(33–94)-YFP and CFP-Syb(33–116)-YFP constructs. The CFP-SNAP-25-YFP sensor was built by using the same method, but using residues 141–206 or full-length rat SNAP-25B as the linker. Substitution of four Cys residues of SNAP-25 to Ala was accomplished by site-directed mutagenesis using PCR, and the fragment was then inserted between CFP and YFP as described above. SNAP-25(83–120)-CFP-SNAP-25(141–206)-YFP was built by first inserting a cDNA fragment that encodes residues 83–120 of SNAP-25 into the XhoI/EcoRI sites of pEYFP-N1, and then subcloning CFP-SNAP-25(141–206) into downstream sites by using EcoRI/BamHI. YFP-Syb(FL)-CFP was built by first inserting a full-length Syb II cDNA into pECFP-C1 by using EcoRI/BamHI sites, and then inserting a full-length YFP cDNA upstream at XhoI/EcoRI sites.
To generate purified recombinant CFP-Syb(33–94)-YFP and CFP-SNAP-25(141–206)-YFP, these fusion proteins were also expressed in Escherichia coli. To this end, we subcloned the sensor constructs from pECFP-YFP to pTrchis (Invitrogen) by using NheI/BamHI sites. All cDNA fragments were generated by PCR, and constructs were confirmed by sequencing.
His6-tagged sensor proteins were purified as described (11). Purified proteins were dialyzed against Hepes buffer (50 mM Hepes, pH 7.1) overnight.
In Vitro Fluorescence Measurements and Toxin Detection. In general, we used 300 nM sensor protein in a total volume of 600 μl of Hepes buffer that also contained 2 mM DTT and 10 μM ZnCl2. The emission spectra from 450 to 550 nm were collected by using a PTI QM-1 spectrofluorometer. The excitation wavelength was 434 nm. For experiments using a plate reader, 300 nM sensor protein or a mixture of unfused CFP and YFP were prepared in a volume of 100 μl per well in a 96-well plate. For each data point, samples were excited twice at 434 nm, and the fluorescence intensity of the YFP emission channel (527 nm) and CFP channel (470 nm) were collected sequentially for each excitation. Samples were either scanned at 30-sec intervals for 90 min (Fig. 1d), or were scanned once at the end of the incubation period (4 or 16 h, Fig. 1e) using a Spectra Max Gemini microplate reader (Molecular Devices). The amplitude of the FRET signal in vitro was affected by many factors, such as buffer composition, salt, Zn2+, and reducing agents (data not shown); therefore, the in vitro experiments hereafter were all carried out under the same conditions (50 mM Hepes/2 mM DTT/10 μM ZnCl2, pH 7.1). DTT (2 mM) and Zn2+ (10 μM) were added to optimize BoNT protease activity (12).
Fig. 1.
CFP/YFP-based sensors for detecting BoNT protease activity in vitro. (a) Schematic representation of the BoNT sensor constructs. CFP and YFP were connected via a fragment of synaptobrevin (amino acids 33–94, Upper), or SNAP-25 (amino acids 141–206, Lower), respectively. The cleavage sites for each BoNT are indicated. (b) Recombinant toxin sensor CFP-SNAP-25(141–206)-YFP yielded a FRET signal in vitro. His6-tagged CFP-SNAP-25(141–206)-YFP was selectively excited at 434 nm, which is optimal for CFP. The emission spectra are shown. The left arrow indicates the CFP emission peak; the right arrow indicates the YFP emission peak that is the result of FRET from the CFP donor. The emission spectra of recombinant his6-tagged CFP and YFP alone, as well as the mixture of these two proteins (1:1), are also shown. These control spectra indicate that YFP yields a small emission signal when directly excited at 434 nm, and that FRET does not occur unless CFP and YFP are linked together. (c) In vitro cleavage of toxin sensors monitored optically and via immunoblot analysis. (Upper) BoNT/A was mixed with CFP-SNAP-25(141–206)-YFP, and the emission spectra were collected at the indicated times after adding toxin. Cleavage of the sensor resulted in decreased FRET and a concurrent increase in the CFP fluorescence. (Lower) Samples (30 μl) were taken from the cuvette after each emission scan (in the experiment shown in Upper) and subjected to SDS/PAGE gel and immunoblot analysis. Cleavage of CFP-SNAP-25(141–206)-YFP was detected by using an anti-his6 antibody that recognizes the his6 tag at the N terminus of the sensor protein. (d) Sensor cleavage kinetics monitored by using a plate reader. CFP-SNAP-25(141–206)-YFP was cleaved with BoNT/A (100 pM) as detailed in Materials and Methods. CFP-SNAP-25(141–206)-YFP alone, as well as the CFP/YFP mixture with added BoNT/A, were analyzed in parallel as controls. (e) Picomolar sensitivity of the toxin sensors. (Left) Experiments were carried out as in d after measuring the emission ratio after 4 h; these values are plotted against the log of the toxin concentration. (Right) The EC50 values for BoNT/A, -E, -B, and -F, when incubated with toxin sensors for 4 or 16 h, were determined as described for Left. Each value represents the mean of three independent determinations.
To detect toxin activity in vitro, BoNT/A, -B, -E, or -F holotoxin was first reduced by incubation with 2 mM DTT (and, to maintain toxin activity, 10 μM ZnCl2) for 30 min at 37°C (12).
Live Cell Imaging and FRET Analysis. PC12 cells were transfected with cDNA constructs by electroporation (Bio-Rad). The fluorescence images were collected by using a Nikon TE-300 microscope with a 1.4 numerical aperture, ×100 oil-immersion objective. CFP/YFP FRET in live cells was quantified by using a three-filter set method (13, 14). In brief, three consecutive images were acquired for each cell, through three different filter sets: CFP filter (excitation, 436/10 nm; emission, 470/30 nm), FRET filter (excitation, 436/10 nm; emission, 535/30 nm), and YFP filter (excitation, 500/20 nm, emission, 535/30 nm). A JP4 beamsplitter (Set ID 86000, Chroma Technology, Rockingham, VT) was used. All images were acquired with exactly the same settings (4 × 4 Binning, 200-msec exposure time). To avoid the concentration-dependent FRET signal that can arise from high expression levels of fluorescent proteins, cells with CFP and YFP intensities less than or equal to half of the maximal 12-bit scale were counted in our experiments (15, 16). The background (from areas that did not contain cells) was subtracted from each raw image. PC12 cells transfected with CFP or YFP alone were first tested to obtain the cross-talk/bleed-through values for these filter sets. The FRET filter channel exhibits ≈56–64% bleed-through for CFP, and ≈24% for YFP. There is virtually no cross-talk/bleed-through for YFP using the CFP filter, or for CFP using the YFP filter, which simplified the FRET calculations. For cells expressing toxin sensors, the “corrected FRET” value was calculated by using the following equation: corrected FRET = FRET – (CFP × 0.60) – (YFP × 0.24), where FRET, CFP, and YFP correspond to the fluorescence intensities of images acquired through FRET, CFP, and YFP filter sets, respectively. The average fraction of cross-talk/bleed-through coming from CFP and YFP fluorescence are 0.6 and 0.24, respectively, when acquiring images through the FRET filter set. Because toxin cleavage of the CFP-SNAP25FL-YFP sensor resulted in dissociation of the YFP-tagged fragment from the membrane, which was degraded in the cytosol (Fig. 2 c and f), the FRET ratio used in our data analysis was calculated by normalizing the “corrected FRET” value to only the CFP fluorescence intensity (corrected FRET/CFP). We note that the CFP intensity in these calculations was an underestimate because of donor quenching if FRET occurred. However, it has been reported that the decrease in CFP fluorescence due to donor quenching is only ≈5–10% (13, 17). All images and calculations were performed by using metamorph software (Universal Imaging, Downington, PA).
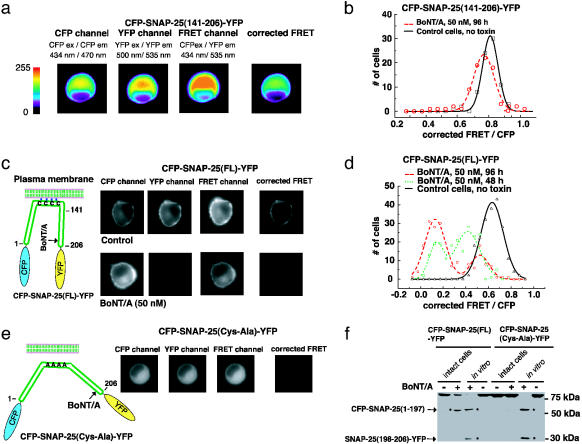
Fig. 2.
Monitoring BoNT/A activity in living cells. (a) Measuring toxin sensor FRET in living cells. CFP-SNAP-25(141–206)-YFP was expressed in wild-type PC12 cells; this protein has a uniform cytosolic distribution. Three images using different filter set (CFP, FRET, and YFP) were taken for each cell sequentially, using exactly the same settings. Images were color coded to reflect the fluorescence intensity in arbitrary units as indicated in the key. The corrected FRET value was calculated by subtracting the cross-talk from both CFP and YFP from the signals collected by using the FRET filter set, as detailed in the Materials and Methods. (b) PC12 cells that express CFP-SNAP-25(141–206)-YFP were used to detect BoNT/A activity. BoNT/A holotoxin was added to the culture medium, and 80 cells were analyzed after 96 h. The corrected FRET signal was normalized to the CFP fluorescence signal and plotted as a histogram with the indicated bins. Control cells, not treated with toxins, were analyzed in parallel. (c) A sensitive toxin biosensor was created by using full-length SNAP-25. (Left) An alternative toxin sensor was built by linking CFP and YFP through full-length SNAP-25 (amino acids 1–206); a schematic representation is shown. Full-length SNAP-25 is anchored to the plasma membrane via the palmitoylation of four cysteine residues. (Right) The CFP-SNAP-25(FL)-YFP sensor was expressed in PC12 cells. BoNT/A holotoxin was added to the culture medium, and the FRET signals of 200 cells were analyzed after 48 and 96 h as described in a. Control cells were not treated with toxins and were analyzed in parallel. Images of representative cells are shown. This sensor yielded significant FRET (upper “corrected FRET”), which was abolished after cells were treated with BoNT/A (96 h, lower “corrected FRET”). Note that one of the cleavage products, the C terminus of SNAP-25 tagged with YFP, was rapidly degraded after toxin cleavage. Thus, the fluorescence signal of YFP was significantly decreased in toxin-treated cells (lower “YFP”). (d) The FRET ratios of cells analyzed in c were plotted as a histogram with indicated bins, as described in b. (e) PC12 cells were transfected with CFP-SNAP-25(Cys-Ala)-YFP (full length SNAP-25 with Cys 85,88,90,92 Ala mutations). This protein was diffusely distributed throughout the cytosol and lacked the strong FRET signal observed for CFP-SNAP-25(FL)-YFP (“corrected FRET” frame, Fig. 5c). (f) PC12 cells were transfected with CFP-SNAP-25(FL)-YFP and CFP-SNAP-25(Cys-Ala)-YFP. Cells were incubated with (+, intact cells) or without (–, intact cells) BoNT/A (50 nM, 72 h), and then harvested and analyzed. In parallel, half of the cell lysates from samples that had not been exposed to BoNT/A were also subsequently incubated with (+, in vitro) or without (–, in vitro) reduced BoNT/A (200 nM) in vitro (30 min, 37°C); these samples served as controls to show the cleavage products that accumulate after the action of the toxin (two cleavage products are indicated by arrows). The same amount of each sample (30-μg cell lysate) was subjected to SDS/PAGE and immunoblot analysis using an anti-GFP antibody, which recognizes both CFP and YFP. Whereas CFP-SNAP-25(FL)-YFP underwent significant cleavage in intact cells, there was no detectable cleavage of CFP-SNAP-25(Cys-Ala)-YFP, indicating that membrane anchoring of SNAP-25 is important for efficient cleavage by BoNT/A in cells. Note that only one cleavage product [CFP-SNAP-25(1–197)] was detected in toxin-treated cells, indicating that the other cleavage product [SNAP-25(198–206)-YFP] was rapidly degraded in cells.
For experiments involving toxin treatment, indicated holotoxins were added to the cell culture media and cells were then analyzed as described above. Control cells were transfected with toxin sensors but not treated with toxins, and they were analyzed in an identical manner.
Immunoblot Analysis. Wild-type PC12 cells or a PC12 cell line that stably expresses synaptotagmin II (18) were transfected with toxin sensor constructs; BoNT was added to the culture medium 24 h after transfection, and cells were incubated for 72 h (BoNT/A) or 48 h (BoNT/B). Cells were then harvested, and cell lysates were subjected to immunoblot analysis as described (18). Control cells were transfected with the same cDNA constructs and assayed in parallel except that they were not treated with toxins. Half of the control cell lysates were treated with toxins in vitro (200 nM BoNT/A or -B, 30 min at 37°C), and subjected to immunoblot analysis. A GFP polyclonal antibody (SC-8334, Santa Cruz Biotechnology) was used to detect toxin sensors and the cleavage products in cell lysates. An anti-his6 antibody (Qiagen, Valencia, CA) was used to assay for cleavage of recombinant sensors (Fig. 1c).
Results
Monitoring BoNT Activity in Vitro By Using FRET-Based Reporters. To build sensors that report toxin activity in vitro, we linked CFP and YFP together by using either a Syb or SNAP-25 fragment that can be cleaved by the appropriate BoNTs (Fig. 1a). If the CFP and YFP are close enough to each other, excitation of the CFP moiety will result in the sensitized emission from the YFP moiety as a consequence of FRET (16, 19). Cleavage of the linker sequence between CFP and YFP separates them and abolishes FRET.
BoNTs are unique zinc-dependent endopeptidases that require relatively long protein fragments as substrates (see ref. 6 for a review). Previous studies indicate that, in addition to toxin cleavage sites, a highly conserved 10-residue sequence on the N-terminal side of the cleavage site, termed the “SNARE motif,” is critical for toxin-substrate recognition and efficient cleavage (20). Moreover, residues on the C-terminal side of the toxin cleavage site also affect cleavage efficiency (21–25). Thus, we used residues 33–94 of Syb and 141–206 of SNAP-25 as the linkers because these fragments are efficiently cleaved by most BoNTs while still bringing CFP and YFP close enough to result in FRET (21–24). These toxin sensors are denoted CFP-Syb(33–94)-YFP and CFP-SNAP-25(141–206)-YFP (Fig. 1a). When CFP was selectively excited at 434 nm, both recombinant sensor proteins exhibited a YFP fluorescence peak at 527 nm because of FRET (Fig. 1b and Fig. 4a, which is published as supporting information on the PNAS web site).
We next examined whether these toxin sensors can be cleaved by BoNTs in vitro. CFP-SNAP-25(141–206)-YFP was mixed with 10 nM BoNT/A, and the emission spectra were collected at the indicated time points (Fig. 1c Upper). Incubation of the toxin sensor with BoNT/A resulted in a decrease in the YFP emission and a concurrent increase in the CFP emission. This loss of FRET over time reflects the cleavage of CFP-SNAP-25(141–206)-YFP by BoNT/A, which was confirmed by immunoblot analysis of the same samples (Fig. 1c Lower). Similar results were obtained by treating this reporter with BoNT/E (data not shown).
We performed similar experiments to assay for cleavage of CFP-Syb(33–94)-YFP by BoNT/B (Fig. 4b) and BoNT/F (data not shown). For each toxin, we observed the same kinetics of sensor cleavage by using both the optical readout and the immunoblot assay. Cleavage of the sensor proteins was specific, because mixing BoNT/B and -F with CFP-SNAP-25(141–206)-YFP or mixing BoNT/A and -E with CFP-Syb(33–94)-YFP did not change the emission spectra (data not shown).
These two toxin sensors can be readily used for large-scale screening of toxin inhibitors in vitro. As evidenced in Fig. 1d, cleavage of toxin sensors can be monitored by using a plate reader in real time in a small volume (300 nM sensor, 100 pM BoNT/A, in 100-μl well volume in a 96-well plate). The sensitivity of this assay using a plate reader was explored by incubating various concentrations of toxin with fixed amounts of their target sensors (300 nM). After 4 h of incubation, half-maximal cleavage of CFP-SNAP-25(141–206)-YFP was observed by using 15 pM BoNT/A and 20 pM BoNT/E (Fig. 1e Left). When the CFP-Syb(33–94)-YFP sensor was used, these values were 242 pM for BoNT/B and 207 pM for BoNT/F. Extension of the incubation period to 16 h increased the detection sensitivity of BoNT/B and BoNT/F activity by 8- and 2-fold, respectively (Fig. 1e Right).
A SNAP-25-Based BoNT Sensor That Can Be Used to Monitor Toxin Activity in Living Cells. One advantage of CFP/YFP based sensors is that, in principle, they can be used to monitor toxin activity in living cells. This is an important issue, because cell-based high-throughput screening assays have the potential to reveal not only agents that can block proteolytic activity of the toxins, but also agents that can block other steps in the action of the toxin such as binding to its cellular receptor(s), light chain translocation across endosomal membranes, and light chain refolding in the cytosol after translocation.
To carry out cell-based studies, we first transfected PC12 cells with the CFP-SNAP-25(141–206)-YFP sensor (Fig. 2a). The FRET signal in living cells was acquired by using an established three-filter set method with an epifluorescence microscope as shown in Fig. 2a (13, 14), details are provided in Materials and Methods. Transfected PC12 cells were treated with 50 nM BoNT/A for 96 h. Their fluorescence images were analyzed, and the normalized FRET ratio (corrected FRET/CFP) was plotted in Fig. 2b. Although the SNAP-25(141–206) fragment was reported to undergo cleavage at a rate similar to the full-length protein in vitro (26), CFP-SNAP-25(141–206)-YFP appeared to be a poor toxin substrate in PC12 cells. Incubation of BoNT/A for 96 h exhibited a small, but significant, shift in the FRET ratio among the cell population. These data indicate that cleavage of this sensor is inefficient in situ. Thus, this reporter may not be suitable for cell-based studies, an issue we address below.
Surprisingly, we found that using full-length SNAP-25 as the linker between CFP and YFP yielded significant levels of FRET when expressed in PC12 cells, despite the fact that SNAP-25 is 206 aa residues long (Fig. 2c and Fig. 5c, which is published as supporting information on the PNAS web site). This FRET signal depends on the membrane anchor of SNAP-25 because mutation of the palmitoylation sites within SNAP-25(Cys 85,88,90,92 Ala) (27–30), which results in the cytosolic distribution of the protein [denoted as CFP-SNAP-25(Cys-Ala)-YFP], significantly reduced the FRET signal (Fig. 2e). This finding suggests that membrane anchoring of SNAP-25 alters its structure, bringing the N and C termini of the protein close to each other. This could be due to conformational changes in SNAP-25, and/or the interaction of membrane-bound SNAP-25 with other molecules.
The full-length SNAP-25-based sensor, denoted as CFP-SNAP-25(FL)-YFP, was expressed in PC12 cells that were then treated with 50 nM BoNT/A. A progressive decrease in the FRET ratio was observed over time (Fig. 2d). We noticed the distribution of the remaining FRET ratios among cells treated with BoNT/A appeared to have two peaks (Figs. 2d and 5b). We did not observe a correlation between the remaining FRET ratios and biosensor expression levels (data not shown). Thus, the two peaks may reflect heterogeneity within the cell population.
BoNT/A cleavage of the sensor resulted in two fragments: an N-terminal fragment of SNAP-25 tagged with CFP that remained on the membrane (Fig. 2c) and a short C-terminal fragment of SNAP-25 tagged with YFP that was expected to redistribute into the cytosol after cleavage. Interestingly, we observed that the signal from the C-terminal cleavage product, SNAP-25(198–206)-YFP, was diminished (Fig. 2c, the “YFP” frame at Right). This observation was confirmed by immunoblot analysis (Fig. 2f), indicating the soluble fragment was degraded much faster than the fragment retained on the membrane.
Localization of SNAP-25 to the Plasma Membrane Is Critical for Efficient Cleavage by BoNT/A. It was recently reported that the BoNT/A light chain contains a membrane localization signal and is targeted to the plasma membrane in differentiated PC12 cells (31). Thus, we investigated the possibility that SNAP-25 must be anchored to the membrane for efficient cleavage by BoNT/A. To directly test this possibility, we expressed FRET sensors that were either anchored to the membrane [sensor = CFP-SNAP-25(FL)-YFP] or distributed throughout the cytosol [sensor = CFP-SNAP-25(Cys-Ala)-YFP] (Fig. 2e) and monitored the cleavage of these sensors using an immunoblot assay when the cells were treated with BoNT/A. As indicated in Fig. 2f, incubation of cells with BoNT/A resulted in significant cleavage of the CFP-SNAP-25(FL)-YFP sensor, whereas there was no detectable cleavage of the CFP-SNAP-25(Cys-Ala)-YFP sensor under the same assay conditions. To confirm this finding, we also targeted the inefficient sensor that was based on SNAP-25(141–206) (Fig. 2 a and b) to the plasma membrane by using a short fragment of SNAP-25 [residues 83–120, which were previously shown to target GFP to plasma membranes (28)] (Fig. 5a). Targeting of the CFP-SNAP-25(141–206)-YFP sensor to the plasma membrane resulted in efficient cleavage by BoNT/A (Fig. 5b). These data indicate that the subcellular localization of SNAP-25 is critical for efficient cleavage by BoNT/A in cells.
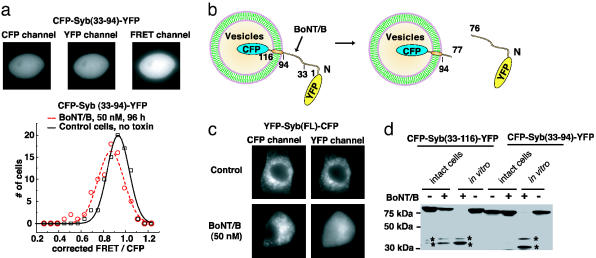
Creating a BoNT Sensor That Can Detect Cleavage of Syb in Living Cells. We next tested whether CFP-Syb(33–94)-YFP could be used to assay BoNT/B activity in cells. For these studies, we used a PC12 cell line that expresses synaptotagmin II, which mediates entry of BoNT/B (18). As shown in Fig. 3a Upper, CFP-Syb(33–94)-YFP is a soluble protein present throughout the cells. Similar to the case of CFP-SNAP-25(141–206)-YFP, BoNT/B (50 nM, 96 h) treatment only slightly decreased the FRET ratio (Fig. 3a Lower), indicating that the cleavage of this sensor is inefficient in cells.
Fig. 3.
Localization of Syb to vesicles is required for efficient cleavage by BoNT/B. (a) CFP-Syb(33–94)-YFP was expressed in a PC12 cell line that stably expresses synaptotagmin II (18). This sensor appears to be soluble inside cells and generates strong FRET signals (Upper). BoNT/B holotoxin was added to the culture medium, and 80 cells were analyzed after 96 h as described in Fig. 2b. Control cells, not treated with toxin, were analyzed in parallel. (b) A schematic description of the YFP-Syb(FL)-CFP toxin sensor. Full-length Syb is 116 aa long and is localized to vesicles through a single transmembrane domain. Cleavage of the sensor by BoNT/B should release the cytoplasmic domain of Syb, tagged with YFP, from the vesicle surface. (c) PC12 cells that stably express synaptotagmin II were transfected with YFP-Syb(FL)-CFP and incubated with (50 nM, 48 h, Lower), or without BoNT/B(Upper). CFP and YFP fluorescence images were collected for each cell, and representative cells are shown. YFP-Syb(FL)-CFP is localized to vesicles, and was excluded from the nucleus in living cells, as evidenced by both CFP and YFP fluorescent signals (Upper). BoNT/B treatment resulted in a redistribution of YFP, which became dispersed throughout the cytosol and entered the nucleus. (d) A truncated version of Syb, residues 33–116, was used to link CFP to YFP. This construct contains the same cytosolic region (residues 33–94) as the Syb fragment in the soluble sensor CFP-Syb(33–94)-YFP, but also contains the transmembrane domain of Syb (residues 95–116). PC12 cells that express synaptotagmin II were transfected with CFP-Syb(33–116)-YFP or CFP-Syb(33–94)-YFP. Cells were then incubated with (+, intact cells) or without (–, intact cells) BoNT/B (50 nM, 48 h) and harvested. Half of the cell lysates from samples that had not been exposed to BoNT/B were subsequently incubated with (+, in vitro) or without (–, in vitro) reduced BoNT/B in vitro. The same amount of each sample (30-μg cell lysate) was subjected to SDS/PAGE and immunoblot analysis using an anti-GFP antibody. Two cleavage products are indicated by asterisks. Whereas CFP-Syb(33–116)-YFP underwent significant cleavage in intact cells, there was no detectable cleavage of CFP-Syb(33–94)-YFP, indicating that the localization to vesicles is important for efficient cleavage by BoNT/B in living cells.
Our experience with the BoNT/A sensor prompted us to investigate whether the subcellular localization of Syb is important for cleavage by BoNT/B. Endogenous Syb is 116 aa residues long, and resides on secretory vesicles through a single transmembrane domain (residues 95–116, Fig. 3b). To ensure proper vesicular localization, we used full-length Syb as the linker between CFP and YFP. Because CFP is relatively resistant to the acidic environment in the vesicle lumen (19), CFP was fused to the C terminus of Syb and is predicted to reside inside vesicles, whereas YFP was fused to the N terminus of Syb and faces the cytosol (Fig. 3b). This sensor is denoted as YFP-Syb(FL)-CFP. Because, in this case, FRET is unlikely to occur between CFP and YFP, a unique approach was taken to monitor the cleavage by BoNT/B. Cleavage of the YFP-Syb(FL)-CFP sensor would generate two fragments: an N-terminal cleavage product tagged with YFP that would be released into the cytosol, and a short C-terminal fragment tagged with CFP that is restricted to the vesicle membrane. Thus, toxin activity would result in the redistribution of YFP fluorescence in cells. As shown in Fig. 3c, treatment with BoNT/B resulted in the dissociation of YFP from the vesicles and its redistribution into the cytosol. We note that the soluble YFP fragment was able to enter the nucleus, where there was no fluorescence signal before toxin treatment (Fig. 3c), providing an area where the YFP redistribution can be readily detected. Unlike the FRET assay, this detection method does not require a short distance between CFP and YFP, thus providing a distinct approach to monitor protease activity in living cells.
Targeting of Syb to Secretory Vesicles Is Critical for Efficient Cleavage by BoNT/B. Finally, we addressed the reason for the inefficient cleavage of Syb(33–94) by BoNT/B: was this caused by deletion of the first 32 residues, or by deletion of the transmembrane domain that anchors Syb to the vesicle membrane? To answer this question, we generated a sensor based on the truncated form of Syb that lacks the N-terminal 32 residues but harbors the transmembrane domain [denoted as CFP-Syb(33–116)-YFP]. Thus, this sensor contains the same cytosolic domain as the inefficient Syb-based sensor (residues 33–94), but now incorporates the transmembrane domain of Syb (residues 95–116), which targets and anchors the reporter to puncta (Fig. 3c) that correspond to secretory vesicles (data not shown). CFP-Syb(33–94)-YFP and CFP-Syb(33–116)-YFP were expressed in PC12 cells and tested for cleavage by BoNT/B. A significant amount of CFP-Syb(33–116)-YFP was cleaved within 48 h, whereas there was no detectable cleavage of CFP-Syb(33–94)-YFP (Fig. 3d). These data indicate that the vesicular localization of Syb is crucial for efficient cleavage by BoNT/B. Interestingly, a recent report indicated that negatively charged lipid mixtures enhance the cleavage rate of Syb by BoNT/B, TeNT, and BoNT/F in vitro (32). It is possible that BoNT/B may bind to secretory vesicle membranes in cells, increasing the chance of encountering Syb. Alternatively, it is also possible that the presence of the transmembrane domain may be critical for maintaining a proper conformational state of Syb that is required for efficient cleavage.
Discussion
Currently, there is a great need for sensitive methods that can be used to rapidly detect BoNT activity, and to make it possible to conduct large-scale screening for potential toxin inhibitors. Here we report recently developed methods, based on labeling toxin substrates with CFP/YFP pairs, to detect BoNT activity in vitro and in living cells. Truncated SNAP-25 or Syb fragments were used to link CFP and YFP. Cleavage of the substrate fragments by BoNTs separates CFP and YFP and thereby abolishes the FRET signal between them, thus providing a sensitive and real-time detection method for assaying BoNT activity in vitro.
We further developed these detection methods to monitor BoNT activity in living cells. We found that the truncated forms of SNAP-25 and Syb, used for toxin detection in vitro, were not efficiently cleaved by BoNT/A and -B, respectively, in living cells. However, by using full-length SNAP-25 and Syb (as linkers between CFP and YFP), we created optical reporters that mirror endogenous substrate cleavage in living cells. In principle, these two reporters should be able to detect all seven BoNTs as well as tetanus neurotoxin (TeNT). The substrate linker sequence can be readily modified to achieve specific detection for individual BoNTs or TeNT by changing the length or mutating other toxin cleavage sites. These toxin biosensors can be used to carry out cell-based screening of toxin inhibitors, thus making it possible to identify agents that inhibit toxicity by disrupting any step in the action of the toxin including binding to the cell surface receptor, translocation, refolding, substrate recognition, or catalysis.
Finally, cell-based reporters should make it possible to gain further insights into toxin substrate recognition and cleavage in cells. For example, we used these optical reporters to examine the substrate requirements of BoNT/A and B action in situ. We found that the subcellular localizations of SNAP-25 and Syb are important for efficient cleavage by BoNT/A and B, respectively. These data explain why the reporters, which were based on truncated fragments of the toxin substrates, were not effective sensors in live cells: they were soluble and were not targeted to the correct subcellular compartment. These are intriguing findings that suggest that BoNT/A and B cleave their target proteins preferentially within particular subcellular regions where their substrates perform their functions.
Supplementary Material
Acknowledgments
We thank M. Goodnough (Metabiologics, Madison, WI), Y. Jing (Towers-Perrin, Weatogue, CT), and the Chapman laboratory for advice or reagents. This work was sponsored by the National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research Program. E.R.C. and E.A.J. acknowledge membership in and support from the Region V “Great Lakes” Regional Center of Excellence Consortium (NIH Award 1-U54-AI-057153). This study was also supported by National Institute of General Medical Sciences (NIH) Grant GM 56827, National Institute of Mental Health (NIH) Grant MH61876, the Milwaukee Foundation (to E.R.C.), and industrial sponsors of the Food Research Institute (to E.A.J.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BoNT, botulinum neurotoxin; SNAP-25, synaptosome-associated protein of 25 kDa; Syb, synaptobrevin; FRET, fluorescence resonance energy transfer; CFP, cyan fluorescent protein; YFP, yellow fluorescent protein.
References
- 1.Arnon, S. S., Schechter, R., Inglesby, T. V., Henderson, D. A., Bartlett, J. G., Ascher, M. S., Eitzen, E., Fine, A. D., Hauer, J., Layton, M., et al. (2001) J. Am. Med. Assoc. 285, 1059–1070. [DOI] [PubMed] [Google Scholar]
- 2.Mahant, N., Clouston, P. D. & Lorentz, I. T. (2000) J. Clin. Neurosci. 7, 389–394. [DOI] [PubMed] [Google Scholar]
- 3.Rossetto, O., Seveso, M., Caccin, P., Schiavo, G. & Montecucco, C. (2001) Toxicon 39, 27–41. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, M. (2003) J. Rehab. Med. Suppl. 41, 56–59. [Google Scholar]
- 5.Borodic, G. E., Acquadro, M. & Johnson, E. A. (2001) Exp. Opin. Invest. Drugs 10, 1531–1544. [DOI] [PubMed] [Google Scholar]
- 6.Schiavo, G., Matteoli, M. & Montecucco, C. (2000) Physiol. Rev. 80, 717–766. [DOI] [PubMed] [Google Scholar]
- 7.Wictome, M. & Shone, C. C. (1998) Symp. Ser. Soc. Appl. Microbiol. 27, 87S–97S. [DOI] [PubMed] [Google Scholar]
- 8.Anne, C., Cornille, F., Lenoir, C. & Roques, B. P. (2001) Anal. Biochem. 291, 253–261. [DOI] [PubMed] [Google Scholar]
- 9.Schmidt, J. J., Stafford, R. G. & Millard, C. B. (2001) Anal. Biochem. 296, 130–137. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt, J. J. & Stafford, R. G. (2003) Appl. Environ. Microbiol. 69, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman, E. R., An, S., Edwardson, J. M. & Jahn, R. (1996) J. Biol. Chem. 271, 5844–5849. [DOI] [PubMed] [Google Scholar]
- 12.Foran, P., Shone, C. C. & Dolly, J. O. (1994) Biochemistry 33, 15365–15374. [DOI] [PubMed] [Google Scholar]
- 13.Gordon, G. W., Berry, G., Liang, X. H., Levine, B. & Herman, B. (1998) Biophys. J. 74, 2702–2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sorkin, A., McClure, M., Huang, F. & Carter, R. (2000) Curr. Biol. 10, 1395–1398. [DOI] [PubMed] [Google Scholar]
- 15.Erickson, M. G., Moon, D. L. & Yue, D. T. (2003) Biophys. J. 85, 599–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyawaki, A. & Tsien, R. Y. (2000) Methods Enzymol. 327, 472–500. [DOI] [PubMed] [Google Scholar]
- 17.Sorkina, T., Doolen, S., Galperin, E., Zahniser, N. R. & Sorkin, A. (2003) J. Biol. Chem. 278, 28274–28283. [DOI] [PubMed] [Google Scholar]
- 18.Dong, M., Richards, D. A., Goodnough, M. C., Tepp, W. H., Johnson, E. A. & Chapman, E. R. (2003) J. Cell Biol. 162, 1293–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsien, R. Y. (1998) Annu. Rev. Biochem. 67, 509–544. [DOI] [PubMed] [Google Scholar]
- 20.Rossetto, O., Schiavo, G., Montecucco, C., Poulain, B., Deloye, F., Lozzi, L. & Shone, C. C. (1994) Nature 372, 415–416. [DOI] [PubMed] [Google Scholar]
- 21.Shone, C. C., Quinn, C. P., Wait, R., Hallis, B., Fooks, S. G. & Hambleton, P. (1993) Eur. J. Biochem. 217, 965–971. [DOI] [PubMed] [Google Scholar]
- 22.Shone, C. C. & Roberts, A. K. (1994) Eur. J. Biochem. 225, 263–270. [DOI] [PubMed] [Google Scholar]
- 23.Yamasaki, S., Baumeister, A., Binz, T., Blasi, J., Link, E., Cornille, F., Roques, B., Fykse, E. M., Sudhof, T. C., Jahn, R., et al. (1994) J. Biol. Chem. 269, 12764–12772. [PubMed] [Google Scholar]
- 24.Yamasaki, S., Binz, T., Hayashi, T., Szabo, E., Yamasaki, N., Eklund, M., Jahn, R. & Niemann, H. (1994) Biochem. Biophys. Res. Commun. 200, 829–835. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt, J. J. & Bostian, K. A. (1995) J. Protein Chem. 14, 703–708. [DOI] [PubMed] [Google Scholar]
- 26.Washbourne, P., Pellizzari, R., Baldini, G., Wilson, M. C. & Montecucco, C. (1997) FEBS Lett. 418, 1–5. [DOI] [PubMed] [Google Scholar]
- 27.Lane, S. R. & Liu, Y. (1997) J. Neurochem. 69, 1864–1869. [DOI] [PubMed] [Google Scholar]
- 28.Gonzalo, S., Greentree, W. K. & Linder, M. E. (1999) J. Biol. Chem. 274, 21313–21318. [DOI] [PubMed] [Google Scholar]
- 29.Koticha, D. K., McCarthy, E. E. & Baldini, G. (2002) J. Cell Sci. 115, 3341–3351. [DOI] [PubMed] [Google Scholar]
- 30.Gonelle-Gispert, C., Molinete, M., Halban, P. A. & Sadoul, K. (2000) J. Cell Sci. 113, 3197–3205. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez-Salas, E., Steward, L. E., Ho, H., Garay, P. E., Sun, S. W., Gilmore, M. A., Ordas, J. V., Wang, J., Francis, J. & Aoki, K. R. (2004) Proc. Natl. Acad. Sci. USA 101, 3208–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caccin, P., Rossetto, O., Rigoni, M., Johnson, E., Schiavo, G. & Montecucco, C. (2003) FEBS Lett. 542, 132–136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.