Abstract
Objective
Insulin signaling plays a unique role in the regulation of energy homeostasis and the impairment of insulin action is associated with altered lipid metabolism, obesity, and Type 2 Diabetes. The main aim of this study was to provide further insight into the regulatory mechanisms governing the insulin signaling pathway by investigating the role of non-proteolytic ubiquitination in insulin-mediated activation of AKT.
Methods
The molecular mechanism of AKT regulation through ubiquitination is first dissected in vitro in 3T3-L1 preadipocytes and then validated in vivo using mice with adipo-specific deletion of GPS2, an endogenous inhibitor of Ubc13 activity (GPS2-AKO mice).
Results
Our results indicate that K63 ubiquitination is a critical component of AKT activation in the insulin signaling pathway and that counter-regulation of this step is provided by GPS2 preventing AKT ubiquitination through inhibition of Ubc13 enzymatic activity. Removal of this negative checkpoint, through GPS2 downregulation or genetic deletion, results in sustained activation of insulin signaling both in vitro and in vivo. As a result, the balance between lipid accumulation and utilization is shifted toward storage in the adipose tissue and GPS2-AKO mice become obese under normal laboratory chow diet. However, the adipose tissue of GPS2-AKO mice is not inflamed, the levels of circulating adiponectin are elevated, and systemic insulin sensitivity is overall improved.
Conclusions
Our findings characterize a novel layer of regulation of the insulin signaling pathway based on non-proteolytic ubiquitination of AKT and define GPS2 as a previously unrecognized component of the insulin signaling cascade. In accordance with this role, we have shown that GPS2 presence in adipocytes modulates systemic metabolism by restricting the activation of insulin signaling during the fasted state, whereas in absence of GPS2, the adipose tissue is more efficient at lipid storage, and obesity becomes uncoupled from inflammation and insulin resistance.
Keywords: Adipose tissue, Obesity, Insulin, AKT, Ubiquitin, GPS2
Graphical abstract
Highlights
-
•
Ubc13-mediated ubiquitination of AKT is required for activation of the insulin signaling pathway.
-
•
GPS2 regulates insulin signaling by inhibiting AKT ubiquitination and activation.
-
•
Adipo-specific deletion of GPS2 results in increased adiposity and altered lipid flux in the adipocytes.
-
•
GPS2-AKO mice have higher levels of circulating adiponectin and are insulin sensitive despite being obese.
1. Introduction
The adipose tissue is an extremely flexible organ that plays a critical role in the regulation of energy homeostasis through both triglyceride storage and adipokine secretion [1], [2]. The health of the white adipose tissue, and thus of the whole body, largely depends on the existence of a proper balance between lipid storage and utilization, which is challenged under conditions of excess food intake. Major pathological consequences of overnutrition and obesity result from an increase in lipid flux to non-adipose organs and the development of insulin resistance [3], [4]. During the normal cycles of daily fasting and feeding, adipose tissue metabolism is tightly regulated to respond to the energetic demands of the organism. Insulin signaling plays a key role in this process by promoting nutrient storage during the fed state via enhanced glucose uptake and triglyceride synthesis, and by inhibiting triglyceride breakdown and fatty acid release through lipolysis [5], [6]. The AKT/PKB kinase is an obligatory mediator of the insulin signaling pathway downstream of phosphatidylinositol 3-kinase (PI3K) [7], [8]. Full AKT activation is promoted by dual phosphorylation by PDK1, a PI3K-dependent kinase, and the mTOR-rictor complex. The key rate-limiting event for AKT activation is not its phosphorylation per se, but rather its translocation to the plasma membrane. Accordingly, full signaling capacity is achieved by tethering AKT to the plasma membrane [9], [10], [11], [12]. Intriguingly, recent evidence indicates that AKT recruitment to the membrane, and thus activation, upon stimulation with the growth factors EGF and IGF is unexpectedly regulated through K63 ubiquitination [13], [14], [15]. However, it is currently unknown whether AKT ubiquitination similarly contributes to the regulation of metabolic homeostasis through modulation of insulin signaling.
Ubiquitination is a reversible modification that is achieved via the sequential actions of several classes of enzymes, including an ubiquitin (Ub)-activating enzyme (E1), an Ub-conjugating enzyme (E2), and an Ub ligase (E3) [16], [17]. Poly-ubiquitination of target proteins with chains of different topology can promote either protein degradation or serve, as in the case of other post-translational modifications, to influence protein function and interactions [18], [19]. The key E2 enzyme for promoting the formation of non-proteolytic K63 ubiquitin chains is Ubc13, which catalyzes the synthesis of ubiquitin chains in complex with the non-catalytic subunits Mms2/Uev1A and specific E3 ligases [20], [21], [22]. Consistent with the flexibility of ubiquitination as a tight regulatory switch for signaling pathways is the existence of multiple strategies to rapidly reverse the active mark. Among them, and best characterized, is the removal of ubiquitin modifications by chain specific deubiquitinases [16], [23], [24], [25], as reported in the case of AKT deubiquitination by CYLD [26], [27]. However, an equally if not more effective strategy, is restricting the deposition of the ubiquitin chains through inhibition of the ubiquitination machinery. Examples of inhibitors behaving in this manner are the ubiquitin thioesterase OTUB1 in the DNA damage response pathway and G-Protein Suppressor 2 (GPS2) in the TNFR1 signaling pathway [28], [29].
GPS2 is a small multifunctional protein that was originally identified while screening for suppressors of Ras activation in the yeast pheromone response pathway [30]. Recent studies by our lab and others indicate that GPS2 plays an important anti-inflammatory role in adipose tissue and macrophages and is required for the expression of genes regulating cholesterol and triglyceride metabolism [29], [31], [32], [33], [34]. Due to multiple functional interactions existing between GPS2 and various transcriptional regulators, GPS2 activity has been studied mainly in the context of its nuclear functions, including both transcriptional repression and activation [29], [31], [32], [34], [35], [36], [37], [38], [39], [40], [41], [42]. However, GPS2 also plays an important non-transcriptional role in the cytosol by regulating JNK activation downstream of TNFR1 [29]. Intriguingly, our findings reveal that GPS2 activity in different cellular compartments relies on a conserved regulatory strategy based on the inhibition of ubiquitin conjugating complexes that are responsible for the formation of K63 ubiquitin chains (TRAF2/Ubc13 in the cytosol and RNF8/Ubc13 in the nucleus) [29], [34]. Furthermore, recent data indicate that GPS2 directly inhibits Ubc13 enzymatic activity (Unpublished data; Lentucci et al., under revision), suggesting that GPS2-mediated regulation might extend to other signaling pathways relying on Ubc13-mediated ubiquitination events. Here, we have investigated the hypothesis that GPS2 is required for restricting the activation of the insulin signaling pathway through inhibition of Ubc13-mediated ubiquitination of AKT.
2. Experimental procedures
2.1. Animals studies
Fat-specific GPS2 knockout mice (GPS2-AKO) were generated using a cre/lox approach and maintained on a mixed 129sv/C57BL6J background. Conditional GPS2 floxed mice were generated by inGenious Targeting Laboratory. The 9.52 kb region used to construct the targeting vector was first sub cloned from a positively identified C57BL/6 (RP23: 91G16) BAC clone into a ∼2.4 kb backbone vector (pSP72, Promega) containing an ampicillin selection cassette. The total size of the targeting construct (including vector backbone and Neo cassette) is ∼13.62 kb. The region was designed such that the short homology arm (SA) extends about 2.55 kb 3′ to exon 6. The long homology arm (LA) ends 5′ to exon 3 and is 6.07 kb long. A pGK-gb2 loxP/FRT Neo cassette is inserted on the 3′ side of exon 6 and the single loxP site is inserted 5′ of exon 3. The target region is 0.90 kb and includes exon 3–6. The targeting vector was linearized by NotI and then transfected by electroporation of BA1 (C57BL/6×129/SvEv) hybrid embryonic stem cells. After selection with G418 antibiotic, surviving clones were expanded for PCR analysis to identify recombinant ES clones and control for retention of the third LoxP site. Secondary confirmation of positive clones was performed by Southern Blotting analysis prior to microinjection into C57BL/6 blastocysts. Resulting chimeras with a high percentage agouti coat color were mated to wild-type C57BL/6J mice to generate F1 heterozygous offspring. The Neo cassette was excised by crossing with FLP mice (Jackson Laboratories). Adipose tissue specific deletion was achieved by crossing Gps2flox/flox mice with heterozygous Adipoq-Cre C57BL/6J transgenic mice expressing Cre recombinase under control of the adiponectin promoter [43]. Male mice and littermate controls were used for all experiments. Mice were maintained on standard laboratory chow diet in a temperature controlled facility on a 12-hour light/dark cycle. All animal studies were approved by the Boston University Institutional Animal Care and Use Committee (IACUC) and performed in strict accordance of NIH guidelines for animal care.
2.2. Body composition analysis and metabolic testing
Mice body composition was assessed by non-invasive MRI scanning on an EchoMRI 700 (BUSM Metabolic Phenotyping core). Glucose Tolerance Test (GTT) and Insulin Tolerance Test (ITT) were performed according to established protocols [44]. Briefly, mice were starved overnight or 4–6 h for GTT and ITT, respectively. Blood glucose levels were measured, at the described time points after glucose (1.5 mg/g body weight) or insulin (Humulin R, Lilly) (0.5 U/kg body weight) IP injection, from tail nicking using a OneTouch Ultra glucometer. Plasma was collected from overnight fasted mice after cardiac puncture and profiled using Milliplex Multiplex Assays (Millipore, BUSM Analytical Core) and Free Fatty Acid Fluorometric Assay (Cayman Chemical).
2.3. H&E staining
Upon harvesting, adipose tissue depots and liver tissues were incubated at 4 °C in Z-fix solution (Anatech LTD) overnight. Tissues were then transferred to 70% ethanol, paraffin embedded, sectioned, and stained with hematoxylin and eosin (Tuft Pathology Core/BNORC Adipose Biology Core). Imaging of adipocyte cell size was performed as described [45], [46].
2.4. Cell culture and isolation
3T3-L1 preadipocytes (American Type Culture Collection) were cultured in high glucose Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum (Hyclone) under 5% CO2. Cells were transfected using Lipofectamine 2000 (Invitrogen) and Opti-MEM reduced serum media (Thermo Fisher Scientific) according to each manufacturer's instructions. Cells were serum starved overnight prior to 100 nM insulin (Life Technologies) treatment for described time points. The following siRNAs were used: MISSION siRNA universal negative control (Sigma), siGPS2 (#s80309, 20 nM, Ambion), siUBC13 (#s123784, 20 nM, Ambion).
For in vitro differentiated adipocytes, the stromal vascular fraction (SVF) was isolated from adipose tissue depots after collagenase type I/dispase II digestion in 4% BSA KRH buffer for 45 min then washed, filtered, and spun down at 900 rpm for 10 min. Cells were cultured in high glucose DMEM with 10% fetal bovine serum (Hyclone) and 1× pen/strep until confluence. Two days later, in vitro adipogenic differentiation was induced using a standard insulin, IBMX (Sigma), and DEX (Sigma) cocktail for 2 days then changed to maintenance media containing high glucose DMEM with 10% fetal bovine serum, pen/strep, and insulin for 12 additional days.
Primary mature adipocytes were isolated from adipose tissue depots using collagenase type I/dispase II (Fisher) digestion in 4% BSA (Sigma) Kreb-Ringer HEPES (KRH) buffer for 45 min, then filtered and separated by floating and washed 3 times in 1% BSA KRH buffer prior to resuspension in lysis buffer for protein extraction or Trizol for RNA analysis.
Akt null cells are UBC-Cre ERT2; AKT1 fl/fl, AKT2 fl/fl in an AKT3 KO background. To generate this line, primary brown adipocyte precursors (bAPC) cells were isolated from P1 neonates and immortalized with pBabe-SV40 Large T antigen. To induce AKT1 and AKT2 deletion UBC-Cre ERT2; AKT1 fl/fl, AKT2 fl/fl cells were treated on 2 consecutive days with 1 μM 4-hydroxy tamoxifen and on the 3rd day the media was changed to regular media to allow protein turnover. On the 4th day, cells were plated during the morning at 80% confluence, serum removed later in the day for overnight starvation prior to 15′ insulin treatment (150 nM) and protein extraction.
2.5. Protein isolation, western blot analysis and immunoprecipitation
Whole cell extracts were prepared from cultured cells or mouse tissue by homogenization in lysis buffer (50 mM Tris HCl pH 8, 250 mM NaCl, 5 mM EDTA, 0.5% NP-40) supplemented with 0.1 mM PMSF, 10 mM NEM (Thermo fisher scientific), 1× protease inhibitors (Pierce), and 1× phosphatase inhibitors (Pierce). Protein concentrations were analyzed using the Bradford assay (Biorad) and normalized for protein loading. Immunoprecipitation was performed on whole cell lysates upon incubation of indicated antibodies overnight followed by collection of immune complexes with Protein A-sepharose 4B conjugate (Thermo fisher scientific), SDS-PAGE electrophoresis, and western blotting as previously described [29]. The following antibodies were used for western blotting: Anti-GPS2-N antibody was generated in rabbit against a specific peptide representing aa 1–11, B-Tubulin (T4026, 1:2000, Sigma), Phospho-HSL ser563 (#4139, 1:1000, Cell Signaling), HSL (#4107S, 1:1000, Cell Signaling), Phospho-PKA R2 ser99 (#ab32390, 1:1000, Abcam), PKA R2 (#ab38949, 1:1000, Abcam), Phospho-(ser/thr) PKA substrates (#9621, 1:1000, Cell Signaling), Phospho-AKT ser473 (#9271, 1:500, Cell Signaling), Phospho-AKT thr308 (#9275, 1:500, Cell Signaling), AKT (#9272, 1:1000, Cell Signaling), Phospho-GSK3B ser9 (#9322, 1:1000, Cell Signaling), GSK3B (#9315, 1:1000, Cell Signaling), Phospho-ACC ser79 (#3661, 1:1000, Cell Signaling), ACC (#3676, 1:1000, Cell Signaling), Phospho-AMPK thr172 (#2535, 1:1000, Cell Signaling), AMPK (#2603, 1:1000, Cell Signaling), Polyubiquitin K63-linkage-specific (HWA4C4, 1:500, Enzo Lifescience), AKT1 (1:1000, Cell Signaling), AKT2 (1:1000, Cell Signaling), S6 (1:1000, Cell Signaling), Phospho-S6 (1:1000, Cell Signaling), HA (1:1000, Cell Signaling), Phospho-JNK thr183/tyr185 (#4668, 1:1000, Cell Signaling), JNK (#sc-571, 1:1000, Santa Cruz).
2.6. RNA isolation and gene expression analysis
Total RNA extracted from cultured cells, primary adipocytes, or mouse tissue was purified using RNeasy plus mini columns (Qiagen) and reverse transcribed using the iScript cDNA Synthesis System (Biorad) according to manufacturers' instructions. PCR reactions were performed using Fast Sybr Master Mix (ABI) on a ViiA7 Real-Time PCR System. Relative mRNA expression was determined after normalization to the housekeeping gene, cyclophilin A. Data are shown as mean ± SEM between the indicated n value. Statistical significance was calculated by two-tailed Student's t test; *represents p value < 0.05 and **p value < 0.01. All primer sequences are available upon request.
2.7. FACS analysis
SVF cells were purified as described above. Flow cytometry analyses were performed using the following anti-mouse antibodies: PerCP anti CD11b (Biolegend), FITC antiF4/80 (BioRAD). Single cell suspensions were stained with Aqua Zombie dye, washed, pre-blocked with mouse FcBlock (Biolegend), and stained with antibody cocktails in the presence of Brilliant Violet buffer (BD Biosciences). Ultracomp beads (eBioscience) stained with abovementioned antibodies and ArC beads (Life Technologies) stained with Aqua Zombie dye were used for compensation in FACSDIVA. All data were acquired on a SORP LSRII (BD Biosciences). At least 20,000 events were collected from each sample. Manual data analysis was performed by a blinded investigator using FlowJo 10. Results presented are from three independent experiments, with statistical significance calculate by two-tailed Student's t test.
2.8. Lipolysis assay
In vitro differentiated cells were washed with PBS then incubated with high glucose DMEM + 2% BSA without serum for 2–3 h, then washed with phenol-free DMEM without serum and incubated in serum free phenol free DMEM + 2% BSA with or without 10 μM isoproterenol (Sigma) for 1 h. Media was collected for measuring the released glycerol using the free glycerol reagent (Sigma). Glycerol concentrations were calculated from a standard curve and values normalized to protein concentration per well assessed by Bradford assay as described above. Results shown include data points from three independent experiments, each time the assay was performed with technical triplicates.
3. Results
3.1. AKT activation in the insulin signaling cascade is regulated by ubiquitination
Non-proteolytic ubiquitination of AKT has been recently described as a key step in the activation cascades downstream of EGF and IGF receptor stimulation [13], [47]. Despite some differences in the enzymatic machineries that are enlisted in the two different pathways, in both cases, K63 ubiquitination plays a critical role in promoting AKT recruitment to the plasma membrane [13], [47]. Membrane recruitment of AKT through anchoring to phosphoinositol-3,4,5-triphosphate (PIP3) is also a limiting step in the insulin signaling pathway [48], [49]. However, it is currently unknown whether ubiquitination is required for the activation of AKT upon insulin stimulation. To address this question, we first asked whether AKT is post-translationally modified by the addition of K63 ubiquitin chains upon insulin stimulation of 3T3-L1 preadipocytes. As shown in Figure 1A, we observed that AKT undergoes K63 ubiquitination within minutes of insulin stimulation. The peak of ubiquitination slightly precedes AKT maximal phosphorylation by the phosphoinositide-dependent protein kinase 1 (PDK1) (Figure 1A), in accord with the proposed function. Next, we addressed whether ubiquitination occurs on the same sites described in the context of EGF/IGF signaling [13], and whether this modification is required for activation of the insulin signaling pathway. To answer these questions, we reconstituted AKT1/2/3 null cells, generated from primary brown adipocyte precursors, with either wild type HA-AKT2 or HA-AKT2 mutants in which we had disrupted either Lys8 (AKT2-K8R), Lys14 (AKT2-K14R), or both (AKT2-K8R/K14R) by targeted mutagenesis. Their overexpression in AKT null cells revealed that the single mutants were still partially phosphorylated upon insulin stimulation, whereas removal of both ubiquitination sites resulted in a complete loss of AKT phosphorylation (Figure 1B). The activation of downstream AKT targets, such as PRAS40 and ribosomal protein S6, was also inhibited in cells reconstituted with the double mutant (Figure 1B). Together, these results indicate that AKT ubiquitination is a required step for full activation of the insulin signaling pathway. To further confirm this conclusion and explore which ubiquitination machinery mediates AKT ubiquitination within the insulin pathway, we downregulated Ubc13, the E2 conjugating enzyme responsible for the synthesis of K63 ubiquitin chains, by transient siRNA transfection. Ubc13, in concert with specific E3 ligases, was previously found responsible for mediating the ubiquitination, and thus activation, of AKT upon stimulation of IGF, EGF and ErbB receptors [13], [47], [50]. In accord with these results, we observed that basal activation of AKT, albeit already very low, is reduced upon Ubc13 downregulation. In addition, our results indicate that disrupting AKT ubiquitination through downregulation of Ubc13 significantly impaired insulin-dependent phosphorylation of AKT (Figure 1C and Supplemental Figure S1A). Together, our data indicate that AKT is ubiquitinated upon insulin stimulation and that Ubc13-mediated K63 ubiquitination is required for full activation of AKT and its downstream targets, hence raising the interesting question of whether AKT ubiquitination can be modulated to regulate insulin signaling.
Figure 1.
AKT ubiquitination and activation upon insulin stimulation is regulated by the opposing actions of the ubiquitin conjugating enzyme Ubc13 and its inhibitor GPS2. (A) Detection of K63 ubiquitination by IP/WB of AKT in 3T3-L1 preadipocytes serum starved overnight prior to stimulation with 100 nM insulin. Phosphorylation of AKT on Thr308 is measured in parallel on whole cell extracts from the same cells and normalized to total AKT levels. (B) Inducible AKT1 and AKT2 deletion upon 4-hydroxy tamoxifen treatment (4OH-Tam) is rescued by transient overexpression of AKT2 WT or ubiquitination mutants. Protein expression levels and insulin mediated activation of AKT2 and downstream effectors PRAS40 and ribosomal protein S6 are assayed by WB in whole cell extracts. (C) WB analysis of phospho-AKT/total AKT on whole cell extracts from siCTRL versus siUBC13 transiently transfected 3T3-L1 preadipocytes. Cells were stimulated with insulin (5′) after overnight serum starvation. (D) WB analysis of phospho-AKT/total AKT and phospho-GSK3B/total GSK3B in cytosolic cell extracts from proliferating 3T3-L1 preadipocytes transiently transfected with siCTRL (WT) or siGPS2 (GPS2-KD). (E) WB analysis of phospho-AKT/total AKT in whole cell extracts from 293T cells. (F) WB analysis of phospho-AKT/total AKT in whole cell extracts from WT and GPS2-KD 3T3-L1 preadipocytes starved overnight prior to insulin stimulation (3′). All western blots are representative images of at least three independent experiments.
Previous studies from our lab show that GPS2 negatively regulates different cellular functions through inhibition of K63 ubiquitination events [29], [34]. Recent work has revealed that this effect is achieved through direct inhibition of Ubc13 enzymatic activity (Unpublished data; Lentucci et al., under revision). Because our data indicate that Ubc13-mediated ubiquitination is required for AKT phosphorylation and activation, we then asked whether GPS2 negatively regulated the activation of the insulin pathway. To investigate this hypothesis, we monitored AKT activation in 3T3-L1 preadipocytes upon modulation of GPS2 expression. First, we downregulated GPS2 by transient siRNA transfection in 3T3-L1 preadipocytes (GPS2-KD) (Supplemental Figure S1B). A significant increase in the basal level of AKT phosphorylation was observed upon GPS2 downregulation (Figure 1D), indicating that GPS2 is required for restricting the constitutive activation of AKT in proliferating cells under basal conditions. Similar results were observed in human 293T cells (Figure 1E), suggesting that GPS2-mediated regulation of AKT is conserved among different cell types. Next, to specifically investigate the relevance of GPS2-mediated inhibition in the context of insulin signaling, we starved GPS2 KD cells and subjected them to insulin stimulation. Again, AKT activation was enhanced in GPS2-KD cells (Figure 1F).
Together, these results reveal that Ubc13-mediated K63 ubiquitination of AKT represents a novel, key regulatory node of the insulin signaling cascade. GPS2, by virtue of its ability to inhibit Ubc13 enzymatic activity, emerges as a novel regulator of the insulin signaling pathway required for restricting the activation of AKT and downstream effectors both in presence and absence of stimulation.
3.2. Constitutive AKT ubiquitination and activation in GPS2-deficient adipocytes
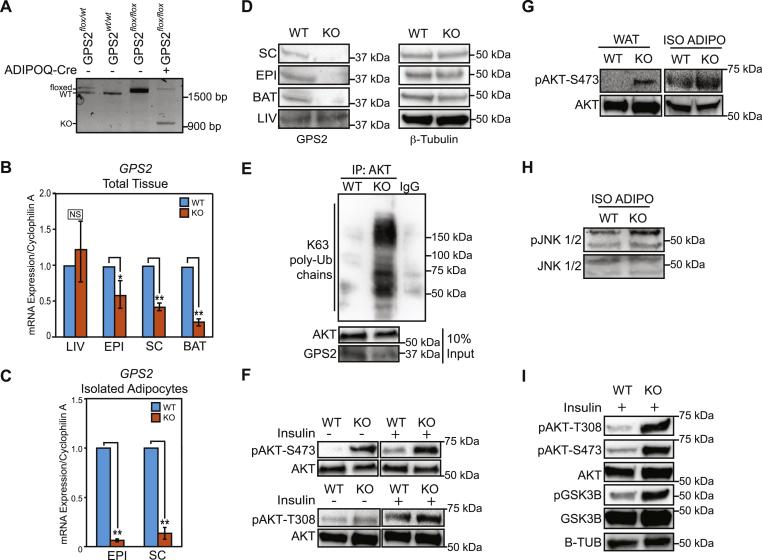
Properly functioning insulin signaling is especially important in adipocytes to maintain cell homeostasis and allow for appropriate responses to metabolic cues. Based on our findings of GPS2 acting as a negative regulator of AKT activation, we hypothesized that GPS2 would play an important physiologic role in adipose tissue. To investigate the physiologic relevance of GPS2 in regulating insulin signaling in the adipose tissue, we generated an adipo-specific knockout mouse model (GPS2-AKO) by crossing GPS2fl/fl mice, carrying LoxP sites flanking exons 3–6, with Adipo-Cre mice (Jackson laboratories) [43]. The efficiency of this deletion strategy was confirmed by genomic PCR, western blotting, and gene expression analysis (Figure 2A–D). Together, these experiments confirmed that GPS2 deletion is specific to mature adipocytes within the adipose tissue. Then, we investigated in in vitro differentiated adipocytes from the stromal vascular fraction (SVF) of either wild type (WT) or mutant (GPS2-AKO) mice the hypothesis that loss of GPS2 results in increased ubiquitination and activation of AKT. The efficiency of differentiation was similar for cells of both genotypes (Supplemental Figure S2). However, when assaying the level of AKT ubiquitination by IP/WB using an antibody specific against K63 ubiquitin chains for detection, we found a striking increase in AKT-associated ubiquitin chains in GPS2-deficient adipocytes compared to their wildtype counterparts (Figure 2E). The increase in ubiquitination was associated with enhanced basal phosphorylation on Ser473 and Thr308 both in absence of stimulation and upon short-term insulin treatment (Figure 2F), confirming that AKT ubiquitination and activation are strongly enhanced in cultured adipocytes depleted of GPS2.
Figure 2.
Sustained AKT activation in Adipo-specific GPS2 Knockout (GPS2-AKO) mice. (A) Confirmation of GPS2 deletion in the adipose tissue of GPS2-AKO mice by genomic DNA analysis. (B) RT-qPCR analysis of GPS2 mRNA expression in white epididymal (EPI), subcutaneous (SC), and brown (BAT) adipose tissue depots and liver (LIV) from AKO versus WT mice, n = 6–7. (C) GPS2 mRNA expression in primary adipocytes isolated from white fat depots, n = 3–4. (D) Confirmation of protein knockout by WB in extracts from WAT, BAT, and LIV tissues. (E) IP/WB analysis of AKT ubiquitination with K63 ubiquitin chains in in vitro differentiated adipocytes from AKO versus WT mice. (F) WB analysis of phospho-AKT/AKT in whole cell extracts from in vitro differentiated adipocytes upon insulin stimulation (3') after overnight serum starvation. (G) WB analysis of phospho-AKT/total AKT on whole cell extracts from WAT tissue and primary adipocytes isolated from the EPI tissue after overnight fast. (H) WB analysis of phospho-JNK/total JNK on whole cell extracts from primary adipocytes. (I) WB analysis of phospho-AKT/total AKT and phospho-GSK3B/total GSK3B in whole cell extracts from WAT after i.p. insulin injection (0.5 U/kg body weight). Results are expressed as mean ∓ SEM. Statistical significance was calculated by two-tailed Student's t test; * represents p value < 0.05 and **p value < 0.01. All western blots are representative images of at least three independent experiments.
Next, we confirmed that insulin-dependent stimulation of AKT is affected by GPS2 deletion in vivo by monitoring AKT activation in adipose tissue under fasted and insulin stimulated conditions. Strikingly, we observed a significant increase in basal phosphorylation of AKT in both the whole subcutaneous adipose tissue depot and in mature adipocytes isolated from the epididymal adipose tissue depot of GPS2-AKO mice following overnight fasting (Figure 2G). In the same conditions, removal of GPS2-mediated inhibition of Ubc13 activity in adipocytes also led to increased activation of JNK (Figure 2H), as expected based on previous findings [29], [39]. In addition, we observed a significant increase in the level of AKT phosphorylation and activation in the subcutaneous adipose tissue when mice were injected with insulin for a short time prior to sacrifice (Figure 2I). Enhanced activation of insulin signaling pathways downstream of AKT in the adipose tissue from GPS2-AKO mice was confirmed by augmented phosphorylation of AKT target substrate GSK3β (Figure 2I). Therefore, together, our results indicate that GPS2 deletion promotes sustained basal and insulin-stimulated phosphorylation and activation of AKT, and downstream effectors of the insulin signaling pathway, in vivo in the adipose tissue.
3.3. Altered regulation of lipolysis and lipogenesis in GPS2-AKO mice causes adipocyte hypertrophy and excessive body adiposity
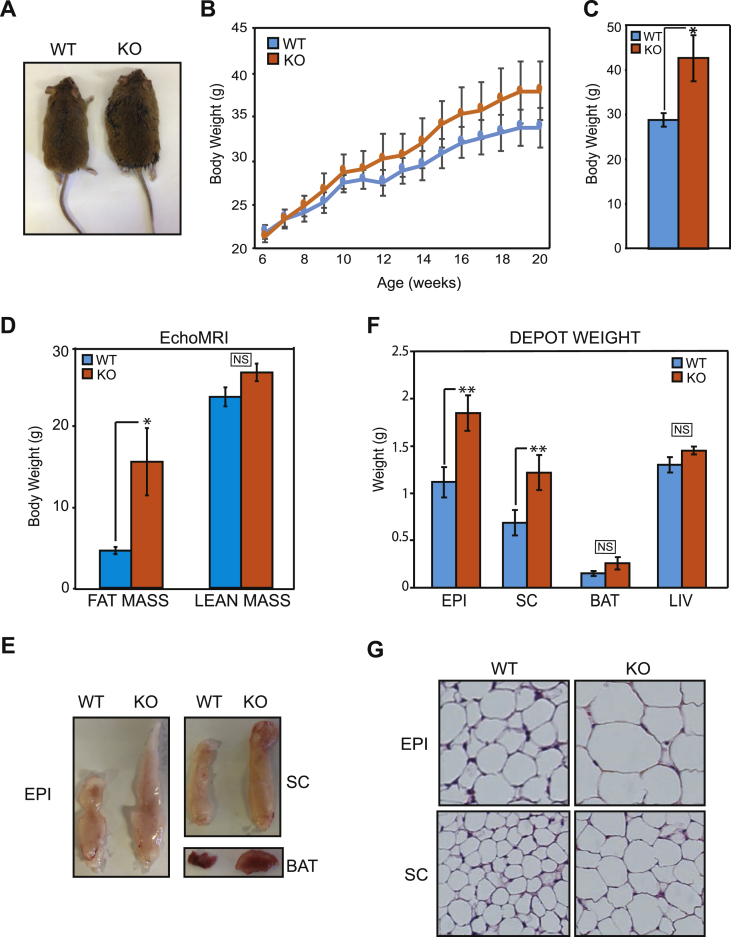
A tight regulation of insulin action in adipose tissue is required for the proper modulation of nutrient storage and mobilization throughout the daily cycles of fasting and feeding [7], [51], [52]. Our in vitro and in vivo results combined suggest that removal of GPS2-mediated regulation is sufficient to promote aberrant activation of the insulin signaling pathway through constitutive phosphorylation and activation of AKT. Thus, we asked whether the fat-specific deletion of GPS2 affects nutrient storage, lipid fluxes and whole body metabolism in vivo. First, we observed that GPS2-AKO mice were healthy and fertile but could be easily distinguished from their wild type (WT) littermates based on their larger size (Figure 3A). The difference in body weight was not apparent at birth but increased gradually with time (Figure 3B) and became significant once mice reached adulthood (Figure 3C). Echo-MRI scans of these mice confirmed that the increase in body mass was attributed to an increase in fat mass, without significant changes in lean mass (Figure 3D). Gross appearance revealed that all adipose tissue depots appeared larger when comparing GPS2-AKO to WT littermates (Figure 3E). Weights of individual white fat depots were significantly higher for GPS2-AKO mice (Figure 3F). The interscapular brown adipose tissue (BAT) also trended to be larger (Figure 3F). The enhanced adiposity correlated with a substantial increase in the size of adipocytes (hypertrophy) within both the SC and EPI depots (Figure 3G). In conclusion, these data indicate that GPS2-depleted adipocytes expand more than their wild type counterparts, resulting in elevated adipose tissue mass and larger body weight, even when mice are fed a normal chow diet.
Figure 3.
Increased body weight, fat mass, and adipocyte cell size in GPS2-AKO mice. (A) Representative image of whole body morphology of adult WT and GPS2-AKO mice. (B) Progressive body weight gain in AKO versus WT mice, WT n = 5–9, KO n = 7–11. (C) Total body weight of 4–6 months old mice, n = 5. (D) Body composition analysis of total fat versus lean mass measured by EchoMRI scanning of WT and AKO mice at 4–6 months old, n = 5. (E) Representative images of gross appearance of EPI, SC, and BAT depots in AKO versus WT mice at 4–6 months old. (F) Individual adipose tissue depot and liver weights in AKO versus WT mice at 4–6 months old, n = 9–11. (G) Representative H&E staining of paraffin-embedded EPI and SC sections imaged at 10× magnification. Results are expressed as mean ∓ SEM. Statistical significance was calculated by two-tailed Student's t test; *represents p value < 0.05 and **p value < 0.01.
Adipocytes are the primary site of energy storage. The balance between lipid storage in the form of triglycerides and mobilization as released free fatty acids is directly regulated by insulin through AKT-dependent as well as AKT-independent pathways [4], [53], [54] In this context, a major physiological function of insulin in the postprandial, anabolic state is to support the accumulation of lipid storage by promoting de novo lipogenesis while restraining lipid mobilization via lipolysis. Based on the enhanced AKT activation we observed in the adipose tissue of both fasted and insulin-stimulated GPS2-AKO mice, we asked whether an impairment of the regulated cycles of lipogenesis and lipolysis was underlying the increased adipocyte cell size and body adiposity of these mice.
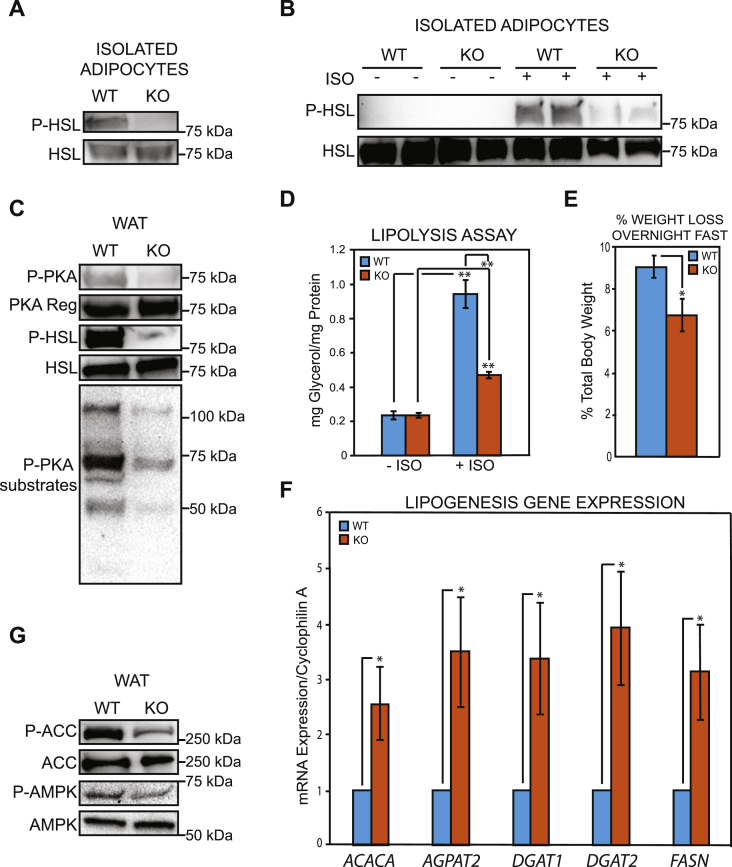
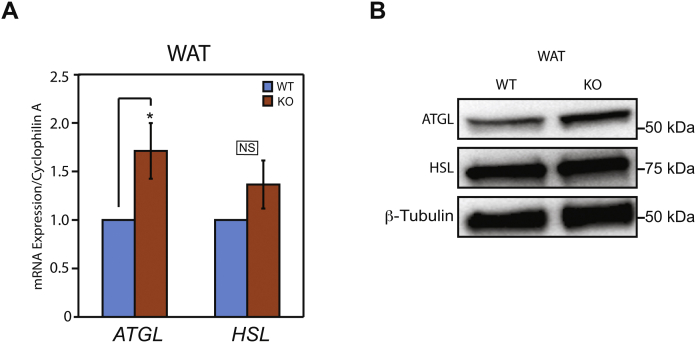
Our previous work showed that GPS2 overexpression in aP2-GPS2 transgenic mice promoted a strong increase in the phosphorylation, and thus activation, of hormone sensitive lipase, HSL [34]. Here, we found that GPS2 deletion promoted the opposite phenotype. In isolated adipocytes from fasted GPS2-AKO mice, HSL phosphorylation was completely blunted, in contrast to the strong signal observed in adipocytes from WT littermates (Figure 4A). Similar results were observed upon isoproterenol stimulation of in vitro differentiated adipocytes from WT or GPS2-AKO mice, with the induced HSL phosphorylation being severely reduced in absence of GPS2 (Figure 4B). In agreement with GPS2 being required for the priming of HSL and ATGL promoters only during the early stages of adipocyte differentiation prior to the expression of adiponectin [34], we did not observe any significant change in the expression of HSL upon GPS2 deletion in adiponectin-expressing, mature adipocytes. However, we unexpectedly observed a trend for increased ATGL mRNA and protein levels, which could reflect an attempt at compensating for the reduced lipolysis (Figure 4B and Supplement Figure S2A and S2B). Upstream regulation of the lipolysis pathway was also impaired, as shown by reduced phosphorylation of PKA and PKA substrates in subcutaneous white adipose tissue homogenates (Figure 4C). To further confirm that lipid mobilization from the adipose tissue is impaired in GPS2-AKO mice, we confirmed that lipolysis is impaired in absence of GPS2 ex vivo and in vivo. Ex vivo, the rate of lipolysis, as assessed by measuring glycerol release from in vitro differentiated adipocytes, was severely reduced in GPS2-deficient cells compared to wild type counterparts (Figure 4D). In vivo, GPS2-AKO mice lost significantly less body weight after an overnight fast than their WT littermates, in accordance with their impaired ability to promote lipid release from triglyceride stores (Figure 4E). Thus, our current and previous work combined indicates that GPS2 is indeed required for lipid mobilization through lipolysis. Interestingly, it appears that GPS2 plays complementary genomic and non-genomic roles in regulating different steps of the lipolysis pathway in differentiating preadipocytes and mature adipocytes.
Figure 4.
Improved lipid storage in the white adipose tissue of GPS2-AKO mice as a result of impaired lipolysis and increased lipogenesis. (A) WB analysis of phospho-HSL/total HSL in whole cell extracts of primary adipocytes isolated from the EPI adipose tissue depot of AKO versus WT mice after overnight fasting. (B) WB analysis of phospho-HSL/total HSL protein levels in whole cell extracts from in vitro differentiated adipocytes in fasted or isoproterenol (10 μM) stimulated conditions. (C) WB analysis for markers of active lipolysis in whole cell extracts from the WAT of fasted mice. (D) Basal and isoproterenol induced lipolysis assay on in vitro differentiated adipocytes. Statistical significance was calculated comparing 3 independent experiments. (E) Percentage of total body weight loss after prolonged overnight fast, n = 7. (F) qPCR analysis of lipogenic markers in WAT of AKO versus WT mice, n = 5–7. (G) WB analysis of phospho-ACC/total ACC and phospho-AMPK/total AMPK in whole cell extracts from the WAT of fasted mice. Results are expressed as mean ∓ SEM. Statistical significance was calculated by two-tailed Student's t test; *represents p value < 0.05 and **p value < 0.01. All western blots are representative images of at least three independent experiments.
Next, we investigated the effect of enhanced insulin signaling on the regulation of lipogenesis. In both adipose tissue and liver, acute stimulation of de novo lipogenesis (DNL) is mainly regulated by insulin and glucose availability through the transcriptional activities of SREBP1, ChREBP and LXRs [5], [55], [56], [57]. Gene expression analysis by RT-qPCR showed a significant increase in the mRNA levels of the lipogenic genes Fasn, Dgat1, Dgat2, AGPAT2, and Acaca in the subcutaneous WAT of GPS2-AKO mice compared to WT littermates (Figure 4F). In addition to the transcriptional regulation of enzyme availability, insulin-mediated activation of AKT impinges upon DNL through AMPK inhibition, resulting in dephosphorylation and activation of the Acetyl-CoA carboxylase (ACC) [58]. In agreement with AKT being constitutively activated in GPS2-AKO mice, we observed both a significant decrease in active AMPK and decreased phosphorylation of ACC in the white adipose tissue of GPS2-AKO mice (Figure 4G). Together, these results are consistent with the hypothesis that both in the fed and fasted state, lipogenesis is enhanced while lipid mobilization is impaired in GPS2-AKO mice, markedly shifting the balance toward fat storage.
3.4. Fat-specific deletion of GPS2 results in improved systemic insulin sensitivity
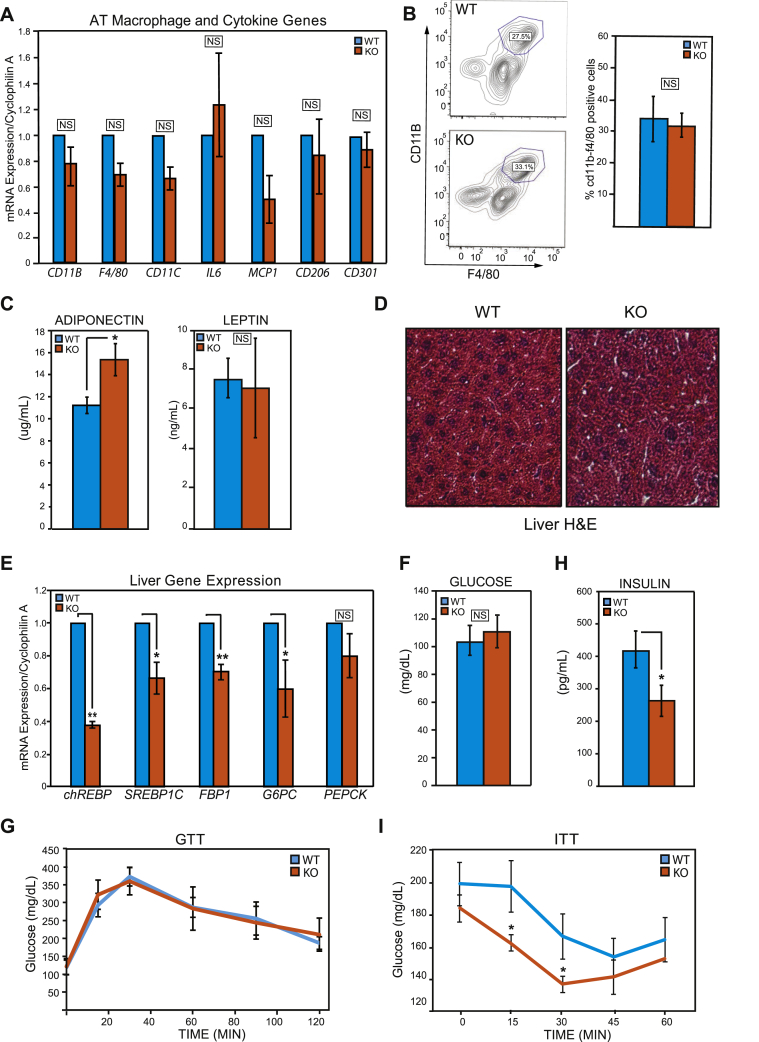
Sustained insulin signaling, improved lipid storing capacity of the adipose tissue, impaired adipose tissue lipolysis and enhanced DNL have all been associated with improved systemic insulin sensitivity [51], [59], [60], [61], [62], suggesting that adipo-specific GPS2 deletion might be beneficial for the organism. However, obesity is generally associated with chronic inflammation and with the development of insulin resistance [63], [64], [65]. To investigate how the removal of GPS2 and the corresponding enhanced insulin signaling in the adipose tissue affect the maintenance of systemic homeostasis, we first assessed the level of local inflammation in the adipose tissue by RT-qPCR and FACS analysis. Surprisingly, given GPS2 previously reported anti-inflammatory role and the obesity that characterize GPS2-AKO mice, no significant differences were observed in either the expression of gene markers of inflammation or in the amount of macrophages present within the adipose tissue (Figure 5A and B).
Figure 5.
Uncoupling of adipose tissue inflammation, obesity and insulin sensitivity in GPS2-AKO mice under chow diet. (A) RT-qPCR analysis of pro-inflammatory genes and macrophage markers in the WAT. (B) Macrophage infiltration in the WAT as measured by FACS analysis with quantification of f4/80 and CD11b positive cells. Statistical significance was calculated comparing 3 independent experiments. (C) Plasma adiponectin and leptin levels after an overnight fast, n = 7. (D) Representative H&E staining of paraffin-embedded liver sections imaged at 10× magnification. (E) Hepatic expression of gluconeogenic and de novo lipogenesis genes by RT-qPCR analysis. (F) Fasting blood glucose levels in AKO versus WT mice after overnight fast, n = 8. (G) Glucose tolerance test following glucose i.p. injection (1.5 mg/g of body weight) after an overnight fast, n = 4. (H) Plasma insulin levels in AKO versus WT mice after overnight fast, n = 5–7. Statistical significance was calculated comparing 3 independent experiments. (I) Insulin tolerance test after a 0.5 U/kg body weight insulin i.p. injection in AKO versus WT mice following a 4 h fast, n = 7–8. Results are expressed as mean ∓ SEM. Statistical significance was calculated by two-tailed Student's t test; *represents p value < 0.05 and **p value < 0.01.
Next, we sampled plasma from fasted WT and GPS2-AKO littermates for the presence of secreted adipokines. Plasma levels of the anti-inflammatory and insulin-sensitizing hormone adiponectin [66], [67] were found to be significantly elevated in GPS2-AKO mice despite the reported increase in body adiposity (Figure 5C), whereas no significant differences were observed in the amount of circulating leptin (Figure 5C). Consistent with an increase in circulating adiponectin and with adiponectin playing an important role in promoting the “healthy expansion” of the adipose tissue [68], [69], we did not observe any evidence of hepatic steatosis in the liver of GPS2-AKO mice despite their increase in total body fat (Figure 5D). Accordingly, the expression of both ChREBP and SREBP1c, key markers of DNL, were found downregulated in the liver of GPS2-AKO mice (Figure 5E). The expression of key gluconeogenic enzymes, such as G6Pase, FBP1, and PEPCK was also reduced in the liver of GPS2-deficient mice (Figure 5E). Furthermore, we sampled circulating free fatty acids (FFA) from GPS2-AKO and WT littermates and found a slight trend to decrease but no significant differences. While this result could initially appear surprising in the face of the decreased lipolysis, it has to be taken into consideration that in a context of increased obesity circulating FFA are usually elevated, thus maintenance of the baseline level actually reflects a decrease compared to the expected phenotype (Supplemental Figure S4A). In conclusion, these results indicate that despite the increase in adiposity, which typically correlates with metabolic dysfunction and peripheral tissue lipid deposition, GPS2 deletion in the adipose tissue appears to have a positive effect on whole body lipid metabolism with reduced spillover to non-adipose tissues such as the liver.
Improved lipid storing capacity of the adipose tissue can contribute to improved systemic insulin sensitivity [3], [4], [59], [60]. Blood glucose levels after overnight food deprivation were comparable between WT and GPS2-AKO matched littermate mice (Figure 5F), and glucose tolerance tests (GTT) were also normal (Figure 5G). However, plasma insulin levels under the same experimental fasting conditions were significantly reduced in GPS2-AKO mice (Figure 5H), consistent with our previous data that AKT is activated in vitro in basal conditions and in vivo under food deprivation upon GPS2 deletion/downregulation. To directly test the insulin sensitivity of GPS2-AKO mice, we injected matching cohorts of WT and GPS2-AKO littermates with a suboptimal dose of insulin after a short morning fast. Baseline insulin levels of GPS2-AKO mice tended to be lower under these experimental conditions as well (Supplemental Figure S4B), and insulin-induced glucose clearance was greater in GPS2-AKO mice than WT littermates (Figure 5I), confirming the predicted improvement in insulin sensitivity. In conclusion, taken together, our results indicate that enhancing insulin signaling in the adipose tissue, through GPS2 deletion and modulation of AKT ubiquitination status, has a positive effect on whole body insulin sensitivity despite the increased adiposity.
4. Discussion
Insulin action is critically required for promoting cell growth, regulating tissue development, and controlling whole body metabolism through modulation of glucose homeostasis and lipid metabolism. Its disruption or reduced functionality leads to the development of insulin resistance, a hallmark of Type 2 Diabetes and other metabolic disorders. The signaling mechanisms that mediate the physiological responses to insulin have been thoroughly studied with the identification of multiple transduction pathways, including both positive and negative regulators. Major players in these pathways are kinases, which are usually counter-regulated by opposing phosphatases (i.e. PI3K and PTEN). Our work has further extended this signaling network by revealing the existence of an additional level of regulation based on the opposing actions of an ubiquitin conjugating enzyme Ubc13 and its endogenous inhibitor GPS2. Insulin-mediated ubiquitination of AKT, via Ubc13-mediated synthesis of K63 ubiquitin chains, is an unexpected, critical step for the activation of downstream signaling events. Non-proteolytic ubiquitination of AKT had been previously reported for growth factor-mediated pathways, and a cancer-associated mutation that affects the ubiquitination of AKT1 (E17K) had been linked to AKT hyperactivation in breast and colon cancer. In this context, the gain of an additional lysine in the PH domain was found to promote enhanced AKT ubiquitination, constitutive recruitment to the membrane, and activation [14], [70], [71]. Interestingly, exome sequencing of patients with severe hypoglycemia but undetectable plasma insulin has recently led to the identification of a homologous activating mutation in AKT2 that might be similarly promoting enhanced ubiquitination. Overexpression of the E17K mutant in 3T3-L1 preadipocytes promoted insulin-independent localization of both AKT and GLUT4 to the plasma membrane and nuclear export of FoxO1, indicating constitutive activation of the AKT signaling pathway [72], [73]. These observations support our findings that regulation of AKT activation by ubiquitination is a conserved feature among the metabolic and growth signaling pathways mediated by different AKT isoforms. They also indicate that the non-proteolytic ubiquitination of AKT represents a key regulatory node for both adipose tissue and systemic homeostasis and metabolism.
In addition to revealing a role for K63 ubiquitination in the insulin pathway, our work has identified GPS2 as an endogenous inhibitor of this regulatory step. We previously characterized GPS2 as an inhibitor of Ubc13/TRAF2-6 activity in the context of pro-inflammatory pathways downstream of TNFα and TLR receptors [29]. Here, we confirm that a significant increase in JNK activation in primary adipocytes is observed upon GPS2 deletion. We also report the novel finding that GPS2 is a critical player in the insulin transduction pathway. In particular, our data reveal that physiological levels of GPS2 in adipocytes are required for preventing the constitutive ubiquitination of AKT, and therefore the restriction of its activation, as well as the activation of downstream signaling events, when insulin levels are low. In accord with these conclusions, we found that GPS2 deletion in mice adipose tissue results in constitutive ubiquitination and activation of AKT, leading to disrupted lipid metabolism and increased adiposity but also to elevated adiponectin levels. Interestingly, overexpression studies in mice indicate that increasing the level of secreted adiponectin contributes to mimicking a state of constant feeding and results in improved insulin sensitivity despite the substantial expansion of the adipose tissue [1], [51], [69]. Similarly, deletion of PTEN, a well-known inhibitor of AKT, promotes sustained insulin signaling and improved insulin sensitivity despite a significant increase in body weight [51]. Our results indicate that the absence of GPS2 similarly favors the retention of triglycerides in the adipose tissue, and thus improvement of insulin sensitivity in mice that become obese when fed a regular laboratory chow diet.
Interestingly, our previous work showed GPS2 playing an important anti-inflammatory role in the regulation of pro-inflammatory signaling pathways. The anti-inflammatory role of GPS2 was then confirmed by macrophage-specific and B-cell-specific deletion studies and by GPS2 overexpression in the adipose tissue (both adipocytes and macrophages) of DIO obese mice [29], [74] (Lentucci et al., under revision). In contrast, we report here that GPS2 adipo-specific deletion has a positive effect on adipocyte insulin signaling and systemic insulin sensitivity in chow-fed mice, and that, under these conditions, inflammation of the adipose tissue is undetectable in GPS2-AKO mice despite their increase in body fat. These observations are in agreement with a recent report that hyperinsulenimia can drive adipose tissue inflammation, whereas a reduction in circulating insulin levels causes a decrease in the expression of pro-inflammatory markers and macrophage expansion in the adipose tissue [75]. Thus, our findings reveal that GPS2 plays an intriguing role at the intersection between inflammatory and metabolic pathways, and suggest that sustained AKT activation through enhanced ubiquitination is likely to overcome the negative effect that the elevated levels of P-JNK might have on insulin signaling via IRS1 phosphorylation. Finally, it is important to note that there are layers to the phenotype associated with the loss of GPS2 function that might depend on the genetic background and the experimental conditions. For example, while the GPS2-AKO mice described here become obese even when fed a chow diet, similar mice generated by Fan and colleagues were not obese at baseline and did not appear to gain extra weight upon HFD feeding [74]. Thus, further studies that explore the interplay between increased activation of pro-inflammatory signaling pathways and enhanced insulin signaling in GPS2-AKO mice will be critical for a comprehensive understanding of the crosstalk between these pathways in condition of diet- or genetic-induced obesity. Interestingly, our results indicate that the synergistic roles played by GPS2 in the regulation of JNK activity and insulin signaling are mediated through a conserved mechanism based on the inhibition of Ubc13 enzymatic activity. This raises the interesting question of whether the ubiquitination machinery responsible for the synthesis of non-proteolytic K63 ubiquitin chains represents a key regulatory node between inflammation and lipid metabolism at the onset of obesity.
Lastly, it is worth noting that there is a large body of work describing GPS2 as a transcriptional cofactor, involved in mediating both gene repression, as part of the NCoR/SMRT complex, and activation through direct interaction with nuclear receptors and other TFs [31], [37], [39], [41], [42], [76]. For example, in the adipose tissue, downregulation of GPS2 was associated with the derepression of inflammatory target genes in human obese patients [32] and with the activation of key enzymes for triglyceride breakdown, HSL and ATGL, in murine differentiating adipocytes [34]. Here, we have confirmed a critical role for GPS2 in the regulation of adipocyte triglyceride turnover. However, the expression of HSL and ATGL was not impaired in GPS2-null primary adipocytes, indicating that lipolysis is likely suppressed due to the sustained activation of insulin signaling even under baseline or fasted conditions, rather than direct transcriptional effects. Thus, our previous and current findings, taken together, indicate that while GPS2 is essential for the early development of preadipocytes, its deletion at later stages does not affect differentiation but GPS2 remains essential for maintaining metabolic homeostasis in mature adipocytes. Intriguingly, increased adiposity and altered lipid mobilization have been previously reported also in the case of mice lacking a binding partner of GPS2, the transcriptional corepressor TBLR1. The phenotype observed upon TBLR1 deletion appears to similarly extend beyond the transcriptional regulation of HSL and ATGL, with TBLR1 being described as required for controlling multiple steps of the lipolytic cascade, including both changes in gene expression and a decrease in the phosphorylation of HSL and other PKA substrates [77]. This suggests that multiple components of the NCoR/SMRT corepressor complex might play complementary genomic and non-genomic functions in the coordinated regulation of cell homeostasis.
Acknowledgements
We thank past and present members of the Varelas, Farmer, Pilch, and Kandror labs for sharing reagents and providing useful suggestions and technical advices during the course of these studies. We are very grateful to Dr. Barbara Corkey for insightful discussions. We thank the BUMC Analytical Core (Dr. Matthew Au), the BUMC Mouse Metabolic Phenotyping Core (Dr. Thomas Balon), the BNORC Adipocyte Core (Drs. Susan Fried/Andrew Greenberg) and the BUMC FACS Core (Drs. Anna Belkina/Jennifer Snyder-Cappione) for their excellent services. CMC is an American Diabetes Association (ADA) fellow and CTC is supported by an NRSA Individual Predoctoral Fellowship from NIH/NIDDK (F31DK108571). This work was supported by awards to MDC from the Boston Nutrition and Obesity Center (BNORC - Pilot and Feasibility Award), and to VP from the Boston Nutrition and Obesity Center (BNORC - Pilot and Feasibility Award), Dr. Peter Paul (Career Development Award) and NIH/NIDDK (R01DK100422).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.10.007.
Conflict of interest
None declared.
Appendix ASupplementary data
The following are the supplementary data related to this article:
Figure S1.
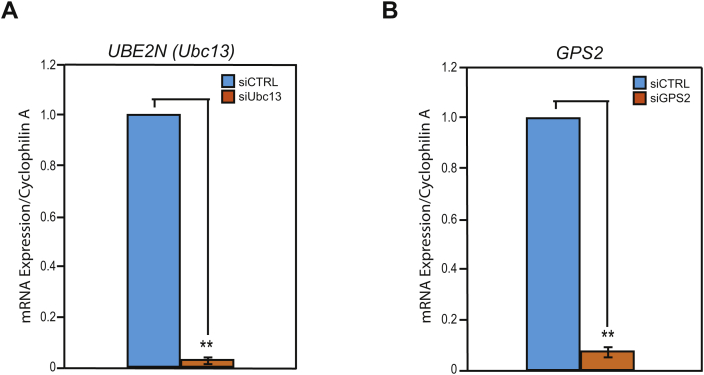
(A) siRNA validation by RT-qPCR of transiently transfected 3T3-L1 preadipocytes comparing siCTRL versus siUBC13 treated cells. (B) siRNA validation by RT-qPCR of transiently transfected 3T3-L1 preadipocytes comparing siCTRL versus siGPS2 treated cells. Results are expressed as mean ∓ SEM from 3 independent experiments and statistical significance was calculated comparing the 3 independent experiments. Statistical significance was calculated by two-tailed Student's t test; *represents p value < 0.05 and **p value < 0.01.
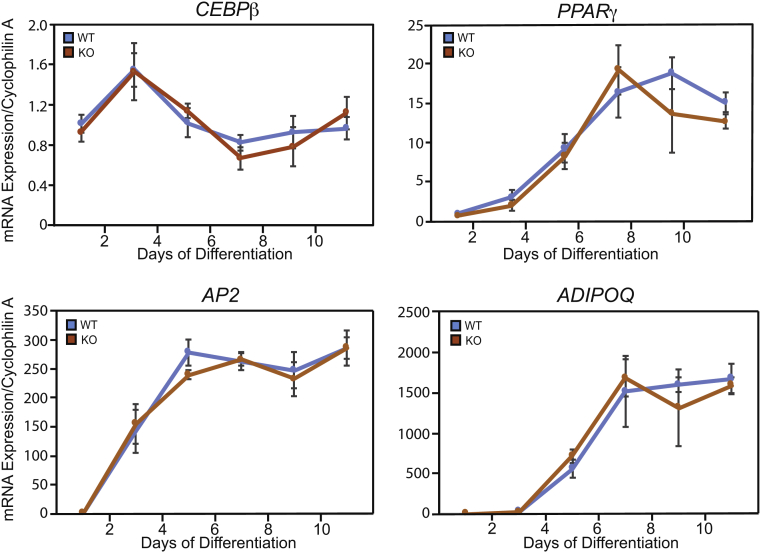
Figure S2.
RT-qPCR analysis of gene markers of adipogenesis in in vitro differentiated adipocytes from WT versus AKO mice at specified time points. Results are expressed as mean ∓ SEM. Statistical significance was calculated by two-tailed Student's t test; *represents p value < 0.05 and **p value < 0.01.
Figure S3.
(A) RT-qPCR analysis of ATGL and HSL, lipolysis markers, in WAT of AKO versus WT mice, n = 4–5. Results are expressed as mean ∓ SEM. (B) Western blot analysis of ATGL and HSL normalized to β-tubulin in whole cell extracts from WAT from AKO versus WT mice in fasting conditions. Statistical significance was calculated by two-tailed Student's t test; *represents p value < 0.05 and **p value < 0.01. All western blots are representative images of at least three independent experiments.
Figure S4.
(A) Plasma non esterified free fatty acid levels in AKO versus WT mice after overnight fast, n = 3. Results are expressed as mean ∓ SEM. (B) Plasma insulin levels in AKO versus WT mice after a 6 h fast, n = 3–6. Results are expressed as mean ∓ SEM. Statistical significance was calculated by two-tailed Student's t test and the p value (p) reported in the figure.
References
- 1.Stern J.H., Rutkowski J.M., Scherer P.E. Adiponectin, leptin, and fatty acids in the maintenance of metabolic homeostasis through adipose tissue crosstalk. Cell Metabolism. 2016;23(5):770–784. doi: 10.1016/j.cmet.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156(1–2):20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Virtue S., Vidal-Puig A. Adipose tissue expandability, lipotoxicity and the metabolic syndrome–an allostatic perspective. Biochimica et Biophysica Acta. 2010;1801(3):338–349. doi: 10.1016/j.bbalip.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Rutkowski J.M., Stern J.H., Scherer P.E. The cell biology of fat expansion. The Journal of Cell Biology. 2015;208(5):501–512. doi: 10.1083/jcb.201409063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czech M.P., Tencerova M., Pedersen D.J., Aouadi M. Insulin signalling mechanisms for triacylglycerol storage. Diabetologia. 2013;56(5):949–964. doi: 10.1007/s00125-013-2869-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaworski K., Sarkadi-Nagy E., Duncan R.E., Ahmadian M., Sul H.S. Regulation of triglyceride metabolism. IV. Hormonal regulation of lipolysis in adipose tissue. The American Journal of Physiology-Gastrointestinal and Liver Physiology. 2007;293(1):G1–G4. doi: 10.1152/ajpgi.00554.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Summers S.A., Whiteman E.L., Birnbaum M.J. Insulin signaling in the adipocyte. International Journal of Obesity and Related Metabolic Disorders. 2000;24(Suppl 4):S67–S70. doi: 10.1038/sj.ijo.0801509. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Z.Y., Zhou Q.L., Coleman K.A., Chouinard M., Boese Q., Czech M.P. Insulin signaling through Akt/protein kinase B analyzed by small interfering RNA-mediated gene silencing. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(13):7569–7574. doi: 10.1073/pnas.1332633100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Segrelles C., García-Escudero R., Garín M.I., Aranda J.F., Hernández P., Ariza J.M. Akt signaling leads to stem cell activation and promotes tumor development in epidermis. Stem Cells. 2014;32(7):1917–1928. doi: 10.1002/stem.1669. [DOI] [PubMed] [Google Scholar]
- 10.Blanco-Aparicio C., Pérez-Gallego L., Pequeño B., Leal J.F., Renner O., Carnero A. Mice expressing myrAKT1 in the mammary gland develop carcinogen-induced ER-positive mammary tumors that mimic human breast cancer. Carcinogenesis. 2007;28(3):584–594. doi: 10.1093/carcin/bgl190. [DOI] [PubMed] [Google Scholar]
- 11.De Vita G., Berlingieri M.T., Visconti R., Castellone M.D., Viglietto G., Baldassarre G. Akt/protein kinase B promotes survival and hormone-independent proliferation of thyroid cells in the absence of dedifferentiating and transforming effects. Cancer Research. 2000;60(14):3916–3920. [PubMed] [Google Scholar]
- 12.Cairns R.A., Harris I.S., Mak T.W. Regulation of cancer cell metabolism. Nature Reviews Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 13.Yang W.L., Wang J., Chan C.H., Lee S.W., Campos A.D., Lamothe B. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325(5944):1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W.L., Wu C.Y., Wu J., Lin H.K. Regulation of Akt signaling activation by ubiquitination. Cell Cycle. 2010;9(3):487–497. doi: 10.4161/cc.9.3.10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Risso G., Blaustein M., Pozzi B., Mammi P., Srebrow A. Akt/PKB: one kinase, many modifications. Biochemical Journal. 2015;468(2):203–214. doi: 10.1042/BJ20150041. [DOI] [PubMed] [Google Scholar]
- 16.Stringer D.K., Piper R.C. Terminating protein ubiquitination: Hasta la vista, ubiquitin. Cell Cycle. 2011;10(18):3067–3071. doi: 10.4161/cc.10.18.17191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagy V., Dikic I. Ubiquitin ligase complexes: from substrate selectivity to conjugational specificity. The Journal of Biological Chemistry. 2010;391(2–3):163–169. doi: 10.1515/bc.2010.021. [DOI] [PubMed] [Google Scholar]
- 18.Pickart C.M., Eddins M.J. Ubiquitin: structures, functions, mechanisms. Biochimica et Biophysica Acta. 2004;1695(1–3):55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 19.Ikeda F., Crosetto N., Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143(5):677–681. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto M., Okamoto T., Takeda K., Sato S., Sanjo H., Uematsu S. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nature Immunology. 2006;7(9):962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- 21.Petroski M.D., Zhou X., Dong G., Daniel-Issakani S., Payan D.G., Huang J. Substrate modification with lysine 63-linked ubiquitin chains through the UBC13-UEV1A ubiquitin-conjugating enzyme. The Journal of Biological Chemistry. 2007;282(41):29936–29945. doi: 10.1074/jbc.M703911200. [DOI] [PubMed] [Google Scholar]
- 22.Andersen P.L., Zhou H., Pastushok L., Moraes T., McKenna S., Ziola B. Distinct regulation of Ubc13 functions by the two ubiquitin-conjugating enzyme variants Mms2 and Uev1A. The Journal of Cell Biology. 2005;170(5):745–755. doi: 10.1083/jcb.200502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sowa M.E., Bennett E.J., Gygi S.P., Harper J.W. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138(2):389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katz E.J., Isasa M., Crosas B. A new map to understand deubiquitination. Biochemical Society Transactions. 2010;38(Pt 1):21–28. doi: 10.1042/BST0380021. [DOI] [PubMed] [Google Scholar]
- 25.Wiener R., Zhang X., Wang T., Wolberger C. The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature. 2012;483(7391):618–622. doi: 10.1038/nature10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim J.H., Jono H., Komatsu K., Woo C.H., Lee J., Miyata M. CYLD negatively regulates transforming growth factor-beta-signalling via deubiquitinating Akt. Nature Communications. 2012;3:771. doi: 10.1038/ncomms1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang W.L., Jin G., Li C.F., Jeong Y.S., Moten A., Xu D. Cycles of ubiquitination and deubiquitination critically regulate growth factor-mediated activation of Akt signaling. Science Signaling. 2013;6(257):ra3. doi: 10.1126/scisignal.2003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S., Juang Y.C. Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature. 2010;466(7309):941–946. doi: 10.1038/nature09297. [DOI] [PubMed] [Google Scholar]
- 29.Cardamone M.D., Krones A., Tanasa B., Taylor H., Ricci L., Ohgi K.A. A protective strategy against hyperinflammatory responses requiring the nontranscriptional actions of GPS2. Molecular Cell. 2012;46(1):91–104. doi: 10.1016/j.molcel.2012.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spain B.H., Bowdish K.S., Pacal A.R., Staub S.F., Koo D., Chang C.Y. Two human cDNAs, including a homolog of Arabidopsis FUS6 (COP11), suppress G-protein- and mitogen-activated protein kinase-mediated signal transduction in yeast and mammalian cells. Molecular and Cellular Biology. 1996;16(12):6698–6706. doi: 10.1128/mcb.16.12.6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakobsson T., Venteclef N., Toresson G., Damdimopoulos A.E., Ehrlund A., Lou X. GPS2 is required for cholesterol efflux by triggering histone demethylation, LXR recruitment, and coregulator assembly at the ABCG1 locus. Molecular Cell. 2009;34(4):510–518. doi: 10.1016/j.molcel.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 32.Toubal A., Clement K., Fan R., Ancel P., Pelloux V., Rouault C. SMRT-GPS2 corepressor pathway dysregulation coincides with obesity-linked adipocyte inflammation. The Journal of Clinical Investigation. 2013;123(1):362–379. doi: 10.1172/JCI64052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venteclef N., Jakobsson T., Ehrlund A., Damdimopoulos A., Mikkonen L., Ellis E. GPS2-dependent corepressor/SUMO pathways govern anti-inflammatory actions of LRH-1 and LXRbeta in the hepatic acute phase response. Genes & Development. 2010;24(4):381–395. doi: 10.1101/gad.545110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardamone M.D., Tanasa B., Chan M., Cederquist C.T., Andricovich J., Rosenfeld M.G. GPS2/KDM4A pioneering activity regulates promoter-specific recruitment of PPARgamma. Cell Reports. 2014;8(1):163–176. doi: 10.1016/j.celrep.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng Y.C., Kuo F., Breiding D.E., Wang Y.F., Mansur C.P., Androphy E.J. AMF1 (GPS2) modulates p53 transactivation. Molecular and Cellular Biology. 2001;21(17):5913–5924. doi: 10.1128/MCB.21.17.5913-5924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng Y.C., Breiding D.E., Sverdrup F., Richard J., Androphy E.J. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. Journal of Virology. 2000;74(13):5872–5879. doi: 10.1128/jvi.74.13.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanyal S., Bavner A., Haroniti A., Nilsson L.M., Lundasen T., Rehnmark S. Involvement of corepressor complex subunit GPS2 in transcriptional pathways governing human bile acid biosynthesis. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(40):15665–15670. doi: 10.1073/pnas.0706736104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee T.H., Yi W., Griswold M.D., Zhu F., Her C. Formation of hMSH4-hMSH5 heterocomplex is a prerequisite for subsequent GPS2 recruitment. DNA Repair (Amst) 2006;5(1):32–42. doi: 10.1016/j.dnarep.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 39.Zhang J., Kalkum M., Chait B.T., Roeder R.G. The N-CoR-HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Molecular Cell. 2002;9(3):611–623. doi: 10.1016/s1097-2765(02)00468-9. [DOI] [PubMed] [Google Scholar]
- 40.Zhang D., Harry G.J., Blackshear P.J., Zeldin D.C. G-protein pathway suppressor 2 (GPS2) interacts with the regulatory factor X4 variant 3 (RFX4_v3) and functions as a transcriptional co-activator. The Journal of Biological Chemistry. 2008;283(13):8580–8590. doi: 10.1074/jbc.M708209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo C., Li Y., Gow C.H., Wong M., Zha J., Yan C. The optimal corepressor function of nuclear receptor corepressor (NCoR) for peroxisome proliferatoractivated receptor gamma requires G-protein pathway suppressor 2. The Journal of Biological Chemistry. 2015;290(6):3666–3679. doi: 10.1074/jbc.M114.598797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng X., Kao H.Y. G protein pathway suppressor 2 (GPS2) is a transcriptional corepressor important for estrogen receptor alpha-mediated transcriptional regulation. The Journal of Biological Chemistry. 2009;284(52):36395–36404. doi: 10.1074/jbc.M109.062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eguchi J., Wang X., Yu S., Kershaw E.E., Chiu P.C., Dushay J. Transcriptional control of adipose lipid handling by IRF4. Cell Metabolism. 2011;13(3):249–259. doi: 10.1016/j.cmet.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ayala J.E., Samuel V.T., Morton G.J., Obici S., Croniger C.M., Shulman G.I. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Disease Models & Mechanisms. 2010;3(9–10):525–534. doi: 10.1242/dmm.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parlee S.D., Lentz S.I., Mori H., MacDougald O.A. Quantifying size and number of adipocytes in adipose tissue. Methods in Enzymology. 2014;537:93–122. doi: 10.1016/B978-0-12-411619-1.00006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berry R., Church C.D., Gericke M.T., Jeffery E., Colman L., Rodeheffer M.S. Imaging of adipose tissue. Methods in Enzymology. 2014;537:47–73. doi: 10.1016/B978-0-12-411619-1.00004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan C.H., Li C.F., Yang W.L., Gao Y., Lee S.W., Feng Z. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149(5):1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kohn A.D., Summers S.A., Birnbaum M.J., Roth R.A. Expression of a constitutively active Akt Ser/Thr kinase in 3T3-L1 adipocytes stimulates glucose uptake and glucose transporter 4 translocation. The Journal of Biological Chemistry. 1996;271(49):31372–31378. doi: 10.1074/jbc.271.49.31372. [DOI] [PubMed] [Google Scholar]
- 49.Pearce L.R., Komander D., Alessi D.R. The nuts and bolts of AGC protein kinases. Nature Reviews Molecular Cell Biology. 2010;11(1):9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 50.Li W., Peng C., Lee M.H., Lim D., Zhu F., Fu Y. TRAF4 is a critical molecule for Akt activation in lung cancer. Cancer Research. 2013;73(23):6938–6950. doi: 10.1158/0008-5472.CAN-13-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morley T.S., Xia J.Y., Scherer P.E. Selective enhancement of insulin sensitivity in the mature adipocyte is sufficient for systemic metabolic improvements. Nature Communications. 2015;6:7906. doi: 10.1038/ncomms8906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bluher M., Michael M.D., Peroni O.D., Ueki K., Carter N., Kahn B.B. Adipose tissue selective insulin receptor knockout protects against obesity and obesity-related glucose intolerance. Developmental Cell. 2002;3(1):25–38. doi: 10.1016/s1534-5807(02)00199-5. [DOI] [PubMed] [Google Scholar]
- 53.Duncan R.E., Ahmadian M., Jaworski K., Sarkadi-Nagy E., Sul H.S. Regulation of lipolysis in adipocytes. Annual Review of Nutrition. 2007;27:79–101. doi: 10.1146/annurev.nutr.27.061406.093734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choi S.M., Tucker D.F., Gross D.N., Easton R.M., DiPilato L.M., Dean A.S. Insulin regulates adipocyte lipolysis via an Akt-independent signaling pathway. Molecular and Cellular Biology. 2010;30(21):5009–5020. doi: 10.1128/MCB.00797-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kersten S. Mechanisms of nutritional and hormonal regulation of lipogenesis. EMBO Reports. 2001;2(4):282–286. doi: 10.1093/embo-reports/kve071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong R.H., Sul H.S. Insulin signaling in fatty acid and fat synthesis: a transcriptional perspective. Current Opinion in Pharmacology. 2010;10(6):684–691. doi: 10.1016/j.coph.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang Y., Wallace M., Sanchez-Gurmaches J., Hsiao W.Y., Li H., Lee P.L. Adipose tissue mTORC2 regulates ChREBP-driven de novo lipogenesis and hepatic glucose metabolism. Nature Communications. 2016;7:11365. doi: 10.1038/ncomms11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berggreen C., Gormand A., Omar B., Degerman E., Goransson O. Protein kinase B activity is required for the effects of insulin on lipid metabolism in adipocytes. American Journal of Physiology – Endocrinology and Metabolism. 2009;296(4):E635–E646. doi: 10.1152/ajpendo.90596.2008. [DOI] [PubMed] [Google Scholar]
- 59.Chen H.C., Stone S.J., Zhou P., Buhman K.K., Farese R.V., Jr. Dissociation of obesity and impaired glucose disposal in mice overexpressing acyl coenzyme a:diacylglycerol acyltransferase 1 in white adipose tissue. Diabetes. 2002;51(11):3189–3195. doi: 10.2337/diabetes.51.11.3189. [DOI] [PubMed] [Google Scholar]
- 60.Shepherd P.R., Gnudi L., Tozzo E., Yang H., Leach F., Kahn B.B. Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. The Journal of Biological Chemistry. 1993;268(30):22243–22246. [PubMed] [Google Scholar]
- 61.Herman M.A., Peroni O.D., Villoria J., Schon M.R., Abumrad N.A., Bluher M. A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature. 2012;484(7394):333–338. doi: 10.1038/nature10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yore M.M., Syed I., Moraes-Vieira P.M., Zhang T., Herman M.A., Homan E.A. Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell. 2014;159(2):318–332. doi: 10.1016/j.cell.2014.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hotamisligil G.S. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- 64.Olefsky J.M., Glass C.K. Macrophages, inflammation, and insulin resistance. Annual Review of Physiology. 2010;72:219–246. doi: 10.1146/annurev-physiol-021909-135846. [DOI] [PubMed] [Google Scholar]
- 65.Chawla A., Nguyen K.D., Goh Y.P. Macrophage-mediated inflammation in metabolic disease. Nature Reviews Immunology. 2011;11(11):738–749. doi: 10.1038/nri3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lihn A.S., Pedersen S.B., Richelsen B. Adiponectin: action, regulation and association to insulin sensitivity. Obesity Reviews. 2005;6(1):13–21. doi: 10.1111/j.1467-789X.2005.00159.x. [DOI] [PubMed] [Google Scholar]
- 67.Okamoto Y., Kihara S., Funahashi T., Matsuzawa Y., Libby P. Adiponectin: a key adipocytokine in metabolic syndrome. Clinical Science (London) 2006;110(3):267–278. doi: 10.1042/CS20050182. [DOI] [PubMed] [Google Scholar]
- 68.Xu A., Wang Y., Keshaw H., Xu L.Y., Lam K.S., Cooper G.J. The fat-derived hormone adiponectin alleviates alcoholic and nonalcoholic fatty liver diseases in mice. Journal of Clinical Investigation. 2003;112(1):91–100. doi: 10.1172/JCI17797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J.Y., van de Wall E., Laplante M., Azzara A., Trujillo M.E., Hofmann S.M. Obesity-associated improvements in metabolic profile through expansion of adipose tissue. Journal of Clinical Investigation. 2007;117(9):2621–2637. doi: 10.1172/JCI31021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carpten J.D., Faber A.L., Horn C., Donoho G.P., Briggs S.L., Robbins C.M. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448(7152):439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 71.Kim M.S., Jeong E.G., Yoo N.J., Lee S.H. Mutational analysis of oncogenic AKT E17K mutation in common solid cancers and acute leukaemias. British Journal of Cancer. 2008;98(9):1533–1535. doi: 10.1038/sj.bjc.6604212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hussain K., Challis B., Rocha N., Payne F., Minic M., Thompson A. An activating mutation of AKT2 and human hypoglycemia. Science. 2011;334(6055):474. doi: 10.1126/science.1210878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonzalez E., McGraw T.E. Insulin-modulated Akt subcellular localization determines Akt isoform-specific signaling. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(17):7004–7009. doi: 10.1073/pnas.0901933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fan R., Toubal A., Goni S., Drareni K., Huang Z., Alzaid F. Loss of the co-repressor GPS2 sensitizes macrophage activation upon metabolic stress induced by obesity and type 2 diabetes. Nature Medicine. 2016;22(7):780–791. doi: 10.1038/nm.4114. [DOI] [PubMed] [Google Scholar]
- 75.Pedersen D.J., Guilherme A., Danai L.V., Heyda L., Matevossian A., Cohen J. A major role of insulin in promoting obesity-associated adipose tissue inflammation. Molecular Metabolism. 2015;4(7):507–518. doi: 10.1016/j.molmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Oberoi J., Fairall L., Watson P.J., Yang J.C., Czimmerer Z., Kampmann T. Structural basis for the assembly of the SMRT/NCoR core transcriptional repression machinery. Nature Structural & Molecular Biology. 2011;18(2):177–184. doi: 10.1038/nsmb.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rohm M., Sommerfeld A., Strzoda D., Jones A., Sijmonsma T.P., Rudofsky G. Transcriptional cofactor TBLR1 controls lipid mobilization in white adipose tissue. Cell Metabolism. 2013;17(4):575–585. doi: 10.1016/j.cmet.2013.02.010. [DOI] [PubMed] [Google Scholar]