Abstract
Objective
Long noncoding RNAs (lncRNAs) are emerging as important regulators of diverse biological processes. Recent work has demonstrated that the inducible lncRNA Blnc1 stimulates thermogenic gene expression during brown and beige adipocyte differentiation. However, whether Blnc1 is functionally conserved in humans has not been explored. In addition, the molecular basis of the Blnc1 ribonucleoprotein complex in thermogenic gene induction remains incompletely understood. The aims of the current study were to: i) investigate functional conservation of Blnc1 in mice and humans and ii) elucidate the molecular mechanisms by which Blnc1 controls the thermogenic gene program in brown adipocytes.
Methods
Full-length human Blnc1 was cloned and examined for its ability to stimulate brown adipocyte differentiation. Different truncation mutants of Blnc1 were generated to identify functional RNA domains responsible for thermogenic gene induction. RNA-protein interaction studies were performed to delineate the molecular features of the Blnc1 ribonucleoprotein complex.
Results
Blnc1 is highly conserved in mice and humans at the sequence and function levels, both capable of stimulating brown adipocyte gene expression. A conserved RNA domain was identified to be required and sufficient for the biological activity of Blnc1. We identified hnRNPU as an RNA-binding protein that facilitates the assembly and augments the transcriptional function of the Blnc1/EBF2 ribonucleoprotein complex.
Conclusions
Blnc1 is a conserved lncRNA that promotes thermogenic gene expression in brown adipocytes through formation of the Blnc1/hnRNPU/EBF2 ribonucleoprotein complex.
Keywords: Brown adipocyte differentiation, Brown fat, Thermogenesis, Blnc1, lncRNA, EBF2
Abbreviations: BAT, brown adipose tissue; RACE, rapid amplification of cDNA ends; Ucp1, uncoupling protein 1; Elovl3, elongation of very long chain fatty acids like 3; Cox7a1, cytochrome c oxidase subunit 7A1; Ppargc1a, peroxisome proliferator-activated receptor gamma coactivator 1-alpha; Pparα, peroxisome proliferator-activated receptor alpha; Prdm16, PR domain zinc finger protein 16; Dio2, deiodinase, iodothyronine type II; PPARγ, peroxisome proliferator-activated receptor gamma; FABP4, fatty acid binding protein 4; EBF2, early B cell factor 2; ATP5A, ATP synthase, H+ transporting, mitochondrial F1 complex, alpha 1; UQCRC2, ubiquinol-cytochrome c reductase core protein II; SDHB, succinate dehydrogenase complex iron sulfur subunit B
Highlights
-
•
The long noncoding RNA Blnc1 is conserved between mice and humans.
-
•
Human Blnc1 stimulates thermogenic gene expression during brown adipocyte differentiation.
-
•
hnRNPU physically interacts with human and mouse Blnc1.
-
•
hnRNPU promotes the assembly and function of the Blnc1/EBF2 ribonucleoprotein complex.
1. Introduction
Adipose tissues play a central role in nutrient and energy homeostasis. White adipose tissue stores energy and plays an important role in endocrine signaling and crosstalk with the immune system [1], [2], [3], [4]. Brown adipose tissue (BAT) contains high mitochondrial content and generates heat via uncoupling protein 1 (UCP1) [5], [6], [7], [8]. Genetic ablation of brown fat renders mice cold-sensitive and prone to the development of obesity [9], whereas activation of brown fat thermogenesis has been linked to increased energy expenditure, reduced adiposity, and lower plasma lipids [10], [11], [12]. We recently demonstrated that brown fat secretes endocrine factors, such as Neuregulin 4, and exerts its effects on metabolic physiology in part through mechanisms independent of UCP1-mediated thermogenesis [13], [14]. These findings support the concept that augmenting brown fat abundance and/or function may provide a potentially highly effective treatment for obesity and its associated metabolic disorders [15], [16], [17].
Previous studies have demonstrated that brown fat is present in adult humans [18], [19], [20], [21], and appears highly responsive to physiological and environmental stimuli [12], [22], [23]. Human brown fat shares key molecular and metabolic characteristics with the classical rodent BAT yet appears to contain both classical and brown-like adipocytes, termed beige/brite adipocytes [24]. Brown and beige adipogenesis is controlled by the concerted action of neural and hormonal signals that culminate in the transcriptional activation of the gene program responsible for fuel oxidation and thermogenesis. A number of transcription factors have been identified to regulate the determination and differentiation of brown and beige adipocytes [7], [25]. In addition, several microRNAs have been demonstrated to regulate different aspects of thermogenic adipocyte development and function [26].
Long noncoding RNAs (lncRNAs) are a unique class of transcripts that share similarities with mRNA with regard to their transcriptional regulation and biogenesis, but lack protein-coding potential [27]. Chromatin immunoprecipitation (ChIP) sequencing and RNA sequencing studies revealed that lncRNA genes are highly regulated and share epigenetic features with protein-coding genes. LncRNAs have been shown to regulate diverse biological processes, including transcriptional regulation, cell differentiation, tissue development, and tumorigenesis/metastasis [28], [29], [30], [31], [32], [33], [34], [35]. LncRNAs exert their effects on cell signaling and gene expression through physical interaction with protein factors and/or modulating microRNAs function. Several lncRNAs have been implicated in the regulation of endocrine signaling, metabolic tissue development, and lipid metabolism [36], [37]. We previously identified Blnc1 as a highly inducible lncRNA that promotes brown and beige adipocyte differentiation [38]. Another lncRNA named lnc-BATE1 was reported to regulate brown adipogenesis [39]. At the molecular level, Blnc1 physically associates with EBF2, a transcription factor that controls brown and beige adipocyte identity and differentiation [40], [41], thereby forming a feedforward loop to drive thermogenic adipocyte differentiation. Despite this, whether Blnc1 is functionally conserved in humans has not been explored. In addition, the molecular basis of the Blnc1 ribonucleoprotein complex in thermogenic gene induction remains to be elucidated. In this study, we demonstrated that Blnc1 is a functionally conserved lncRNA that stimulates thermogenic gene expression via the Blnc1/hnRNPU/EBF2 ribonucleoprotein complex.
2. Materials and methods
2.1. Adipocyte differentiation
Brown preadipocytes were immortalized using an established protocol [42], maintained in DMEM supplemented with 10% fetal bovine serum (FBS), and subjected to adipocyte differentiation, as previously described [43], [44]. Briefly, confluent preadipocyte culture was switched to differentiation medium (DMEM, 10% FBS, 20 nM insulin, and 1 nM T3) containing 0.5 mM IBMX, 125 μM indomethacin, and 1 μM dexamethasone for a period of 2 days. The cells were subsequently maintained in differentiation medium for up to 6 days. Differentiated brown adipocytes were treated with vehicle or isoproterenol (Iso) for 4 h before RNA isolation or 6 h before protein lysate preparation. Mitochondrial content and lipid accumulation were examined following staining with MitoTracker and Oil Red O, respectively.
2.2. Plasmid constructs
The streptavidin-binding RNA aptamer (StA; 5′-ACCGACCAGAATCATGC AAGTGCGTAAGATAGTCGCGGGCCGGG-3′) was previously described [45], [46]. The StA tag was fused 5′ end to mouse Blnc1 cDNA for affinity precipitation of the Blnc1 ribonucleoprotein complexes. Mouse Blnc1 truncation mutants were generated by PCR. T1 mutant contains nucleotide 1–559 of mBlnc1 cDNA. T2 and T3 mutants contain a deletion of 252–356 and 351–534 of mBlnc1 cDNA, respectively. These truncation mutants were cloned into the pMSCV retroviral vector. hBlnc1 cDNA was amplified from human liver using Rapid Amplification of cDNA End (RACE). For RNA Domain 1 vectors, mBlnc1 RD1 (560–965) and hBlnc1 RD1 (269–675) were amplified from corresponding full-length cDNA. For hnRNPU knockdown, two different shRNA sequences were designed (#1: ACAGAAAGGTGGAGATAAA and #2: GGGAGAAGTTTGATGAAAA) and cloned into pMSCV retroviral vector.
2.3. Mitochondrial DNA content and respiration
Total DNA was isolated from differentiated brown adipocytes for the quantitation of mitochondrial DNA content. Real-time qPCR primers for mitochondrial DNA (mtND1 and mtCox1) and nuclear DNA (PECAM) are listed in Supplementary Figure S3. Mitochondrial oxygen consumption rate in differentiated brown adipocytes was measured using the Mitocell MT200 miniature respirometer (Strathkelvin Instruments). Briefly, adipocytes were resuspended in 400 μL differentiation medium and transferred into the chamber. Oxygen levels in the mixing chamber were recorded for 5 min followed by FCCP injection (25 μM). Oxygen consumption rate was analyzed using the software provided by the manufacturer (782 Oxygen System version 4.0) and normalized to total protein content.
2.4. Gene expression analyses
Sybr Green-based qPCR was performed in QuantStudio™ 6 Flex Real-Time PCR System. The ribosomal protein 36B4 (Rplp0) was used as a normalization control. For immunoblotting, total cell lysates were prepared in a lysis buffer containing 50 mM Tris (pH = 7.5), 137 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100, and freshly added protease inhibitors. Protein samples were separated by SDS-PAGE and transferred onto PVDF membrane. The membrane was blocked with 5% milk in 1× TBST, incubated with primary and secondary antibodies, and visualized using enhanced chemiluminescence. Primary antibodies against UCP1 (Alpha Diagnosis), PPARγ, Flag-tag (Santa Cruz Biotechnology), FABP4 (Cell Signaling Technology), mitochondrial OXPHOS proteins (MitoSciences), and Myc-tag and Tubulin (Sigma–Aldrich) were used.
2.5. RNA-protein interaction assays
HEK293 cells cultured on 6-well plates were transiently transfected with Flag-hnRNPU, Myc-EBF2, and Blnc1 alone or in combinations. Total lysates were prepared in a lysis buffer that contains 20 mM Tris (pH = 7.5), 100 mM KCl, 5 mM MgCl2, 1% NP40, and freshly added protease inhibitors and RNAse inhibitor (S1402S, New England Biolabs). The lysates were immunoprecipitated using anti-Flag or anti-Myc agarose beads for 90 min at 4 °C, followed by washing with wash buffer (50 mM Tris (pH = 7.5), 150 mM NaCl, 1 mM MgCl2, 0.5% NP40) three times. For IP/qPCR, RNA remaining on the affinity beads was extracted using Trizol, treated with RNase-free DNase, and analyzed by qPCR. For RNA pull-down, total lysates were incubated with Streptavidin agarose beads. After washing, proteins associated with agarose beads were analyzed by immunoblotting.
2.6. Statistics
Data were analyzed using two-tailed Student's t-test. A p-value of less than 0.05 was considered statistically significant.
3. Results
3.1. Identification of the human Blnc1 ortholog
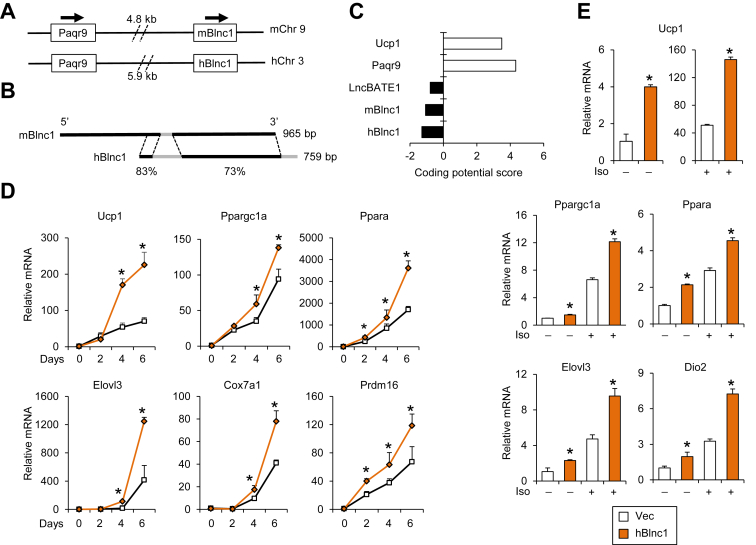
The extent to which lncRNAs are functionally conserved among species remains largely unknown. We have shown that mouse Blnc1 is located 3′ to the protein-coding gene Progestin and AdipoQ Receptor family member 9 (Paqr9) on chromosome 9 [38]. Blnc1 is independently transcribed from its own promoter and undergoes polyadenylation (Figure 1A). To explore whether Blnc1 is conserved between mice and humans, we analyzed genomic DNA sequence adjacent to human Paqr9; the latter is located in a syntenic region on human chromosome 3. We identified two segments that share high sequence similarity to mouse Blnc1 (mBlnc1). This genomic sequence conservation suggests that the ortholog of mBlnc1 may be present in the human genome and actively transcribed. We performed 5′ and 3′ RACE to obtain the full-length cDNA sequence of human Blnc1 (hBlnc1) (Figure S1). Sequence alignment analysis indicated that hBlnc1 and mBlnc1 contain two highly conserved segments, sharing 83% and 73% sequence identity, respectively (Figure 1B and Figure S2). Mouse Blnc1 appears to have a 5′ end extension that is absent in its human counterpart. We performed coding potential analysis using Coding Potential Calculator (CPC), an algorithm that can discriminate protein-coding from noncoding transcripts with high accuracy (http://cpc.cbi.pku.edu.cn) [47]. Our analysis indicated that, like the known lncRNAs mBlnc1 and lnc-BATE1, hBlnc1 was predicted to be a noncoding transcript (Figure 1C). In contrast, Ucp1 and Paqr9 were predicted to be protein-coding genes. Similar results were obtained using Coding Potential Assessing Tool (http://lilab.research.bcm.edu/cpat/) [48].
Figure 1.
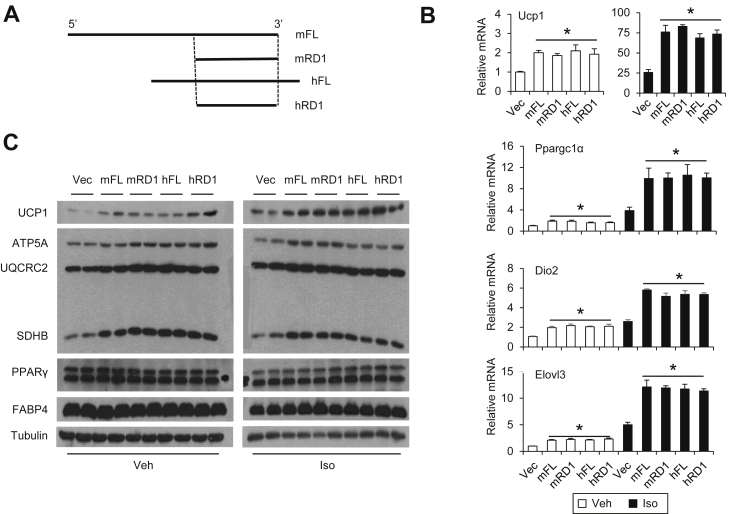
Role of human Blnc1 in brown adipogenesis. (A) Genomic structure of the mouse and human Paqr9/Blnc1 locus (not drawn to scale). (B) Sequence conservation between mBlnc1 and hBlnc1. (C) Coding potential analysis using CPC. Note that negative values reflect a low probability of protein-coding. (D) qPCR analyses of gene expression during differentiation of brown preadipocytes transduced with vector (open) or hBlnc1 (brown) retrovirus. (E) Gene expression of differentiated brown adipocytes treated without (−) or with (+) isoproterenol for 4 h. Data represent mean ± sd. *p < 0.05, hBlnc1 vs. vector.
3.2. Conserved function of human Blnc1 in thermogenic gene induction
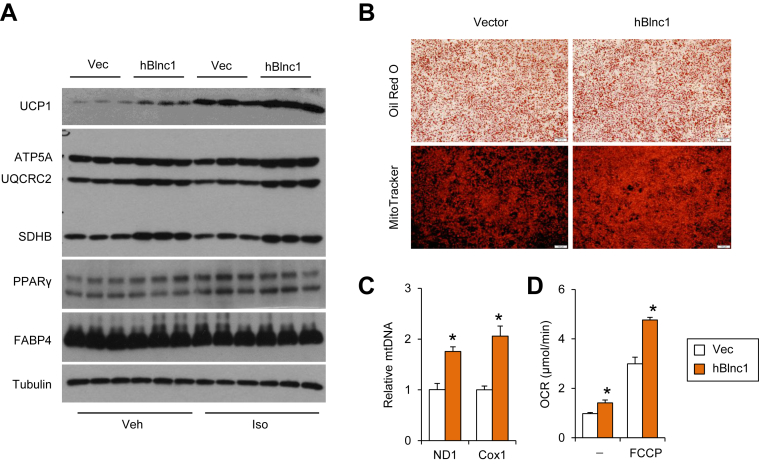
To determine whether hBlnc1 is functionally conserved in the regulation of thermogenic gene expression, we transduced immortalized mouse brown preadipocytes with control or a retroviral vector expressing hBlnc1 and performed gene expression analysis during differentiation. Compared to control, adipocytes overexpressing hBlnc1 exhibited significantly increased mRNA expression of Ucp1, Elovl3, and Cox7a1, genes associated with brown and beige adipocyte thermogenesis (Figure 1D). Interestingly, the mRNA levels of several transcriptional regulators of thermogenic gene program, including Ppargc1a, Pparα, and Prdm16, were also significantly increased by hBlnc1. We performed adrenergic stimulation with isoproterenol (Iso) in differentiated brown adipocytes and assessed hormonal induction of thermogenic gene expression. As shown in Figure 1E, mRNA expression of Ucp1, Elovl3, Dio2, Ppargc1a, and Pparα was induced by hBlnc1 both at the basal state and following Iso treatment. Protein expression of UCP1 and mitochondrial OXPHOS proteins was also increased by hBlnc1 (Figure 2A). In contrast, expression of the adipogenic markers PPARγ and FABP4 appeared similar in two groups. While hBlnc1 overexpression had a modest effect on lipid accumulation in differentiated brown adipocytes, it markedly increased mitochondrial content, as revealed by MitoTracker staining and mitochondrial DNA measurements (Figure 2B–C). Further, oxygen consumption studies indicated that hBlnc1 increased respiratory capacity in transduced brown adipocytes (Figure 2D). We conclude from these results that hBlnc1 is functionally conserved in its regulation of brown adipocyte gene expression.
Figure 2.
Human Blnc1 augments mitochondrial respiration in brown adipocytes. (A) Immunoblots of total lysates from brown adipocytes treated with vehicle (Veh) or isoproterenol (Iso) for 6 h. (B) Oil Red O and MitoTracker staining of differentiated brown adipocytes. Scale bar = 100 μm. (C) Mitochondrial DNA content measured by qPCR in brown adipocytes overexpressing vector (open) and hBlnc1 (brown). (D) Oxygen consumption rate (OCR) in the absence or presence of FCCP. Data represent mean ± sd. *p < 0.05, hBlnc1 vs. vector.
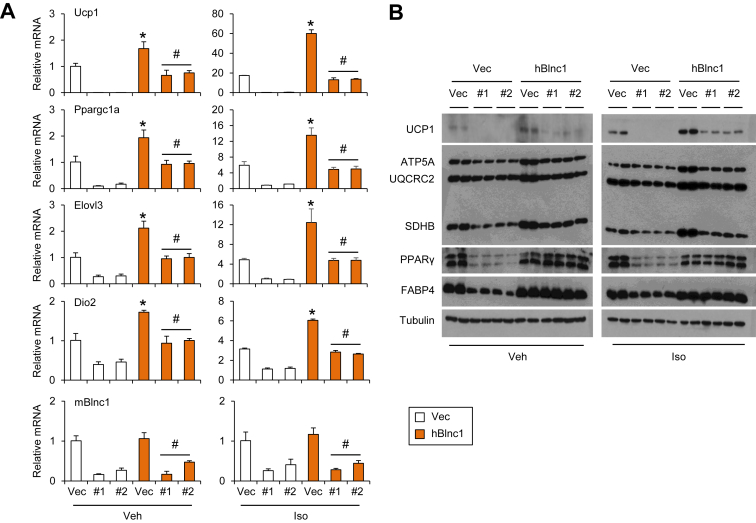
3.3. hBlnc1 rescues RNAi knockdown of mBlnc1
To further assess the extent of functional conservation between hBlnc1 and mBlnc1, we performed rescue studies in mouse brown preadipocytes stably expressing control or shRNA targeting mBlnc1. Consistent with previous studies [38], RNAi knockdown of mBlnc1 severely impaired the induction of thermogenic genes during differentiation (Figure 3A). Compared to control, mRNA expression of Ucp1, Elovl3, Dio2, and Pparα was markedly reduced by retroviral expression of shRNA targeting mBlnc1. Retroviral hBlnc1 overexpression returned mRNA expression of these genes back to control levels. Similar results were observed on protein expression of UCP1 and mitochondrial OXPHOS genes (Figure 3B). The ability of hBlnc1 to rescue the defects of thermogenic gene expression as a result of RNAi knockdown of mBlnc1 indicated that they are functionally conserved in the context of brown adipocyte differentiation.
Figure 3.
Human Blnc1 rescues RNAi knockdown of mouse Blnc1. (A) Differentiated brown adipocytes expressing control (Vec) or shRNA targeting mBlnc1 (#1 and #2) were treated with vehicle (Veh) or isoproterenol (Iso) for 4 h. Gene expression was analyzed by qPCR. (B) Immunoblots of brown adipocyte lysates. Data represent mean ± sd. *p < 0.05, hBlnc1 vs. Vec; #p < 0.05, hBlnc1/siBlnc1 vs. Vec/siBlnc1.
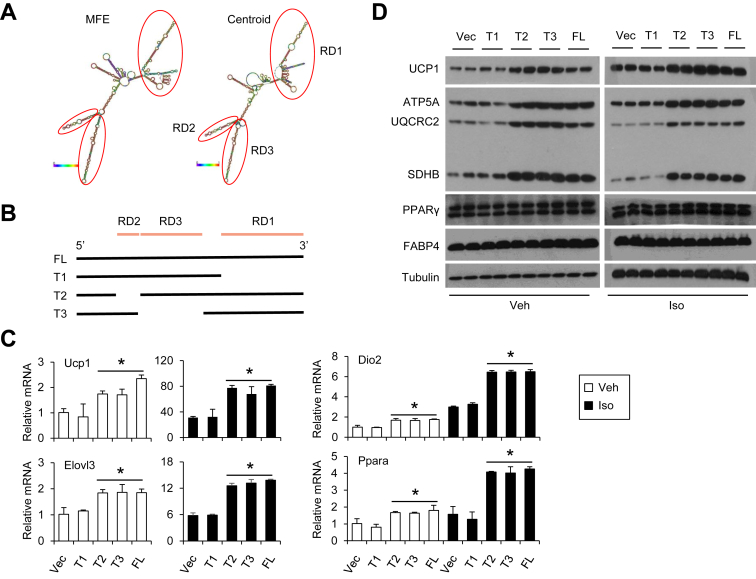
3.4. Functional role of the Blnc1 RNA domains
LncRNAs are known to form complex secondary structure as a result of base pairing. We performed secondary structure prediction for mBlnc1 using the RNAfold web server (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). Both minimum free energy (MFE) and Centroid algorithms predicted mBlnc1 to form several stem-loop structures, as indicated by RNA Domain 1 (RD1), RD2, and RD3 (Figure 4A). Among these, RD1 represents the conserved region between mBlcn1 and hBlnc1. To dissect the functional importance of these three RDs, we generated mBlnc1 truncation mutants that lack each of these domains, denoted T1, T2, and T3 (Figure 4B), and assayed their ability to promote brown adipocyte differentiation. As expected, retroviral overexpression of full-length mBlnc1 (mFL) increased mRNA and protein expression of Ucp1 and mitochondrial OXPHOS genes (Figure 4C–D). In addition, adipocytes overexpressing mBlnc1 exhibited elevated expression of thermogenic genes following Iso stimulation. While deletion of RD2 (T2) and RD3 (T3) appeared to have modest effects on mBlnc1 function, RD1 deletion (T1) rendered mBlnc1 essentially inactive in stimulating the expression of thermogenic genes. These results demonstrate that the conserved RD1 is required for the biological function of mBlnc1 in inducing thermogenic genes in brown adipocytes.
Figure 4.
Functional analysis of mouse Blnc1 domains. (A) Predicted secondary structure of mBlnc1 using RNAfold web server. Three segments predicted to form stem-loop structures were indicated by red cycles. (B) Diagram of mBlnc1 truncation mutants. (C) Gene expression analysis of differentiated brown adipocytes expressing vector (Vec), full-length mBlnc1 (FL), or individual truncation mutants treated with Veh or Iso. (D) Immunoblots of brown adipocyte lysates. Data represent mean ± sd. *p < 0.05, vector vs. Vec.
We next examined whether RD1 of mBlnc1 and hBlnc1 is sufficient to promote thermogenic gene expression in brown adipocytes. We transduced brown preadipocytes with control or retroviral vectors overexpressing full-length mBlnc1 or hBlnc1 and the respective RD1 constructs (Figure 5A). Similar to the full-length construct, expression of mouse RD1 was sufficient to stimulate the expression of thermogenic genes, including Ucp1, Elovl3, and Dio2, and Ppargc1a, under basal and Iso-stimulated conditions (Figure 5B–C). Overexpression of human RD1 also increased the expression of these genes to similar extent as full-length hBlnc1. These results suggest that RD1 contains the functional elements of Blnc1 responsible for the transcriptional activation of brown adipocyte genes.
Figure 5.
A conserved RNA domain in mouse and human Blnc1 promotes brown adipogenesis. (A) Diagram showing the FL and RD1 Blnc1 constructs. (B) qPCR analysis of differentiated brown adipocytes expressing vector (Vec), full-length mBlnc1 (mFL) or hBlnc1 (hFL), or respective RD1 fragments (mRD1 and hRD1). (C) Immunoblots of brown adipocyte lysates. Data represent mean ± sd. *p < 0.05, vs. Vec.
3.5. Blnc1 physically interacts with hnRNPU
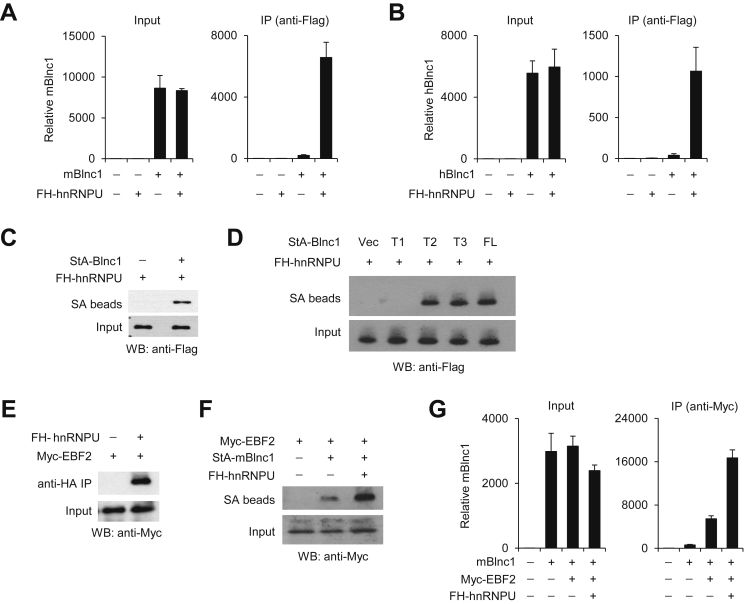
We have demonstrated that Blnc1 is physically associated with EBF2, a transcription factor that has been implicated in the regulation of brown and beige adipocyte development. As EBF2 lacks a discernable RNA-binding domain, we postulated that Blnc1 may interact with EBF2 via additional RNA-binding factors. Recent studies demonstrated that heterogeneous nuclear ribonucleoprotein U (hnRNPU) interacts with lncRNAs and regulates brown adipogenesis [39], [49]. To test whether hnRNPU physically interacts with Blnc1 and EBF2, we transiently transfected HEK293 cells with constructs expressing individual factors and performed immunoprecipitation/qPCR (IP/qPCR) analysis. Both mBlnc1 and hBlnc1 strongly associated with the hnRNPU immunocomplexes (Figure 6A–B). Reciprocal IP studies using streptavidin-binding aptamer-tagged mBlnc1 (StA-Blnc1) indicated that hnRNPU was enriched in the Blnc1 ribonucleoprotein complexes precipitated using streptavidin beads (Figure 6C).
Figure 6.
Physical interaction between Blnc1 and hnRNPU. (A–B) IP/qPCR analyses of mBlnc1 (A) or hBlnc1 (B) in RNA isolated from anti-Flag immunocomplexes or input from transiently transfected HEK293 cells. (C–D) Immunoblots of total lysates and precipitated proteins on streptavidin beads (SA) from transiently transfected HEK293 cells. (E) Immunoblots of total lysates and anti-HA immunocomplex. (F) Immunoblots of Myc-EBF2 in lysates and streptavidin beads (SA) from transfected HEK293 cells. (G) IP/qPCR analyses of mBlnc1 in RNA isolated from anti-Myc immunocomplex or input from transiently transfected HEK293 cells.
We next determined the RNA and protein domains important for physical interaction between Blnc1 and hnRNPU. Similar to full-length Blnc1, T2 and T3 truncation mutants exhibited strong interaction with hnRNPU (Figure 6D). In contrast, T1 mutant that lacks RD1 failed to interact with hnRNPU in this assay, consistent with the important role of RD1 in thermogenic gene induction. These results suggest that hnRNPU may function to recruit Blnc1 to EBF2 and facilitate the formation of the Blnc1 ribonucleoprotein transcriptional complexes.
3.6. hnRNPU facilitates the formation of EBF2/Blnc1 ribonucleoprotein complex
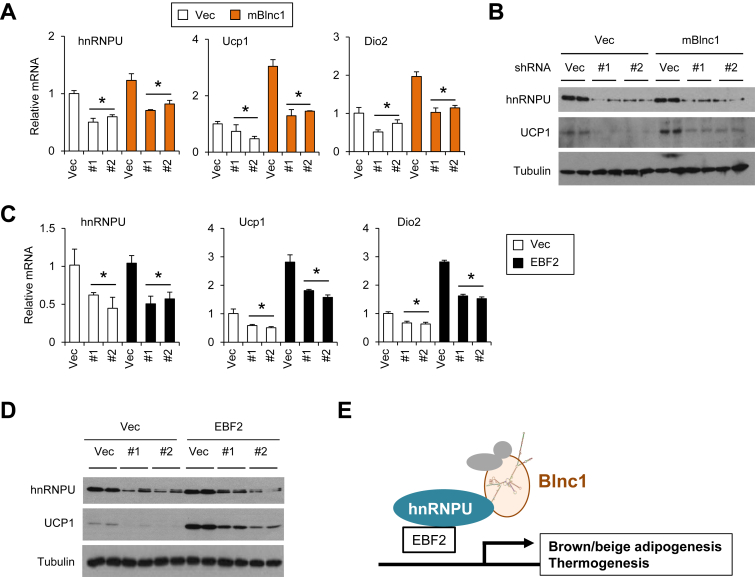
We next performed co-IP experiments to determine whether hnRNPU physically interacts with EBF2. As shown in Figure 6E, EBF2 was abundantly present in the precipitated EBF2 immunocomplex (anti-HA). To determine whether hnRNPU facilitates the recruitment of Blnc1 to EBF2, we transiently transfected HEK293 cells with expression constructs for Myc-tagged EBF2 and StA-Blnc1 in the absence or presence of hnRNPU plasmid and performed RNA IP using streptavidin beads. Immunoblotting analysis revealed that hnRNPU augmented the association between EBF2 and Blnc1 (Figure 6F). IP/qPCR studies indicated that the recruitment of mBlnc1 to EBF2 was significantly enhanced by the addition of hnRNPU (Figure 6G). These results suggest that hnRNPU may serve a critical role in mediating the transcriptional function of Blnc1 in brown adipocytes. To test this, we performed RNAi knockdown of hnRNPU in brown preadipocytes and examined the ability of Blnc1 to stimulate thermogenic gene expression. RNAi knockdown of hnRNPU significantly impaired the induction of Ucp1 and Dio2 in response to mBlnc1 overexpression (Figure 7A–B). Similarly, hnRNPU knockdown diminished the stimulation of Ucp1 and Dio2 expression in response to EBF2 overexpression in differentiated brown adipocytes (Figure 7C–D). Together, these results demonstrate that hnRNPU is required for the induction of thermogenic genes by Blnc1.
Figure 7.
HnRNPU facilitates the assembly of the Blnc1/EBF2 transcriptional complex. (A) qPCR analysis of differentiated brown adipocytes transduced with Vector (Vec) or shRNAs targeting hnRNPU (#1 and #2) in combination of vector (open) or mBlnc1 (brown) overexpression. (B) Immunoblots of brown adipocyte lysates. (C) qPCR analysis of differentiated brown adipocytes transduced with Vector (Vec) or shRNAs targeting hnRNPU (#1 and #2) in combination of vector (open) or EBF2 (brown) overexpression. Data represent mean ± sd. *p < 0.05 si#1 and #2 vs. Vec. (D) Immunoblots of brown adipocyte lysates. (E) Model depicting the induction of thermogenic gene program by the Blnc1 ribonucleoprotein transcriptional complex.
4. Discussion
Proteins and microRNAs have been established as the major products of the information encoded in the genome. More recently, lncRNAs are emerging as a new class of regulatory factors that impinge on diverse biological processes, including metabolic tissue development and function. However, the extent to which lncRNAs are functionally conserved among species remains largely unknown. In this study, we demonstrated that Blnc1 is a highly conserved lncRNA that drives the induction of thermogenic gene program during brown adipocyte differentiation. In addition, both mouse and human Blnc1 physically interacts with the RNA-binding protein hnRNPU and EBF2 to form a ribonucleoprotein complex that regulates the transcription of genes involved in thermogenesis (Figure 7E).
The molecular and functional conservation of Blnc1 is supported by several observations. First, hBlnc1 and mBlnc1 share extensive sequence conservation, particularly the 3′ RNA domain (RD1) with over 73% sequence identity. This fragment is predicted to form a stem-loop structure that likely serves as an interface for RNA-protein interaction. In support of this, the physical interaction between Blnc1 and hnRNPU was essentially abolished in the Blnc1 truncation mutant lacking RD1. Second, retroviral overexpression of hBlnc1 is sufficient to induce the expression of genes involved in thermogenesis and mitochondrial fuel oxidation and increase respiration in differentiated mouse brown adipocytes. The transcriptional activation function of Blnc1 is mediated through RD1, which is both required and sufficient for thermogenic gene induction. Finally, hBlnc1 rescued the defects of thermogenic gene expression in brown adipocytes expressing shRNAs that knockdown endogenous mBlnc1. Interestingly, hBlnc1 overexpression was unable to drive the expression of thermogenic genes to the levels observed in control adipocytes, suggesting that certain aspects of mBlnc1 function may not by fully complemented by hBlnc1.
The molecular mechanisms that mediate the induction of thermogenic genes in brown adipocytes remain incompletely understood. We previously demonstrated that Blnc1 is associated with EBF2. As EBF2 lacks a canonical RNA-binding domain, it is likely that its interaction with Blnc1 may be indirect and mediated by additional proteins. In support of this, we found that hnRNPU physically interacts with both EBF2 and Blnc1. RNA-protein interaction studies indicated that hnRNPU augments the recruitment of Blnc1 to EBF2, suggesting that hnRNPU may serve as a scaffold for the assembly of the Blnc1 ribonucleoprotein complex that is important for transcriptional activation of thermogenic genes in brown adipocytes. The critical role of hnRNPU in mediating the induction of thermogenic genes by Blnc1 is further illustrated by RNAi knockdown studies. It is possible that distinct ribonucleoprotein complexes containing Blnc1 may exist to regulate the differentiation and function of thermogenic adipocytes. In addition, lncRNAs have been shown to recruit chromatin-remodeling proteins to regulate the epigenetic state of their target genes. The nature of the epigenetic regulators recruited by Blnc1 remains to be elucidated. Future studies using proteomic tools will provide important insights into the molecular nature of the Blnc1 ribonucleoprotein complexes.
Acknowledgements
We thank Dr. David Engelke for providing the streptavidin-binding aptamer construct. This work was supported by NIH (DK102456, J.D.L.), American Diabetes Association (1-15-BS-118, J.D.L.), and National Natural Science Foundation of China (U1201213, G.S.). L.M. was supported by a predoctoral fellowship from the Chinese Scholarship Council (201506300141). X.Y.Z. was supported by NIH Pathway to Independence Award (DK106664).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.10.010.
Contributor Information
Gongshe Yang, Email: gsyang999@hotmail.com.
Jiandie D. Lin, Email: jdlin@umich.edu.
Conflict of interest
The authors declare no conflict of interest.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Gregor M.F., Hotamisligil G.S. Inflammatory mechanisms in obesity. Annual Review of Immunology. 2011;29:415–445. doi: 10.1146/annurev-immunol-031210-101322. [DOI] [PubMed] [Google Scholar]
- 2.Odegaard J.I., Chawla A. Pleiotropic actions of insulin resistance and inflammation in metabolic homeostasis. Science. 2013;339:172–177. doi: 10.1126/science.1230721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern J.H., Rutkowski J.M., Scherer P.E. Adiponectin, leptin, and fatty acids in the maintenance of metabolic homeostasis through adipose tissue crosstalk. Cell Metabolism. 2016;23:770–784. doi: 10.1016/j.cmet.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kajimura S., Saito M. A new era in brown adipose tissue biology: molecular control of brown fat development and energy homeostasis. Annual Review of Physiology. 2014;76:225–249. doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 7.Harms M., Seale P. Brown and beige fat: development, function and therapeutic potential. Nature Medicine. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 8.Townsend K.L., Tseng Y.H. Brown fat fuel utilization and thermogenesis. Trends in Endocrinology and Metabolism. 2014;25:168–177. doi: 10.1016/j.tem.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lowell B.B., S-Susulic V., Hamann A., Lawitts J.A., Himms-Hagen J., Boyer B.B. Development of obesity in transgenic mice after genetic ablation of brown adipose tissue. Nature. 1993;366:740–742. doi: 10.1038/366740a0. [DOI] [PubMed] [Google Scholar]
- 10.Yoneshiro T., Aita S., Matsushita M., Kayahara T., Kameya T., Kawai Y. Recruited brown adipose tissue as an antiobesity agent in humans. The Journal of Clinical Investigation. 2013;123:3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartelt A., Bruns O.T., Reimer R., Hohenberg H., Ittrich H., Peldschus K. Brown adipose tissue activity controls triglyceride clearance. Nature Medicine. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- 12.van der Lans A.A., Hoeks J., Brans B., Vijgen G.H., Visser M.G., Vosselman M.J. Cold acclimation recruits human brown fat and increases nonshivering thermogenesis. The Journal of Clinical Investigation. 2013;123:3395–3403. doi: 10.1172/JCI68993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang G.X., Zhao X.Y., Lin J.D. The brown fat secretome: metabolic functions beyond thermogenesis. Trends in Endocrinology and Metabolism. 2015;26:231–237. doi: 10.1016/j.tem.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G.X., Zhao X.Y., Meng Z.X., Kern M., Dietrich A., Chen Z. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nature Medicine. 2014;20:1436–1443. doi: 10.1038/nm.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Enerback S. Human brown adipose tissue. Cell Metabolism. 2010;11:248–252. doi: 10.1016/j.cmet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Nedergaard J., Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell metabolism. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 17.Tseng Y.H., Cypess A.M., Kahn C.R. Cellular bioenergetics as a target for obesity therapy. Nature Reviews Drug Discovery. 2010;9:465–482. doi: 10.1038/nrd3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B. Identification and importance of brown adipose tissue in adult humans. The New England Journal of Medicine. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nedergaard J., Bengtsson T., Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. American Journal of Physiology. Endocrinology and Metabolism. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 20.van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D. Cold-activated brown adipose tissue in healthy men. The New England Journal of Medicine. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 21.Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T. Functional brown adipose tissue in healthy adults. The New England Journal of Medicine. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 22.Ouellet V., Labbe S.M., Blondin D.P., Phoenix S., Guerin B., Haman F. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. The Journal of Clinical Investigation. 2012;122:545–552. doi: 10.1172/JCI60433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orava J., Nuutila P., Lidell M.E., Oikonen V., Noponen T., Viljanen T. Different metabolic responses of human brown adipose tissue to activation by cold and insulin. Cell Metabolism. 2011;14:272–279. doi: 10.1016/j.cmet.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Wu J., Cohen P., Spiegelman B.M. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes & Development. 2013;27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kajimura S., Seale P., Spiegelman B.M. Transcriptional control of brown fat development. Cell Metabolism. 2010;11:257–262. doi: 10.1016/j.cmet.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trajkovski M., Lodish H. MicroRNA networks regulate development of brown adipocytes. Trends in Endocrinology and Metabolism: TEM. 2013;24:442–450. doi: 10.1016/j.tem.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annual Review of Biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonasio R., Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annual Review of Genetics. 2014;48:433–455. doi: 10.1146/annurev-genet-120213-092323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J.T. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 30.Cesana M., Cacchiarelli D., Legnini I., Santini T., Sthandier O., Chinappi M. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 2011;147:358–369. doi: 10.1016/j.cell.2011.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guttman M., Donaghey J., Carey B.W., Garber M., Grenier J.K., Munson G. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Batista P.J., Chang H.Y. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panzitt K., Tschernatsch M.M., Guelly C., Moustafa T., Stradner M., Strohmaier H.M. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132:330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 35.Huarte M., Guttman M., Feldser D., Garber M., Koziol M.J., Kenzelmann-Broz D. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Knoll M., Lodish H.F., Sun L. Long non-coding RNAs as regulators of the endocrine system. Nature Reviews Endocrinology. 2015;11:151–160. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao X.Y., Lin J.D. Long noncoding RNAs: a new regulatory code in metabolic control. Trends in Biochemical Sciences. 2015;40:586–596. doi: 10.1016/j.tibs.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X.Y., Li S., Wang G.X., Yu Q., Lin J.D. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Molecular Cell. 2014;55:372–382. doi: 10.1016/j.molcel.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alvarez-Dominguez J.R., Bai Z., Xu D., Yuan B., Lo K.A., Yoon M.J. De Novo reconstruction of adipose tissue transcriptomes reveals long non-coding RNA regulators of brown adipocyte development. Cell Metabolism. 2015;21:764–776. doi: 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajakumari S., Wu J., Ishibashi J., Lim H.W., Giang A.H., Won K.J. EBF2 determines and maintains brown adipocyte identity. Cell Metabolism. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang W., Kissig M., Rajakumari S., Huang L., Lim H.W., Won K.J. Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14466–14471. doi: 10.1073/pnas.1412685111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klein J., Fasshauer M., Klein H.H., Benito M., Kahn C.R. Novel adipocyte lines from brown fat: a model system for the study of differentiation, energy metabolism, and insulin action. BioEssays: News and Reviews in Molecular, Cellular and Developmental Biology. 2002;24:382–388. doi: 10.1002/bies.10058. [DOI] [PubMed] [Google Scholar]
- 43.Li S., Yu Q., Wang G.X., Lin J.D. The biological clock is regulated by adrenergic signaling in brown fat but is dispensable for cold-induced thermogenesis. PLoS One. 2013;8:e70109. doi: 10.1371/journal.pone.0070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G.X., Cho K.W., Uhm M., Hu C.R., Li S., Cozacov Z. Otopetrin 1 protects mice from obesity-associated metabolic dysfunction through attenuating adipose tissue inflammation. Diabetes. 2014;63:1340–1352. doi: 10.2337/db13-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srisawat C., Engelke D.R. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA. 2001;7:632–641. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker S.C., Good P.D., Gipson T.A., Engelke D.R. The dual use of RNA aptamer sequences for affinity purification and localization studies of RNAs and RNA-protein complexes. Methods in Molecular Biology. 2011;714:423–444. doi: 10.1007/978-1-61779-005-8_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kong L., Zhang Y., Ye Z.Q., Liu X.Q., Zhao S.Q., Wei L. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Research. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Research. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hacisuleyman E., Goff L.A., Trapnell C., Williams A., Henao-Mejia J., Sun L. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nature Structural & Molecular Biology. 2014;21:198–206. doi: 10.1038/nsmb.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.