Abstract
We present here an efficient alternative to the N-methylation for the purpose of morphing protein-binding peptides into more serum stable and cell permeable compounds. This involves the incorporation of a ”cycloalanine (CyAla)” into a peptide in a way that avoids difficult coupling steps. We demonstrate the utility of this chemistry in creating a cell permeable derivative of a high affinity HIV Rev protein-binding peptide.
Keywords: Peptidomimetics, HIV Rev, Cell Permeability, Cyclic Peptide, Antiviral
Graphical Abstract
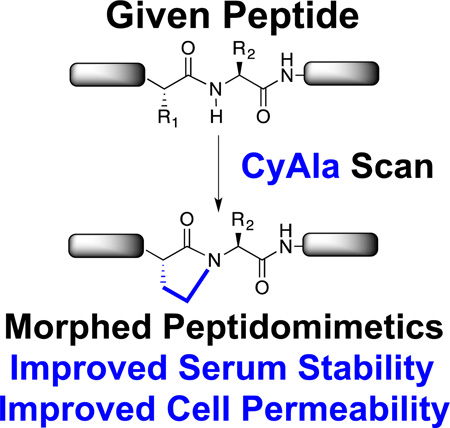
Cycloalanine: Introducing sites of N-alkylation into peptides improves serum stability and cell permeability. The most common strategy, N-methylation, requires difficult, forcing peptide coupling reactions. Here we introduce an alternative strategy involving the efficient incorporation of “cycloalanine” residues into peptides. This approach is employed to create a cell permeable version of a HIV Rev peptide antagonist
Protein surfaces that are difficult to target with traditional small molecules can often be recognized by peptides. These peptide ligands can be discovered by screening libraries or, in some cases, designed by examination of the crystal structure of the protein of interest in complex with a native binding partner or antibody. Unfortunately, peptides are non-optimal drug leads, particularly for intracellular targets, due to poor serum stability and cell permeability. Many efforts have been made to develop general methods to morph peptides into molecules with improved pharmacokinetic properties without compromising binding. One of the most common is N-methylation.[1] Peptidases do not recognize tertiary amide substrates and replacement of the polar N-H unit of the peptide bond with an N-alkyl moiety increases passive membrane permeability greatly.[2] A limitation of this approach is that acylation of N-methyl amino acids is difficult (Fig. 1A), requiring highly activated acylating reagents, and often proceeds in only modest yield.[3] Our laboratory is interested in the creation of high-quality, DNA-encoded one bead one compound (OBOC) libraries of novel oligomeric molecules,[4] including those containing N-alkylated peptides. These harsh, DNA-damaging[5] couplings are particularly problematic in this context.
Fig. 1.
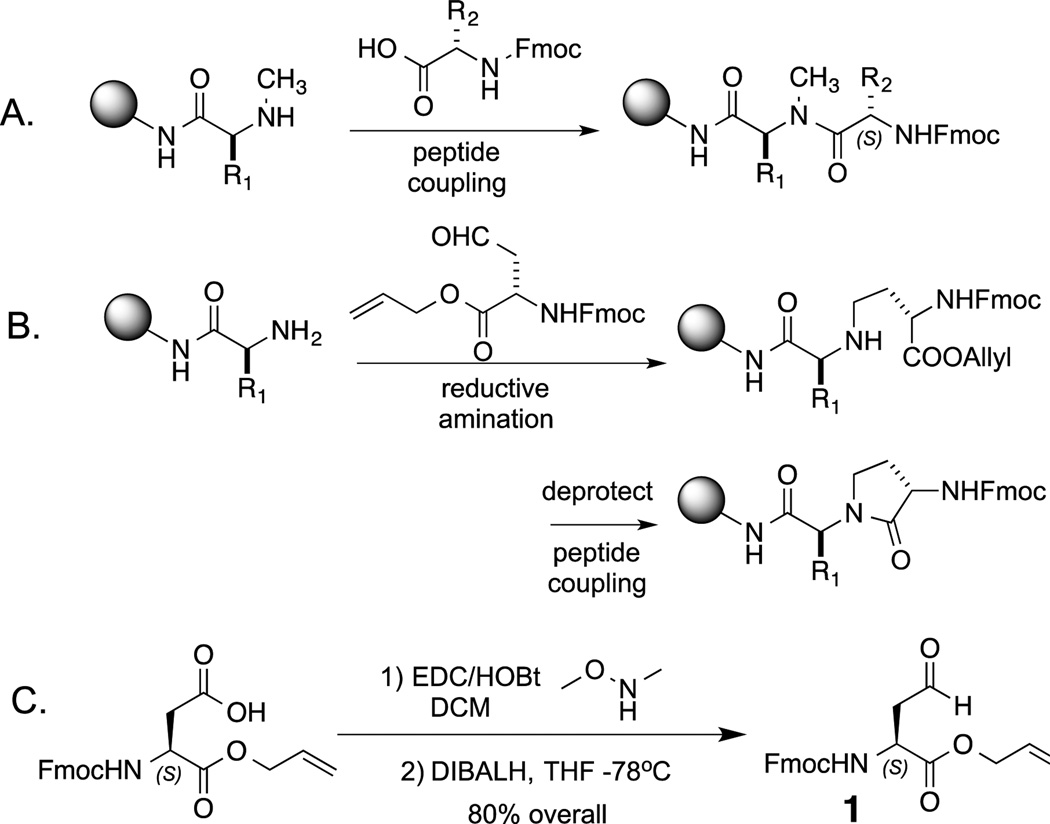
A new strategy for morphing peptides into more serum-stable, cell permeable molecules via N-alkylation. A. The traditional insertion of N-methyl amino acids into peptides necessitates a difficult acylation of the secondary amine. B. The CyAla strategy described here renders the acylation of the secondary amine an intramolecular reaction. C. Synthesis of the building block used to insert a CyAla residue into a peptide.
In this report, we describe a new strategy to produce peptide analogues with improved properties that avoids difficult acylation reactions. The chemistry involves reductive amination of the N-terminal amino acid with the aldehyde[6] that represents the side chain-reduced form of aspartic acid,[7] followed by an efficient, intramolecular acylation reaction (Fig. 1B). Thus, sterically demanding intermolecular peptide coupling is avoided. We call this unit a “cycloalanine” (CyAla).[8]
The Fmoc- and Alloc-protected building block 1 (Fmoc-Asp(H)-OAll, Fig. 1C) was synthesized in gram quantities from commercially available Fmoc-Asp-OAll by conversion to the Weinreb amide and reduction following the published procedure of Kessler and co-workers,[7]
To optimize conditions for solid-phase synthesis, a 1:1 mixture of Fmoc-Gly and Cbz-Gly was loaded onto 2-CTC resin. The Fmoc group was then removed. 1 was linked to the Gly-NH2 via reductive amination. After deprotection of the carboxylate with Pd(PPh3)4, amide bond formation was achieved by treatment with PyAOP/HOAt and DIPEA in DMF. After cleavage from the beads, LC-MS analysis of the crude products (with Cbz-Gly as an internal standard) showed that the desired GlyCyAla-Fmoc dipeptide was created in high yield (>90%) and purity (>90%) (Supp. Fig. S1).
With these conditions in hand, the efficacy of this CyAla substitution strategy was evaluated in the context of a peptide (H2N-YPAASYR) that is a paratope derived from the light chain CDR3 loop of a Fab that binds tightly to the HIV Rev protein.[9] A crystal structure of the Fab-Rev complex (Supp. Fig. S2) showed these seven residues make many of the key contacts with Rev.[9b, 10] Consistent with the structure, Wingfield and colleagues demonstrated that a synthetic peptide containing these residues bound to Rev with moderate affinity.[9b] All possible single CyAla-containing derivatives of NH2-YPAASYR (henceforth called Rev-Binding peptide (RBP) were prepared by solid-phase synthesis, except for substitution of the N-terminal tyrosine (since the following residue is proline, this was not possible). All were obtained in good yield and purity (see supplementary material). The crude products were purified by preparative HPLC.
The affinity of each derivative for Rev protein relative to the parent peptide (KD = 217 ± 22 nM; Supp. Fig. S3A) was tested using a competitive fluorescence polarization assay in which a complex of the Rev protein with the fluorescein-labeled parent peptide was challenged with increasing concentrations of unlabeled molecules (supplementary Fig. S3B and S3C). In the self-competition, unlabeled RBP exhibited an IC50 of 179 nM. Several of the CyAla-containing derivatives had IC50s within two-fold of that exhibited by the parent peptide, with the exception of substitution of the tyrosine at position 6 (Table 1). In particular, substitution of the proline and two alanines appeared to be particularly well tolerated.
Table 1.
Results of competitive binding assays in which fluorescein-labeled RBP was mixed with Rev protein, then competed with increasing amounts of the compounds indicated. See Supp. Fig. S3 for the primary data. The cyclic compounds have an amide bond between the N-terminal amine and the C-terminal carboxylate.
| Name | Sequence | IC50 [nM] |
|---|---|---|
| RBP | YPAASYR | 179 ± 15 |
| CyAla2 | YcAAASYR | 209 ± 17 |
| CyAla3 | YPcAASYR | 218 ± 15 |
| CyAla4 | YPAcASYR | 257 ± 17 |
| CyAla5 | YPAAcAYR | 293 ± 9 |
| CyAla6 | YPAAScAR | >650 |
| CyAla7 | YPAASYcA | 323 ± 20 |
| CyAla3,4 | YPcAcASYR | 205 ± 15 |
| cRBP | cyclo-YPAASYR | 28 ± 5 |
| cCyAla3,4 | cyclo-YPcAcASYR | 28 ± 2 |
| scr cCyAla3,4 | cyclo-YScAYcARP | >5000 |
| FLAG | DYKDDDDK | >5000 |
Based on these data, we synthesized a peptide in which both of the alanine residues were replaced by CyAla units (since Pro is an N-alkyl amino acid, there was no reason to modify it). For comparison purposes, we also made the analogous di-N-Me alanine-substituted peptide using the BTC coupling conditions that have been reported to be the most efficient method of driving the acylation of N-methyl amino acids.[3b] HPLC analysis of the products revealed that the crude di-CyAla compound was produced with superior yield and purity compared to the di-N-methylalanine-containing peptide (Supp. Fig. S4). To further examine the relative efficiencies of incorporating N-methyl or CyAla residues in peptides, a model solid-phase synthesis was also carried out and showed that the di-CyAla compound was formed with >95% purity. In contrast the crude di-N-methylalanine compound was less than 50% pure (Supp. Fig. S6).
The purified peptide containing CyAla residues at position 3 and 4 (CyAla3,4) exhibited an affinity for Rev protein in the competitive binding assay similar to that of the parent peptide (Table 1). We also created a head to tail cyclized version of CyAla3,4 (cCyAla3,4) since it was shown that a cyclic peptide containing the RBP bound Rev with higher affinity than the linear molecule, as expected from the loop-like conformation seen in the crystal structure (Supplementary Fig. S2).[9b] As positive and negative controls the analogous head to tail cyclized RBP (cRBP) and a scrambled version of cCyAla3,4 (scrcCyAla3,4) were also made. As expected, cyclization increased the affinity of both RBP and CyAla3,4 significantly (IC50 = 28 nM; Table 1 and Supplementary Fig. S3C).
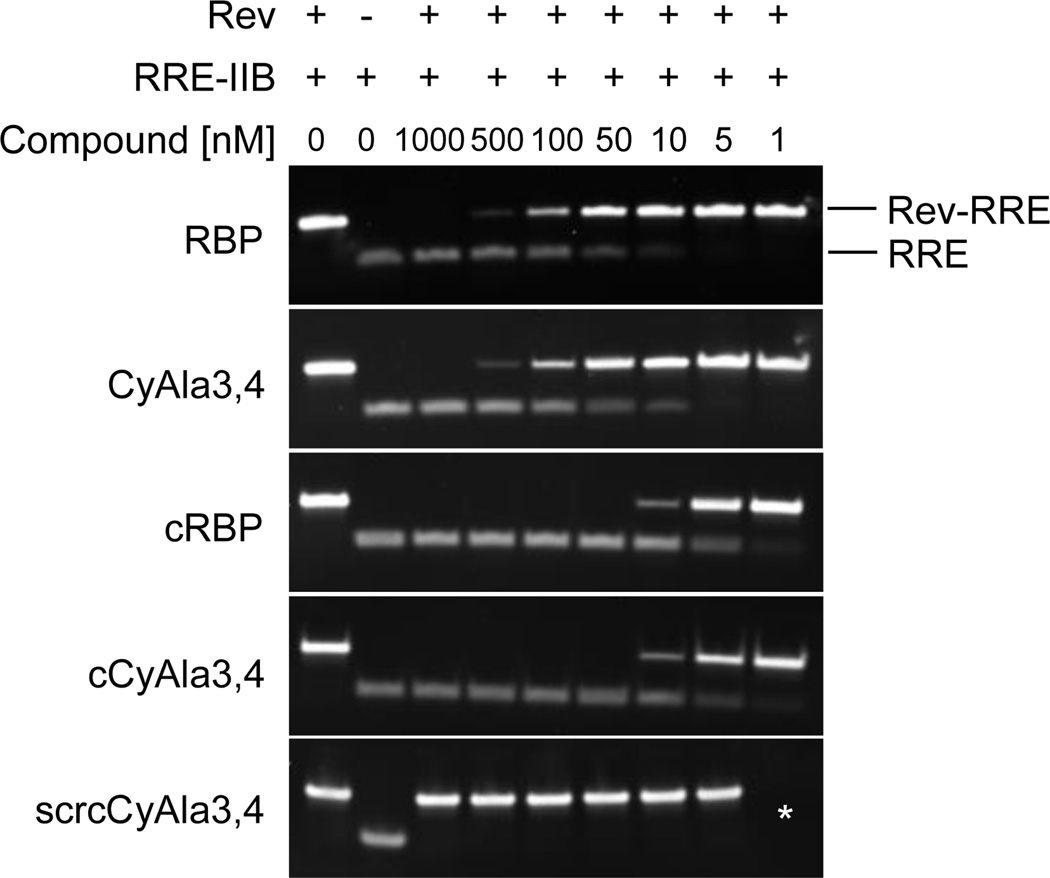
Rev is an RNA-binding protein that is required for export of unspliced or incompletely spliced viral messages from the nucleus to the cytoplasm.[11] This event is essential for viral replication and thus is a therapeutic target of interest, though currently no good Rev inhibitors exist. Rev associates with a specific RNA sequence known as the RRE (Rev response element). Therefore, we examined whether RBP or CyAla-containing analogues could interfere with Rev-RRE binding using a previously established gel shift assay. As shown in Fig. 2, both the linear and cyclic forms of RBP and CyAla3,4 potently inhibit Rev-RRE binding. The linear molecules exhibited IC50s of 62 and 68 nM, respectively, while the cyclic compounds were about ten times more potent. scrCyAla3,4 had no effect on complex formation It is unusual for small molecules to antagonize protein-nucleic acid interactions, particularly with this level of potency. Note that these molecules do not bind to the RNA-binding surface of Rev, and thus must be acting via an allosteric mechanism.
Fig. 2.
Effect of RBP and the RBP mimics indicated on the association of Rev with the RRE RNA as monitored by native gel electrophoresis.
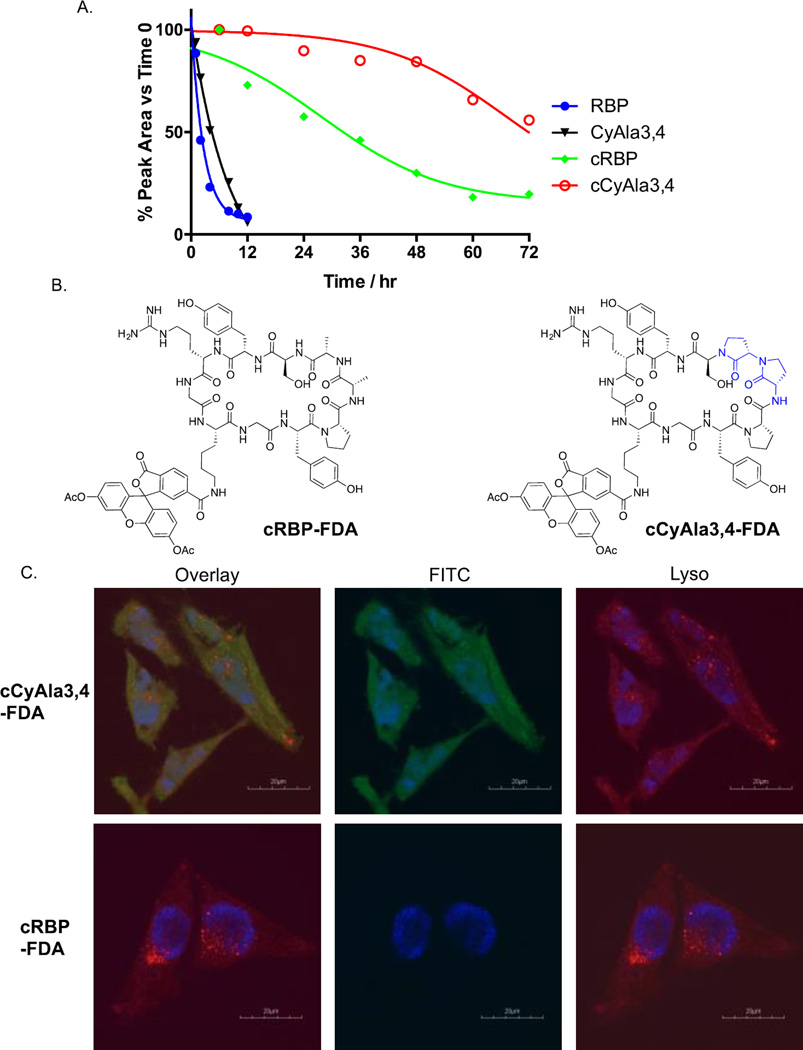
Given this promising result, we evaluated the suitability of these molecules as Rev antagonists in living cells. First, the serum stability was evaluated. As shown in Fig. 3A and Supplementary Fig. S5, RBP had a half-life of approximately two hours when incubated with human serum. CyAla3,4 exhibited an improved half-life of about five hours. Cyclization of either of these species dramatically improved serum stability. cRBP was still 75% intact after a 12 hour incubation, while the level of cCyAla3,4 was essentially unchanged in the same time frame. Indeed, longer serum incubations revealed cRBP had a half-life of approximately 30 hours, and cCyAla3,4 more than 72 hours (Fig 3A, and Supp. S7). Given the superior binding properties and serum stability of the cyclic molecules, we focused on these exclusively for the remainder of the study.
Fig. 3.
Assessment of the serum stability and cell permeability of RBP and selected analogues. A. The compounds indicated were incubated with human serum. At the times indicated the samples were analysed by HPLC and the fraction of the intact peptide remaining was quantified. B. Structures of the FDA conjugates used in part C. C. Live cell confocal fluorescent images of HeLa cells treated with the compounds indicated and stained with Hoechst 33342 (blue; DNA stain) and LysoTracker Red DND-99 (red).
The critical issue of cell permeability was addressed next. Derivatives of cRBP and cCyAla3 were created with a lysine inserted between the N- and C-terminal residues. A fluorescein diacetate (FDA) moiety was tethered to the Lys side chain (Fig 3B). FDA is dim, but the fluorophore “lights up” when the acetyl groups are hydrolyzed, an event catalyzed by intracellular esterases. Thus, if the molecules can access the cytoplasm, the cells become fluorescent.[12]
As expected, incubation of the cRBP-FDA conjugate with HeLa cells did not result in the generation of intracellular fluorescence (Fig. 3C), reflecting the generally poor cell permeability of peptides of this size. While cyclization can increase the permeability of some peptides, this requires the formation of intracellular amide hydrogen bonds that act to desolvate the molecule.[13] But this is not a general phenomenon and apparently cRBP does not adopt such a conformation. In stark contrast, incubation of HeLa cells with cCyAla3,4-FDA produced clear green fluorescence inside the cells (Fig. 3C). While some of the green color was localized to punctate regions, most of it appeared to be spread throughout the cytoplasm. To further address the concern that the peptide analogue may have accessed the cell by endocytosis or some related mechanism, we also stained the cells with Lysotracker dye (red). The overlay of the red and green channels showed no correlation between the localization of lysosomes and the cCyAla3,4-Fluorescein, consistent with most of the peptidomimetic entering the cells by passive diffusion. The huge difference between the cyclic peptide and the diCyAla-substituted molecule validates the utility of this novel N-alkylation strategy as a method to improve cell permeability. A cyclic di-N-methylalanine analogue (cMeAla3,4-FDA) was also made with an FDA fusion. Cells incubated with this compound showed less cytoplasmic fluorescence than those incubated with cCyAla3,4-FDA compound under the same experimental condition (Fig. S8), suggesting that the CyAla strategy may provide more cell permeable molecules than N-methylation.
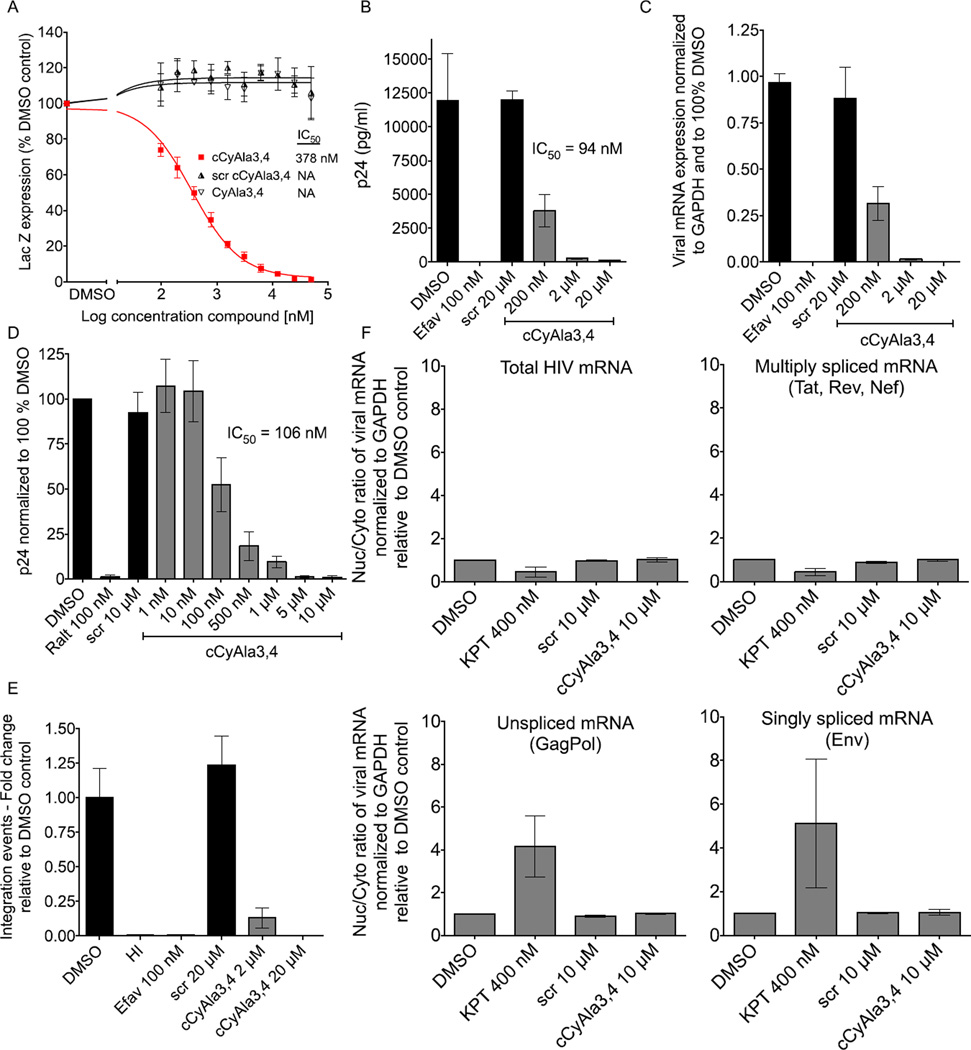
With a serum stable and cell permeable antagonist of the Rev-RNA interaction in hand, we proceeded to evaluate its effect on HIV replication in living cells. These experiments employed a reporter cell line that stably expresses the β-galactosidase (LacZ) gene, driven by the HIV-1 5’ long terminal repeat (LTR), which responds to the viral protein Tat expressed by an incoming virus. The measured β-gal activity correlates directly with viral replication.[14] Thus, HeLa-CD4-LTR-LacZ cells were infected with the HIV-1 isolate NL4-3, a X4 tropic virus, in the presence of increasing concentrations of the linear (CyAla3,4) or cyclic (cCyAla3,4) CyAla-containing analogues of RBP, or, as a control ScrcCyAla3,4. cCyAla3,4, but not the linear compound or the scrambled control, inhibited viral replication, with an IC50 of 378 nM (Fig. 4A). We confirmed the efficacy of cCyAla3,4 to inhibit HIV-1 replication by measuring other replication readouts such as viral capsid p24 in the supernatant by p24 ELISA[14] and cell-associated viral RNA by qPCR (Fig. 4B, and 4C).[15] Efavirenz,[15] a reverse transcription inhibitor, was used as positive control in these assays. We also assessed the activity of cCyAla3,4 to inhibit replication of an R5 tropic HIV-1 strain, YU2. Infection of CCR5+ GHOST cells with this R5 tropic virus was inhibited by cCyAla3,4 with an IC50 of 106 nM (Fig. 4D). The integrase inhibitor Raltegravir[15] was used as a positive control.
Fig. 4.
cCyAla3,4 inhibits early events of the HIV-1 replication cycle. A) The cyclic peptide cCyAla3,4, but not its linear version, inhibits HIV-1 replication. HeLa-CD4 LTR-LacZ cells were infected with the HIV-1 X4 tropic NL4-3 isolate in presence of the indicated compounds and concentrations. Beta-galactosidase activity was quantified 72 h later. scr: scr cCyAla3,4; NA: not applicable. B) cCyAla3,4 inhibits HIV-1 p24 capsid production. HeLa-CD4 cells were infected with NL4-3 for 5 h. Cells were washed three times and fresh media added with the indicated compounds. Capsid p24 in the supernatant was assessed 72 h later by p24 ELISA. Efav: Efavirenz. Error bars in A) and B) represent standard deviation of three independent experiments done in duplicate (n = 3). C) cCyAla3,4 inhibits viral mRNA expression. Experiment done as in B). Total RNA was extracted 96 h post infection and analyzed by quantitative RT-qPCR. Total HIV mRNA normalized to GAPDH and to 100% DMSO control. Error bars represent standard deviation of two independent experiments done in duplicate (n = 2). D) cCyAla3,4 inhibits acute R5 tropic HIV-1 replication. GHOST cells were infected with the HIV-1 R5 tropic YU2 isolate for 24 h in presence of the indicated compounds and concentrations. Cells were washed and fresh media with compounds added. p24 ELISA was performed 72 h later. Ralt: Raltegravir. Error bars represent standard deviation of four independent experiments (n = 4). E) cCyAla3,4 inhibits an early event hindering proviral integration. HeLa-CD4 cells were infected in presence of the indicated compounds. gDNA was extracted 18 h later and integration events were determined by Alu-PCR followed by nested quantitative qPCR. HI: Heat inactivated. Error bars represent standard deviation of at least three independent experiments done in duplicate (n = 4 except cCyAla3,4 scr n = 3). F) cCyAla3,4 does not inhibit Rev-RRE/CRM1 dependent viral mRNA export. HeLa-CD4 cells were infected for 48 h. Cells were washed and compounds added for 24 h. After this period of time, cytoplasmic and nuclear RNA were extracted and analyzed by quantitative RT-qPCR using primers recognizing total HIV mRNA, multiply spliced (Tat, Rev, Nef), singly spliced (Env) or unspliced mRNA (GagPol). All mRNA normalized to GAPDH. The ratio nuc/cyt RNA represents the fold enrichment (accumulation) of viral mRNA in the nucleus with the DMSO control set to 1. KPT-330 is a selective CRM1-mediated nuclear mRNA export inhibitor. Error bars represent standard deviation of two independent experiments done in duplicate (n = 2).
While these results were encouraging, the IC50 values measured, while respectable, were not as low as we might have expected given the low nM potency displayed by cCyAla3,4 in antagonizing Rev-RRE interactions in vitro (Fig. 2). Since cCyAla3,4 appears to be highly cell permeable (Fig. 3C), it seemed unlikely that this difference could be explained by only a fraction of the molecule added to the cells traversing the membrane. Therefore, more detailed assays were undertaken to determine if the inhibition of HIV replication by cCyAla3,4 was indeed due to a blockade of Rev-mediated RNA transport from the nucleus to the cytoplasm.
First, proviral integration events were quantified to determine whether cCyAla3,4 targets an early or late event in the HIV-1 life cycle. Integration is an early event that precedes any Rev-mediated steps in the viral life cycle. This was done by using an Alu-PCR followed by a nested qPCR directed at the HIV-1 promoter region.[15] To our surprise, and contrary to what would be expected from an inhibitor of Rev activity, cCyAla3,4 showed a dose-dependent inhibition of this early stage, Rev-independent proviral integration event (Fig. 4E).
A Rev inhibitor should promote accumulation of unspliced and singly spliced viral mRNAs in the nucleus, while the ratio of the multiply spliced mRNA between the nucleus and cytoplasm remains unchanged. To determine if cCyAla3,4 has an effect on these ratios, viral mRNA was isolated from both the nucleus (nuc) and the cytoplasm (cyt) of cells treated for 24 h with KPT (a selective CRM1-mediated nuclear mRNA transport inhibitor)[16], cCyAla3,4 or scrcCyAla3,4. Quantitative RT-qPCR was then used to measure the relative amounts of unspliced (GagPol), singly spliced (Env) and multiply spliced mRNA (Tat, Rev and Nef) (Fig. 4F). Treatment with KPT, induced a 3- to 5-fold increase in the nucleus of GagPol mRNA and a 3 to 7-fold of Env mRNA. In contrast, the ratio between the nuclear and cytosolic levels of these mRNAs did not change when the cells were treated with 10 µM cCyAla3,4 (Fig. 4F). Together these data rule out the possibility that cCyAla3,4 antagonizes CRM1/Rev-RRE viral mRNA export from the nucleus.
Why cCyAla3,4 cannot engage Rev in HIV-infected cells is unknown. The live cell fluorescence images show that cCyAla3,4-Fluorescein is absent from the nucleus (Fig. 3C). Perhaps it is thus unable to engage Rev at a point at which blocking its interaction with RNA is possible. It seems extremely unlikely that the lack of anti-Rev activity in living cells is due to poor cell permeability of cCyAla3,4. First, the live cell fluorescence imaging indicates that substantial levels of the compound can enter the cell. Moreover, the compound does exhibit significant anti-viral activity (Fig. 4), with IC50’s in the mid-nM region depending on the viral strain. This argues the compound recognizes one or more intracellular off-targets. More work will be required to track down these off-targets.
It may be that the selectivity of the peptide for Rev is insufficient to allow engagement in the cell. It is worthwhile to remember that the RBP, which represented the starting point of this probe development project, comes from a paratope of a Rev-binding Fab antibody fragment that was isolated from a phage display library to aid in obtaining the crystal structure of Rev. Thus, the RBP sequence has not been required by evolution, either natural or applied, to bind to Rev selectively in a complex milieu like the cytoplasm, as would be the case if the parent peptide came from a native binding partner. While binding in vitro is tight, high affinity does not always go hand in hand with high selectivity. We note that a fusion of the Rev-binding Fab with a Tat cell penetrating peptide was shown to have anti-viral activity in cells, as does cCyAla3,4, but the mechanism of action was not probed in detail to determine if this activity was indeed due to Rev inhibition. We speculate that it was not, though this would have to be determined experimentally.
Returning to the major focus of this study, which was to evaluate the CyAla substitution as an alternative to N-methylation for improving the properties of peptide ligands, the data are encouraging. This chemistry is considerably cleaner and more efficient than the placement of N-methyl amino acids in a peptide (Supp. Figs. S4 and S6). Replacing two native peptide residues with CyAla moieties in the seven-residue RBP increased the serum stability of the peptide by about 2.5-fold (Fig. 3A) and increased the cell permeability tremendously (Fig. 3C), perhaps to even greater degree than did the corresponding N-methyl units. All of this without affecting the affinity of the peptide for Rev (Table 1). Another advantage of using CyAla over N-methyl is that the latter substitution results in a mixture of cis and trans peptide bonds, while CyAla is expected to have the same trans-dominated equilibrium as a standard peptide. Thus, the CyAla chemistry should be a useful tool for morphing interesting peptide ligands into cell permeable probe molecules in the future. It should be emphasized however, that this substitution eliminates the native side chain of a peptide, unlike N-methylation, so can be employed only at positions where the side chain does not make a critical contact with the protein.
Finally, while RBP may not have been an optimal starting point for the development of a selective Rev inhibitor, the fact that we were able to develop a serum stable and highly cell permeable molecule that antagonized Rev-RRE binding with low nM potency in vitro is cause for optimism for future probe/drug development against this key HIV protein. It is clearly possible to engage an allosteric site on the protein with high affinity given a molecule of the size of a cyclic heptapeptide. This suggests that a de novo screen of large libraries of molecules of this type under conditions that demand high selectivity, as well as affinity, would be a productive route to interesting Rev inhibitors.
Experimental Section
Additional experimental details are provide in the supplementary material.
Synthesis of CyAla-containing peptides
Fmoc-Gly-OH and Cbz-Gly-OH (1:1 mixture) were loaded onto 2-CTC resin. After removal of the Fmoc group, 1 was condensed onto the amine by reductive amination. The allylic ester was cleaved and the resultant free acid was cyclized in an intramolecular fashion to the free amine. Fmoc chemistry was used during the solid phase synthesis with modifications. The synthetic procedure for each type of chemical reaction conducted is provided below.
Amino acid to amino acid coupling[17]
Fmoc amino acid (3.0 eq to resin loading), Ethyl (hydroxyimino)cyanoacetate (Oxyma) or Ethyl (hydroxyimino)cyanoacetate potassium salt (K-Oxyma, 3.0 eq to resin loading) and N,N’-Diisopropylcarbodiimide (DIC, 3.0 eq to resin loading) were mixed together in dimethylformamide (DMF) for 10 min, before the mixture was added to resin. After 2 h reaction, the resin was washed by DMF 3×/DCM 3×/DMF 3×.
Fmoc deprotection
The Fmoc group was subsequently deprotected by 20% piperidine in DMF for 15 min twice. After deprotection, the resin was extensively washed by DMF 5×/dichloromethane (DCM) 5×/DMF 5×.
Reductive amination
After Fmoc deprotection, resin was treated with compound 4 (10.0 eq) in DMF for 1 h at room temperature, followed by washing with 1:1 DMF/DCM. Subsequently, excess NaBH4 (greater than 10.0 eq) was added in DCM/MeOH. The reaction was carried for 2 h at room temperature. A positive chloranil test indicated the desired secondary amine was generated.
Ally deprotection
The allyl group was removed by incubating the resin with Pd(PPh3)4 (3.0 eq) and PhSiH3 (12.0 eq) in anhydrous DCM with continuous shaking at room temperature for 30 min. The resin was washed with DCM, DMF, 1.0 % DIPEA in DMF, and 1.0 % sodium diethyldithiocarbamate trihydrate in DMF 3× and then DMF 3×.
Cyclization
Cyclization was performed by incubating resin with PyAOP (3.0 eq), 2.04 mg of HOAt (3.0 eq), and DIPEA (6.0 eq) in DMF with rotation at room temperature for 6 hr. Resin was washed with DMF 1× and incubated in DMF with rotation at room temperature overnight.
Cleavage
If no subsequent macrocyclization is needed, the cleavage should be done together with global deprotection of acid-labile protecting groups, The compound was cleaved from chlorotrityl chloride resin by freshly prepared cleavage cocktail: trifluoroacetic acid (TFA)/ triisopropylsilane (TIPS)/H2O (95:2.5:2.5) for 1 hr.
Alternatively, if macrocyclization is needed, the compound was cleaved by a cocktail containing trifluoroethanol(TFE)/acetic acid (HOAc)/DCM (1:1:3). After 1 h reaction at room temperature, the cleavage cocktail was evaporated, followed by ether precipitation, and preparative HPLC purification.
Supplementary Material
Acknowledgments
We thank Dr. Stephen J. Stahl (NIAMS, National Institutes of Health) for the generous gift of HIV-Rev protein. We thank Prof. Phil Dawson (TSRI) for bringing ref. 8 to our attention.
References
- 1.a) Chatterjee J, Gilon C, Hoffman A, Kessler H. Acc Chem Res. 2008;41:1331–1342. doi: 10.1021/ar8000603. [DOI] [PubMed] [Google Scholar]; b) Kessler H, Chatterjee J, Doedens L, Operrer F, Biron E, Hoyer D, Schmid H, Gilon C, Hruby VJ, Mierkes DF. Adv Exp Med Biol. 2009;611:229–231. doi: 10.1007/978-0-387-73657-0_106. [DOI] [PubMed] [Google Scholar]; c) Fiacco SV, Roberts RW. Chembiochem. 2008;9:2200–2203. doi: 10.1002/cbic.200800208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu aP, Liu B, Kodadek T. Nature biotechnology. 2005;23:746–751. doi: 10.1038/nbt1099. [DOI] [PubMed] [Google Scholar]; Ovadia bO, Greenberg S, Chatterjee J, Laufer B, Opperer F, Kessler H, Gilon C, Hoffman A. Mol Pharm. 2011;8:479–487. doi: 10.1021/mp1003306. [DOI] [PubMed] [Google Scholar]
- 3.a) Teixido M, Albericio F, Giralt E. J Pept Res. 2005;65:153–166. doi: 10.1111/j.1399-3011.2004.00213.x. [DOI] [PubMed] [Google Scholar]; b) Thern B, Rudolph J, Jung G. Angewandte Chemie (International ed. 2002;41:2307–2309. doi: 10.1002/1521-3773(20020703)41:13<2307::AID-ANIE2307>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]; c) Videnov GI, Kaiser D, Kempter C, Jung G. Angew. Chem. Intl. Ed. 1996;35:1503–1506. [Google Scholar]
- 4.MacConnell AB, McEnaney PJ, Cavett VJ, Paegel BM. ACS Comb Sci. 2015;17:518–534. doi: 10.1021/acscombsci.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malone ML, Paegel BM. ACS Comb Sci. 2016 doi: 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pels K, Kodadek T. ACS Comb. Sci. 2015;17:152–155. doi: 10.1021/acscombsci.5b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayer T, Riemer C, Kessler H. J Pept Sci. 2001;7:250–261. doi: 10.1002/psc.306. [DOI] [PubMed] [Google Scholar]
- 8.This unit has been inserted into peptides previously using a low yield, two-step protocol in which a methionine residue is methylated, then treated with base to deprotonate the amide C-terminal to the methionine, leading to cyclization (see: Martin V, Legrand B, Vezenkov LL, Berthet M, Subra G, Calmes M, Bantignies JL, Martinez J, Amblard M. Angewandte Chemie (International ed. 2015;54:13966–13970. doi: 10.1002/anie.201506955. The conformational preferences of peptides containing this unit were studied, but no data on their cell permeability was presented.
- 9.a) Stahl SJ, Watts NR, Rader C, DiMattia MA, Mage RG, Palmer I, Kaufman JD, Grimes JM, Stuart DI, Steven AC, Wingfield PT. Journal of molecular biology. 2010;397:697–708. doi: 10.1016/j.jmb.2010.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zhuang X, Stahl SJ, Watts NR, DiMattia MA, Steven AC, Wingfield PT. Journal of Biological Chemistry. 2014;289:20222–20233. doi: 10.1074/jbc.M114.581090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiMattia MA, Watts NR, Stahl SJ, Rader C, Wingfield PT, Stuart DI, Steven AC, Grimes JM. Proc Natl Acad Sci U S A. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.a) Cao Y, Liu X, De Clercq E. Curr HIV Res. 2009;7:101–108. doi: 10.2174/157016209787048564. [DOI] [PubMed] [Google Scholar]; b) Cullen BR, Malim MH. Trends in biochemical sciences. 1991;16:346–350. doi: 10.1016/0968-0004(91)90141-h. [DOI] [PubMed] [Google Scholar]
- 12.Jones KH, Senft JA. J Histochem Cytochem. 1985;33:77–79. doi: 10.1177/33.1.2578146. [DOI] [PubMed] [Google Scholar]
- 13.a) Ahlbach CL, Lexa KW, Bockus AT, Chen V, Crews P, Jacobson MP, Lokey RS. Future Med Chem. 2015;7:2121–2130. doi: 10.4155/fmc.15.78. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rezai T, Yu B, Millhauser GL, Jacobson MP, Lokey RS. J Amer. Chem. Soc. 2006;128:2510–2511. doi: 10.1021/ja0563455. [DOI] [PubMed] [Google Scholar]; c) White TR, Renzelman CM, Rand AC, Rezai T, McEwen CM, Gelev VM, Turner RA, Linington RG, Leung SS, Kalgutkar AS, Bauman JN, Zhang Y, Liras S, Price DA, Mathiowetz AM, Jacobson MP, Lokey RS. Nature Chem Biol. 2011;7:810–817. doi: 10.1038/nchembio.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mousseau G, Clementz MA, Bakeman WN, Nagarsheth N, Cameron M, Shi J, Baran P, Fromentin R, Chomont N, Valente ST. Cell Host Microbe. 2012;12:97–108. doi: 10.1016/j.chom.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mousseau G, Kessing CF, Fromentin R, Trautmann L, Chomont N, Valente ST. MBio. 2015;6:e00465. doi: 10.1128/mBio.00465-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.a) Etchin J, Sanda T, Mansour MR, Kentsis A, Montero J, Le BT, Christie AL, McCauley D, Rodig SJ, Kauffman M, Shacham S, Stone R, Letai A, Kung AL, Thomas Look A. Br. J. Haematol. 2013;161:117–127. doi: 10.1111/bjh.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Boons E, Vanstreels E, Jacquemyn M, Nogueira TC, Neggers JE, Vercruysse T, van den Oord J, Tamir S, Shacham S, Landesman Y, Snoeck R, Pannecouque C, Andrei G, Daelemans D. EBioMedicine. 2015;2:1102–1113. doi: 10.1016/j.ebiom.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subirós-Funosas R, Prohens R, Barbas R, El-Faham A, Albericio F. Chemistry--A European Journal. 2009;15:9394–9403. doi: 10.1002/chem.200900614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.