Abstract
Biosynthesis of sakuranetin, a flavonoid anti-fungal phytoalexin that occurs in rice, is highly dependent on jasmonic acid (JA) signalling and induced by a variety of environmental stimuli. We previously identified OsNOMT, which encodes naringenin 7-O-methyltransferase (NOMT); NOMT is a key enzyme for sakuranetin production. Although OsNOMT expression is induced by JA treatment, the regulation mechanism that activates the biosynthetic pathway of sakuranetin has not yet been elucidated. In this study, we show that JA-inducible basic helix-loop-helix transcriptional factor OsMYC2 drastically enhances the activity of the OsNOMT promoter and is essential for JA-inducible sakuranetin production. In addition, we identified 2 collaborators of OsMYC2, OsMYC2-like protein 1 and 2 (OsMYL1 and OsMYL2) that further activated the OsNOMT promoter in synergy with OsMYC2. Physical interaction of OsMYC2 with OsMYL1 and OsMYL2 further supported the idea that these interactions lead to the enhancement of the transactivation activity of OsMYC2. Our results indicate that JA signalling via OsMYC2 is reinforced by OsMYL1 and OsMYL2, resulting in the inductive production of sakuranetin during defence responses in rice.
Jasmonic acid (JA) and its derivatives, the so called jasmonates, have been extensively characterized as the central players during biotic stress resistance, for example, emission of volatiles to attract the natural enemies of herbivores or production of toxic compounds to repel invaders1,2,3,4. In rice, jasmonates induce the production of anti-microbial specialized metabolites called phytoalexins, which consist of 16 diterpenoids and 1 flavonoid5,6. Among them, the flavonoid sakuranetin is considered to be the biologically important phytoalexin in terms of its high accumulation in blast-infected rice leaves and high anti-fungal activity7,8. Furthermore, its biosynthesis requires JA signalling, while the biosynthesis of the major diterpenoid phytoalexins is regulated by both JA-required and non-JA-required pathways9,10. Besides, blast-induced accumulation of sakuranetin is known to be compromised in a JA-deficient mutant cpm2, while accumulation of diterpenoid phytoalexins remained9. Therefore, we focused on the biosynthetic pathway for sakuranetin as a model for JA-required defence responses. In the final step of sakuranetin biosynthesis, the precursor naringenin is converted to sakuranetin by naringenin 7-O-methyltransferase (NOMT), which is encoded by OsNOMT11. We recently reported that OsNOMT expression is induced by JA treatment12, and jasmonoyl-l-isoleucine (JA-Ile), an active form of jasmonate, is required for the production of sakuranetin10. We have previously shown that CuCl2 is a potent elicitor to induce sakuranetin production in wild-type plants, while such CuCl2-inductive production of sakuranetin is specifically compromised in a JA-Ile deficient mutant osjar110. These findings prompted us to speculate that the elucidation of the regulation mechanism underlying OsNOMT expression would help in understanding the mechanism underlying JA-required sakuranetin production.
For the production of specialized metabolites, diverse transcriptional factors (TFs) have been characterized as hormone-inducible regulators in various plant species4,13,14. In particular, several TFs have been well-studied and reported to contribute to the production of diterpenoid phytoalexins or expression of their biosynthetic genes in rice15,16,17,18. However, regulatory factors responsible for sakuranetin production and/or OsNOMT expression remain unclear.
OsMYC2, a basic helix-loop-helix (bHLH) TF in rice, is a homologue of AtMYC2, a well-studied global regulator of JA signalling in Arabidopsis19,20,21. OsMYC2 is involved in JA-required spikelet development by the direct activation of OsMADS122. Another study revealed that OsMYC2 interacts with almost all of 14 JASMONATE ZIM-domain (JAZ) proteins in rice via the JAZ-interacting domain, one of which represses JA-inducible OsMYC2 expression. Furthermore, overexpression of OsMYC2 causes upregulation of early JA-responsive genes, leading to JA-hypersensitive phenotypes such as bacterial blight resistance23. However, the mechanism underlying the activation of the downstream pathway of OsMYC2 remains largely unknown.
Here, we report that JA-induced expression of OsNOMT and production of sakuranetin are under the control of OsMYC2. Moreover, we identified two bHLH proteins, OsMYC2-like protein 1 (OsMYL1) and OsMYL2, as interacting partners of OsMYC2. Both OsMYL1 and OsMYL2 enhance the transactivation activity of OsMYC2 by physical interaction, leading to further activation of the OsNOMT promoter. Our findings will help in understanding how rice plants modulate the activity of OsMYC2 during JA signalling, leading to the inductive production of sakuranetin as a defence response.
Results
Selection of regulatory factor candidates for OsNOMT by using transcriptome analysis of JA-deficient plants
To identify JA-dependent transcriptional factors (TFs), which could be possible candidates for the OsNOMT regulation in response to JA, we performed transcriptome analysis using CuCl2-elicited cpm2, a JA-deficient mutant9, in comparison with wild-type plants (WT). We used CuCl2 as an elicitor to induce sakuranetin production via JA signaling because CuCl2 treatment is also known to induce JA production and reveals good reproducibility to elicit these phenomena. Since OsNOMT was also shown to be induced within 24 h after CuCl2 treatment (see Supplementary Table S1), JA-dependent regulatory factors for OsNOMT were expected to precede the upregulation of OsNOMT. Therefore, we selected TFs that fulfilled the following criteria: upregulated over 2-fold in either 2 or 6 h after CuCl2 treatment in WT and downregulated under 0.5-fold in either 2 or 6 h after CuCl2 treatment in cpm2 when compared with WT. We also used a parameter Q value to obtain a significant number of genes along with an acceptable false discovery rate (FDR) less than 0.05. As a result, various types of 41 TFs comprising bHLH, MYB, NAC, WRKY, and bZIP families were selected (see Supplementary Table S2), and they included the JA-inductive bHLH factor OsMYC2 and MYB factor OsJAmyb. The expression of OsMYC2 is induced by JA treatment23. Several MYB proteins activate flavonoid biosynthetic genes by interacting with bHLH TFs24. Thus, we focused on the MYB and bHLH proteins among the selected TFs.
OsMYC2 functions as the transcriptional activator of the OsNOMT promoter
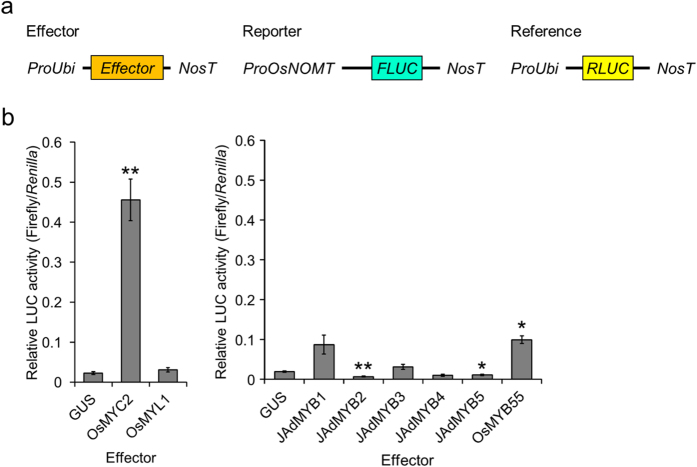
To investigate the effects of the selected TFs, we analysed their transactivation activity by using the OsNOMT promoter-luciferase (LUC) reporter assay. Candidate TFs (OsMYC2, OsMYL1, JA-dependent [JAd] MYB1–MYB5, OsMYB55, and OsJAmyb) were driven by the maize ubiquitin promoter as effectors for their constitutive expression. The luciferase activity assay without the OsNOMT promoter revealed that all the effectors, except OsJAmyb, did not activate the reporter activity in a promoter-independent manner (see Supplementary Fig. S1). Then, we performed the luciferase activity assay with one of the selected proteins, except OsJAmyb, as the effector. As a result, 3 proteins, OsMYC2, JAdMYB1, and OsMYB55, were found to activate the promoter activity of OsNOMT (Fig. 1). Among these TFs, OsMYC2 exhibited the most robust transactivation activity.
Figure 1. Activation of the OsNOMT promoter by JA-induced transcription factors.
(a) Schematic representation of the vectors used for the transient expression assays. FLUC: firefly luciferase, RLUC: renilla luciferase, ProOsNOMT: 5′ 1000-bp upstream region of the transcription start site of OsNOMT, ProUbi: maize ubiquitin promoter, NosT: nopaline synthase terminator. (b) Relative luciferase activities in the bombarded rice leaves. Data are presented as FLUC/RLUC ± standard error. n = 3–5. *P < 0.05, **P < 0.01, compared to the data with GUS as the effector.
Impacts of modification of OsMYC2 expression on OsNOMT expression and phytoalexin production
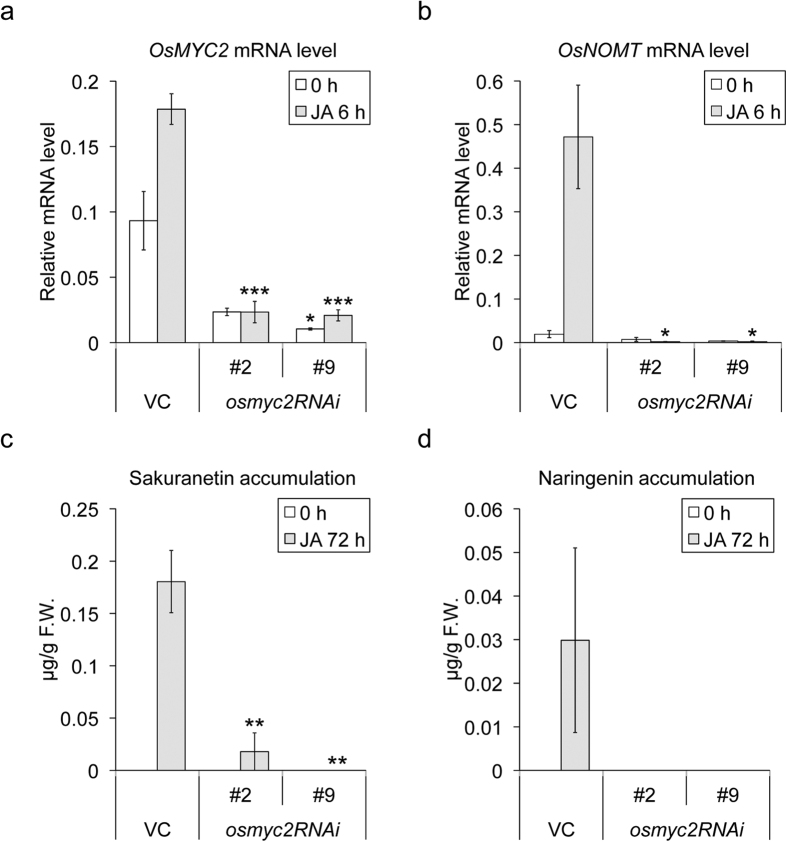
MYC2 is known as a central regulator of JA-signalling in some plants20,25. In rice, OsMYC2 has also been shown to be involved in JA-signalling through interaction with JAZ repressor proteins23. The luciferase activity assay clearly demonstrated that OsMYC2 has a positive impact on the expression of OsNOMT. To further dissect the effects of OsMYC2 on the expression of OsNOMT and production of sakuranetin in planta, we generated OsMYC2-RNAi knockdown plants (henceforth osmyc2RNAi) along with empty vector control (VC) plants. Under basal conditions, the expression level of OsMYC2 was repressed by 75% and 89% in 2 independent osmyc2RNAi lines (#2 and #9), respectively, with no visible phenotypes, compared with the VC plants. Expression of OsMYC2 was induced by JA treatment in the VC plants, as shown by Uji et al.23. The induction of OsMYC2 by JA treatment was not observed in osmyc2RNAi lines (Fig. 2a). We next investigated the expression of OsNOMT after elicitation. Expression of OsNOMT was induced by JA treatment in the VC plants. In the osmyc2RNAi lines, JA-inducible expression of OsNOMT was totally abolished (Fig. 2b). Quantification of phytoalexin contents revealed that the production of sakuranetin and precursor naringenin induced by JA was repressed in osmyc2RNAi lines (Fig. 2c,d). These data indicate that JA-induced induction of OsNOMT expression and production of sakuranetin are under the control of OsMYC2 in rice.
Figure 2. Gene expression and phytoalexin accumulation in the OsMYC2-knockdown plants (osmyc2RNAi).
(a,b) Expression levels of OsMYC2 (a) and OsNOMT (b). Total RNA from the rice leaves after 6 h of 500 μM JA treatment (grey bars) or untreated rice leaves (open bars) was extracted. qRT-PCR analysis was performed using the cDNA samples synthesized from the total RNA. (c,d) Contents of sakuranetin (c) and naringenin (d) in the rice leaves after 6 h of 500 μM JA treatment (grey bars) or no treatment (open bars). Data are presented as mean ± standard error values. (a,b) n = 3–4, (c,d) n = 4–6. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the VC data for each treatment.
Transcriptional factors that work with OsMYC2 to activate the OsNOMT promoter
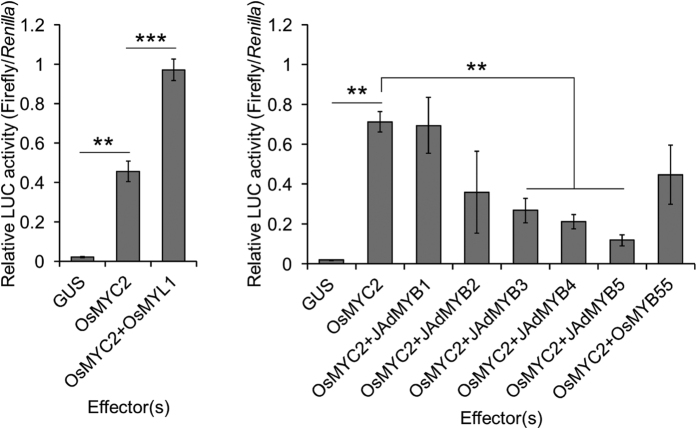
The interaction of bHLH and MYB TFs often enhances the transcription of flavonoid biosynthetic genes24. To test this point, we next performed transient expression assays with dual effectors. MYB proteins or OsMYL1 was co-introduced with OsMYC2 as effectors. As a result, OsMYL1 further activated the transactivation of OsNOMT when co-introduced with OsMYC2, while any MYB TF did not activate transactivation, or rather repressed (Fig. 3). These results indicate that OsMYC2 and OsMYL1, both bHLH proteins, synergistically enhance the transactivation of OsNOMT.
Figure 3. Luciferase activity assay with dual effectors.
Relative luciferase activities in the bombarded rice leaves. Data are presented as FLUC/RLUC ± standard error. n = 3–5. **P < 0.01, ***P < 0.001.
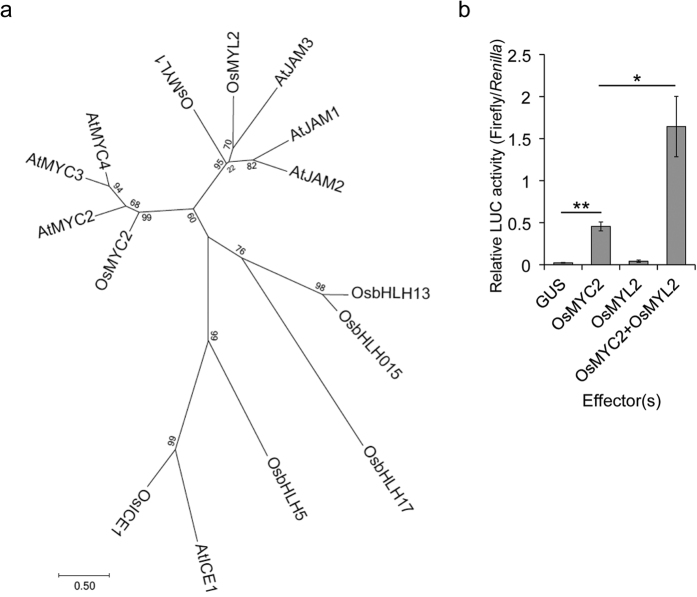
We found another OsMYL protein designated as OsMYL2 that is highly homologous to both OsMYC2 and OsMYL1 (Fig. 4a). Although the expression of OsMYL2 was not affected by CuCl2 in WT in our transcriptome analysis (see Supplementary Table S2), we included this TF in our further analysis because of its similarity to OsMYL1. The luciferase activity assay with OsMYC2 and/or OsMYL2 revealed that OsMYL2 also activated the transactivation of OsNOMT when co-introduced with OsMYC2, just as in the case of OsMYL1 (Fig. 4b). We also confirmed that any combination did not affect the reporter activity in an OsNOMT promoter-independent manner (see Supplementary Fig. S2).
Figure 4. Transactivation activity of OsMYL2.
(a) Phylogenetic tree of the bHLH transcription factors. We used the JTT+G model to construct the phylogenetic tree. (b) Relative luciferase activities in the bombarded rice leaves. Data are presented as FLUC/RLUC ± standard error. n = 5. *P < 0.05, **P < 0.01.
OsMYL1 and OsMYL2 as interacting partners of OsMYC2
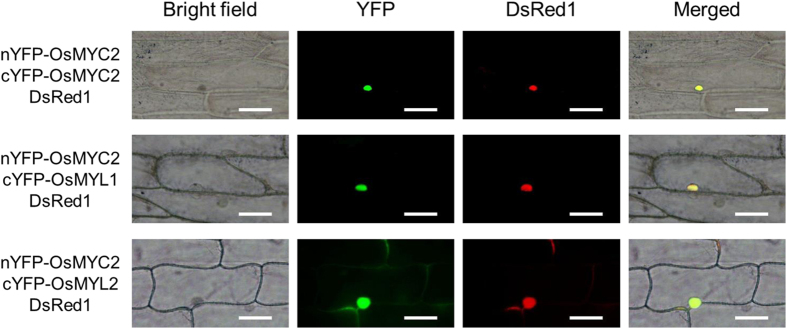
Since the helix-loop-helix motif in bHLH proteins has been reported to form dimers26, we speculated that OsMYC2 interacts with OsMYL1 and OsMYL2, leading to further activation of the OsNOMT promoter. To validate this hypothesis, we performed bimolecular fluorescence complementation (BiFC) assays. Vectors expressing nEYFP-fused protein, cEYFP-fused protein, and DsRed1 (pTH121R17) under the control of the CaMV 35S promoter were introduced into onion epidermal cells by particle bombardment. With the co-expression of nYFP-OsMYC2 and cYFP-OsMYC2, c-YFP-OsMYL1, or cYFP-OsMYL2, YFP fluorescence and DsRed1 fluorescence were detected, especially in the nucleus (Fig. 5). However, with the co-expression of nYFP-OsMYC2/cYFP, nYFP/cYFP-OsMYC2, nYFP/cYFP-OsMYL1, or nYFP/cYFP-OsMYL2, only DsRed1 fluorescence was detected (see Supplementary Fig. S3). These data indicate that OsMYC2 interacts with OsMYL1, OsMYL2, and OsMYC2 alone in the nucleus.
Figure 5. BiFC visualization of OsMYC2-OsMYC2/OsMYL1/OsMYL2 interactions.
Onion epidermal cells were co-transfected with constructs encoding the nEYFP-fused proteins, cEYFP-fused proteins, and DsRed1. Images were obtained 24 h after bombardment by using the BX53 microscope system (Olympus). Bars = 100 μm.
Next, we performed in vitro protein-protein interaction assays by using the amplified luminescent proximity homogeneous assay (AlphaScreen). Since we failed to express tagged OsMYC2 translated from original coding sequence (CDS), we used optimized CDS by synonymous substitution using codon usage of Arabidopsis thaliana (see Optimized synthetic CDS of OsMYC2 online). His-Bls-OsMYC2 (Bls: biotin ligase site27) and FLAG-tagged OsMYC2, OsMYL1, and OsMYL2 were expressed using the wheat germ expression system (see Supplementary Fig. S4). The AlphaScreen assay using biotinylated His-Bls-tagged and FLAG-tagged proteins revealed that OsMYC2 physically interacts with OsMYL1, OsMYL2, and OsMYC2 alone in vitro (Fig. 6).
Figure 6. AlphaScreen-based evaluations of the OsMYC2-OsMYC2/OsMYL1/OsMYL2 interactions.
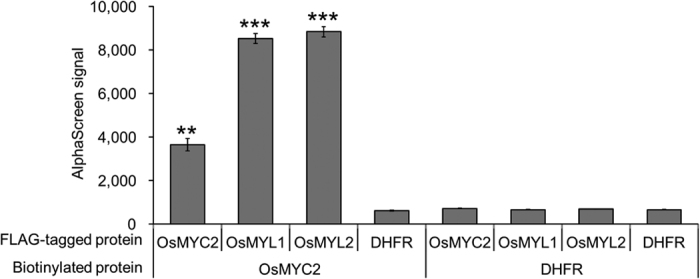
Crude translation mixtures of biotinylated His-Bls-OsMYC2 and FLAG-tagged proteins, OsMYC2, OsMYL1, or OsMYL2, were used for the assay. Biotinylated dihydrofolate reductase (DHFR) and FLAG-tagged DHFR were used as the negative controls. Data are presented as mean ± standard error values. n = 3. **P < 0.01, ***P < 0.001, compared to the data for biotinylated DHFR and FLAG-tagged DHFR.
Transactivation activity of OsMYC2 enhanced by interaction with OsMYL1 or OsMYL2
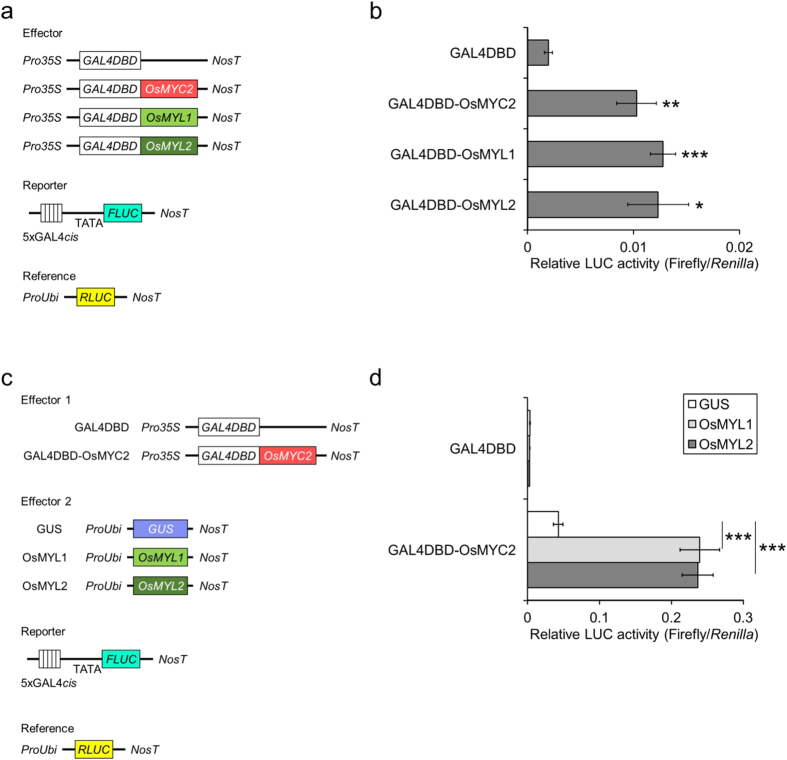
Protein-protein interaction sometimes strikingly affects the function of proteins. This prompted us to test whether the interaction of OsMYC2 with OsMYL1 and OsMYL2 is the casual event of the enhanced transactivation. We investigated how OsMYL1 and OsMYL2 act on the enhancement by using the artificial GAL4 DNA-binding domain (DBD)-fused system28, that is, TFs were forced to interact with the promoter region (Fig. 7a). When OsMYL1 or OsMYL2 was fused to GAL4DBD, they exhibited transactivation activity similar to the activity detected in OsMYC222 (Fig. 7b). Furthermore, co-expression of OsMYL1 or OsMYL2 with GAL4DBD-OsMYC2 resulted in obvious enhancement of the transactivation ability of OsMYC2 (Fig. 7c,d). These data indicate that OsMYL1 and OsMYL2 synergistically activate the transactivation activity of OsMYC2 by physical interaction.
Figure 7. Transcriptional activities of OsMYC2, OsMYL1, and OsMYL2.
(a) Schematic representation of the vectors used for the transient expression assays. GAL4DBD: GAL4 DNA-binding domain, FLUC: firefly luciferase, RLUC: renilla luciferase, Pro35S, CaMV 35 S promoter, ProUbi: maize ubiquitin promoter, NosT: nopaline synthase terminator. (b) Relative luciferase activities in the bombarded rice leaves. Data are presented as FLUC/RLUC ± standard error. n = 6. *P < 0.05, **P < 0.01, ***P < 0.001, compared to the data for GAL4DBD. (c) Schematic representation of the vectors used for the transient expression assays with GAL4DBD-OsMYC2 co-transfected with GAL4DBD-free OsMYL1/OsMYL2. (d) Relative luciferase activities in the bombarded rice leaves. Data are presented as FLUC/RLUC ± standard error. n = 6. ***P < 0.001.
Discussion
Jasmonic acid (JA) and its derivatives contribute greatly to defence responses in the plant kingdom, including production of defence-related specialized metabolites that are strictly regulated by various transcriptional factors (TFs) through the JA signalling pathway. Here, we demonstrated that the bHLH TF OsMYC2 plays a pivotal role in the JA-induced expression of OsNOMT that is enhanced by physical interaction with OsMYL1 and OsMYL2, presumably leading to the consequent accumulation of a flavonoid phytoalexin, sakuranetin, during rice defence responses. Repression of OsMYC2 expression strikingly impaired JA-induced OsNOMT expression and production of sakuranetin and naringenin (Fig. 2). We also found that CuCl2-induced sakuranetin production was impaired in osmyc2RNAi lines, although CuCl2-induced expression of OsNOMT itself was comparable to that in the VC plants (see Supplementary Fig. S5). These results imply that the sakuranetin biosynthetic pathway, specifically the upper flavonoid biosynthetic pathway, would be highly dependent on OsMYC2, while the knockdown of OsMYC2 did not affect the inductive expression of OsNOMT after CuCl2 treatment. Hence, the expression of OsNOMT would also be induced in an OsMYC2-independent manner.
OsMYL1 and OsMYL2 have higher homology to jasmonate-associated MYC2-like proteins (AtJAMs) 1 to 3, rather than OsMYC2 or AtMYC2 (Fig. 4a). OsMYC2 and AtMYC2 are categorized as IIIe group bHLH TFs, while OsMYL1, OsMYL2, and AtJAMs are categorized as IIId group bHLH TFs29. AtJAMs are negative regulators of JA signalling30. Another study revealed that AtJAM1 is a transcriptional repressor that competes with AtMYC2 by sharing the same binding elements. In addition, AtJAM1 does not interact with AtMYC231. Meanwhile, we found that OsMYL1 and OsMYL2 are the collaborators of OsMYC2 that enhance the transactivation activity of OsMYC2 by physical interaction (Figs 3, 4, 5, 6 and 7). Probably, OsMYL1 and OsMYL2 enhance the transactivation activity of OsMYC2 by forming heteromers (Figs 5, 6 and 7), which may lead to the further activation of JA signalling. However, the actual forms of the homomer and heteromers that may be critical for their functions remain unclear. Our effector assay revealed that OsMYL1 and OsMYL2 themselves were not able to activate the OsNOMT promoter without being forced to bind to DNA as the GAL4DBD-fused proteins, indicating that formation of the OsMYC2 and OsMYL heteromer is important for their transactivation ability, possibly by activating OsMYC2 by altering protein structure.
The relationships between AtMYC2 and AtJAMs are totally different from those between OsMYC2 and OsMYL1 or OsMYL2, which may cause the differences in the regulatory mechanisms of the JA-induced defence responses between monocots and dicots. Identification of the important region(s) of OsMYC2, OsMYL1, and OsMYL2 protein for transcriptional activation activities and protein-binding abilities would be required to elucidate these differences in the future. Besides, we suggest apparent difference between OsMYL1 and OsMYL2 on the basis of our transcriptome analysis; expression of OsMYL1 as well as OsMYC2 was induced in a JA-dependent manner, while expression of OsMYL2 was constant (see Supplementary Table S2). Whether optimal use of the OsMYC2-OsMYL1 and OsMYC2-OsMYL2 heteromers in planta is required for appropriate induction of OsNOMT is an interesting question.
DNA-binding ability is one of the most essential functions of the TFs, as well as transactivation activity. To evaluate the DNA-binding ability of OsMYC2, or the heteromers formed with OsMYL1 or OsMYL2, we also tried to find the promoter region of OsNOMT responsible for OsMYC2-dependent activation in this study. However, even though most of the promoter region, except for the TATA box located at −34 to −27 bp from the transcriptional start site, was deleted, there was still OsMYC2-dependent activation in the deletion series; consequently, we could not define the cis-element bound by OsMYC2 (see Supplementary Fig. S6 and Supplementary Methods). Since the DNA-binding ability of OsMYC2 would be affected by physical interaction with OsMYL1 and OsMYL2, co-introduction of OsMYC2 and OsMYLs should be tested further. Uji et al. reported that OsMYC2 binds to a specific E-box sequence of the OsJAZ10 promoter in vivo by using the chromatin immunoprecipitation strategy23. Therefore, in vivo experimental approaches would be required to understand OsMYC2 binding on the OsNOMT promoter. Alternatively, it might be possible that OsMYC2-mediated induction of OsNOMT expression is not regulated by TF(s) directly binding to the promoter region but is regulated in the post-transcriptional stage.
Sakuranetin, a biologically important phytoalexin in rice, is a good subject for elucidating JA signalling because of its high JA-required accumulation, whereas production of diterpenoid phytoalexins occurs via both JA-required and non-JA-required pathways9,10. Deciphering JA signalling mediated by OsMYC2, OsMYL1, and OsMYL2 would help in understanding the regulation of the flavonoid biosynthetic pathway for naringenin and sakuranetin. It could be possible to generate plants that overproduce sakuranetin by heterologous expression of OsNOMT, and further attempts to use OsMYC2 and its collaborators, OsMYL1 and OsMYL2, for enhancing OsNOMT expression and the early flavonoid biosynthetic pathway could be a potential method of generating sakuranetin-biofortified plants.
Methods
Plant materials
For the luciferase activity assays, we used the japonica type rice Oryza sativa L. ‘Nipponbare’. For the transcriptome analysis, we used a JA-deficient mutant, cpm29, and its genetic background, Oryza sativa L. ‘Nihonmasari’ as WT. For the gene expression analyses and phytoalexin quantification analyses, we used RNAi-based OsMYC2-knockdown plants (osmyc2RNAi) under the control of the Zea mays polyubiquitin promoter and VC plants transformed with the empty vector (pANDA32), with their genetic background, Oryza sativa L. ‘Nihonmasari’, as WT.
To grow the rice plants, the seeds were sterilized with 70% ethanol for 2 min, followed by treatment with 1% sodium hypochlorite solution (Kanto Chemical) for 20 min; the seeds were then washed with autoclave-sterilized water. The surface-sterilized seeds were incubated on 0.5% agar medium in a chamber (14-h light and 10-h darkness at 28 °C). Rice transformation was performed according to the method reported by Toki et al.33.
Cloning
Standard methods for cloning were used. PCR-amplified DNA fragments were sequenced after cloning into the vectors. The vectors used in this study and primers used for the cloning are listed in Supplementary Tables S3 and S4, respectively.
pUbi-RLUC
The CDS of renilla luciferase from pRL (Promega, WI, USA) was PCR-amplified and cloned into the pENTR/D-TOPO vector (Invitrogen, CA, USA). The CDS was then inserted into the pUbi-RfA-Tnos vector15 by using the LR reaction (Gateway®).
pGL4-OsNOMT1000
The 5′ 1-kb upstream region of the OsNOMT promoter, extending from the transcriptional start site, was PCR-amplified and cloned into the pZErO2 vector (Invitrogen, CA, USA). The promoter region was extracted and inserted into multi-cloning sites of pGL4.10-Tnos, which the SV40 late poly (A) signal of pGL4.10 (Promega, WI, USA) was converted to the nopaline synthase terminator (Tnos) at the BamHI and XbaI sites, to obtain pGL4-OsNOMT1000.
pUbi-JA-dependent MYB1, pUbi-JA-dependent MYB2, pUbi-JA-dependent MYB3, pUbi-JA-dependent MYB4, pUbi-JA-dependent MYB5, pUbi-OsJAmyb, pUbi-OsMYB55, pUbi-OsMYC2, pUbi-OsMYL1, and pUbi-OsMYL2
Each CDS was PCR-amplified and cloned into the pENTR/D-TOPO vector. The CDS was then inserted into the pUbi-RfA-Tnos vector by using the LR reaction. The CDS of OsMYL1 was 39 bp longer than the RAP-DB annotation (see CDS of OsMYL1 described in this article online).
pANDA-OsMYC2
The 748-bp trigger region of OsMYC2, which consists of 336 bp from the 3′ UTR and 412 bp from the 3′ CDS, was PCR-amplified and cloned into the pENTR/D-TOPO vector. The trigger region was then inserted into the pANDA vector32 by using the LR reaction.
pnYOsMYC2, pcYOsMYC2, pcYOsMYL1, and pcYOsMYL2
Each CDS, cloned into pENTR/D-TOPO as described above, was inserted into the pnYGW or pcYGW vector34 by using the LR reaction.
pEU-E01-6His-Bls-GW-STOP
We performed the inverse overlap extension PCR by using pEU-E01-GW as the template35. Amplified DNA fragments were self-ligated using the In-Fusion system (TaKaRa Bio USA, CA, USA) to obtain pEU-E01-His-Bls-GW-STOP.
pEU-E01-6His-Bls-optOsMYC2 and pEU-E01-FLAG-optOsMYC2
The optimized CDS of OsMYC2 was synthesized using the GeneArt® Strings™ DNA Fragments service (Invitrogen, CA, USA), PCR-amplified in twice, and cloned into the pENTR/D-TOPO vector. The CDS was then inserted into the pEU-E01-6His-Bls-GW-STOP or pEU-E01-FLAG-GW-STOP35 vector by using the LR reaction.
pEU-E01-FLAG-OsMYL1 and pEU-E01-FLAG-OsMYL2
Each CDS, cloned into pENTR/D-TOPO as described above, was inserted into the pEU-E01-FLAG-GW-STOP vector by using the LR reaction.
p35S-GAL-OsMYC2, p35S-GAL-OsMYL1, and p35S-GAL-OsMYL2
Each CDS was PCR-amplified and inserted into 430T1.236 by using the In-Fusion system (TaKaRa Bio USA, CA, USA).
Luciferase activity assay
Six- or 7-day-old rice leaves were excised from the sheath, acclimated overnight, and then used for particle bombardment.
Effector plasmids were used to constitutively express the TFs as effectors under the control of the Z. mays polyubiquitin promoter in the rice leaves. pUbi-GUS15 was used as the control. Constructs, in which the promoter region was fused to the firefly luciferase (FLUC) gene, were used as reporter plasmids. pUbi-RLUC, in which renilla luciferase (RLUC) was under the control of the Z. mays polyubiquitin promoter, was used as the internal control. The PDS-1000 He Biolistic Particle Delivery System (Bio-Rad, CA, USA) was used for the particle bombardment.
For particle bombardment, gold particles (0.6 mg and 1.6 μm in diameter; Bio-Rad, CA, USA) were coated with 1 μg of the reporter plasmid, 0.5 μg of pUbi-RLUC, and effector plasmid(s) (0.5 μg each). The plasmid-coated particles were bombarded at 1100 psi and 9-cm distance. After the bombardment, the rice leaves were incubated for 24 h in a chamber (24-h light at 28 °C) before the luciferase assays. The assays were performed as described previously17. The ratio of luciferase activity (Firefly LUC/Renilla LUC) was calculated to normalize the values in each assay.
Transcriptome analysis
Total RNA was extracted from 1 mM of the CuCl2-treated plants by using the RNeasy plant mini kit (QIAGEN). Microarray analyses were performed as described previously15. Three biological replicate sample sets were analysed. Data normalization and statistical analyses were performed using the Subio Platform with Basic Plug-in (Subio Inc., Kagoshima, Japan). Transcriptome data have been deposited in the Gene Expression Omnibus in NCBI (ID: GSE87698).
Expression analyses
Prior to the experiments, the transgenic plants were selected by PCR amplification of the hygromycin phosphotransferase (HPT) transgene with the primers 5′-ATGAAAAAGCCTGAACTCACCGCGACGTCTGTC-3′ and 5′-CTATTCCTTTGCCCTCGGACGAGTGCTG-3′. Ten-day-old rice leaves were excised from the sheath, acclimated overnight (light at 28 °C), and then used for the subsequent experiments. The rice leaves were elicited with 500 μM of JA or CuCl2 for 6 h. Leaves collected before elicitation were used as the control. Total RNA extracted using the Maxwell® 16 LEV Plant RNA Kit (Promega, WI, USA) was reverse-transcribed with the PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) (TaKaRa BioInc., Japan). The synthesized cDNA was used to perform qRT-PCR. qRT-PCR was performed using the Power SYBR® Green PCR Master Mix (Applied Biosystems, CA, USA) and Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems). To calculate the relative mRNA levels, raw data from qRT-PCR analysed using the standard curve method were normalized by the raw data of the ubiquitin domain-containing protein gene (OsUBQ). Sequences of the primers used for the expression analyses are provided in Supplementary Table S5.
Quantification of phytoalexins and naringenin accumulated in the leaves
Prior to the experiments, the transgenic plants were selected by PCR amplification of the HPT transgene with the primers described above. Ten-day-old rice leaves were excised from the sheath, acclimated overnight (light at 28 °C), and then used for the subsequent experiments. The rice leaves were elicited with 500 μM of JA or CuCl2 for 72 h, soaked in phytoalexin extraction solvent (ethanol:water:acetonitrile:acetic acid, 79:13.99:7:0.01, v/v), and incubated for 24 h at 4 °C to extract the phytoalexins and naringenin. Leaves collected before elicitation were used as the control. The samples were then centrifuged (4 °C, 15 min, and 16,000 g). The supernatant was collected and subjected to phytoalexin and naringenin measurement by using liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) as described previously37.
BiFC assay
Constructs encoding the nEYFP-fused proteins, cEYFP-fused proteins, and DsRed1 were introduced into onion epidermal cells by particle bombardment with the PDS-1000 He Biolistic Particle Delivery System (Bio-Rad, CA, USA). Gold particles (0.6 mg and 1.6 μm in diameter; Bio-Rad, CA, USA) were coated with 0.5 μg of each vector for each bombardment. The plasmid-coated particles were bombarded at 1,100 psi and 9-cm distance. The bombarded cells were incubated at 25 °C in darkness for 24 h. YFP and DsRed1 fluorescence was visualized using the BX53 microscope system (Olympus, Japan). The U-FBNA filter cube (excitation filter, BP470–495 nm; dichromatic mirror, DM505 nm; and suppression filter, BA510–550 nm) and U-FGW cube (excitation filter, BP530–550 nm; dichromatic mirror, DM570 nm; and suppression filter, BA575IF nm) were used for detecting YFP and DsRed1 fluorescence, respectively.
Expression of recombinant proteins
We used the wheat germ cell-free system to express the recombinant proteins38,39. For expression, the CDS of OsMYC2 was optimized by synonymous substitution using codon usage of Arabidopsis thaliana and synthesized. The expressed His-Bls-OsMYC2 protein was biotinylated before testing the interaction with FLAG-tagged protein for detection.
AlphaScreen-based protein-protein interaction assays
The AlphaScreen assays with the recombinant proteins described above were performed as described previously40, with several modifications. The AlphaScreen assays were performed using a total volume of 15 μL containing 100 mM Tris-HCl (pH8.0), 100 mM NaCl, 0.1% Tween20, 1 mg/mL BSA, 1 μL biotinylated proteins, and FLAG-tagged proteins at 25 °C for 1 h in a 384-well Optiplate (PerkinElmer). Using the AlphaScreen IgG (ProteinA) detection kit (PerkinElmer) instruction manual, 10 μL of the detection mixture containing 100 mM Tris-HCl (pH 8.0), 100 mM NaCl, 0.1% Tween20, 1 mg/mL BSA, 5 μg/mL anti-DYKDDDDK antibody (1E6, Wako Pure Chemical Industries, Ltd. Osaka, Japan), 0.1 μL streptavidin-coated donor beads, and 0.1 μL Protein A-coated acceptor beads were added to each well of the 384-well Optiplate, followed by incubation at 25 °C for 1 h. Luminescence was analysed using the AlphaScreen detection program.
Statistical analysis
Data are presented as mean ± standard error values. For all the experiments, except transcriptome analysis, statistical analysis was performed using the two-tailed, unpaired Welch’s t-test. Significance was defined as P < 0.05 (*), P < 0.01 (**), or P < 0.001 (***). For the transcriptome analysis, statistical testing was performed as described in Transcriptome analysis. Significance was defined as false discovery rate (FDR) <0.05.
Additional Information
Accession codes: RAP-DB (http://rapdb.dna.affrc.go.jp/) gene IDs for the sequences described in this study are as follows: JAdMYB1 (Os01g0702700), JAdMYB2 (Os12g0564100), JAdMYB3 (Os01g0192300), JAdMYB4 (Os05g0429900), JAdMYB5 (Os08g0549000), OsJAmyb (Os11g0684000), OsMYB55 (Os05g0553400), OsMYC2 (Os10g0575000), OsMYL1 (Os01g0705700), OsMYL2 (Os01g0235700), OsNOMT (Os12g0240900), and OsUBQ (Os10g0542200).
How to cite this article: Ogawa, S. et al. OsMYC2, an essential factor for JA-inductive sakuranetin production in rice, interacts with MYC2-like proteins that enhance its transactivation ability. Sci. Rep. 7, 40175; doi: 10.1038/srep40175 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by JSPS KAKENHI grant no. 26450136 to KO and Ministry of Education, Culture, Sports, Science, and Technology-Supported Program for the Strategic Research Foundation at Private Universities to HY (2013–2017, project number S131052A01). We are grateful to Prof. Dr. Moritoshi Iino (Osaka City University, Japan) for providing seeds of the cpm2 mutant; Prof. Dr. Masaru Takagi (Saitama University, Japan) for the pRL, 430T1.2, and GAL4-TATA-LUC-NOS vectors; the late Prof. Dr. Ko Shimamoto (Nara Institute of Science and Technology, Japan) for the pANDA vectors; Prof. Dr. Tsuyoshi Nakagawa (Shimane University, Japan) for the pnYGW and pcYGW vectors; and Ms. Tomoko Sakazawa (Teikyo University, Japan) for technical assistance.
Footnotes
Author Contributions S.O., K.M., T.S., H.Y., H.N., and K.O. designed the study. S.O., K.M., and K.N. performed the experiments and analysed the data. S.O. and K.O. interpreted the data and wrote the manuscript.
References
- Hu P. et al. JAV1 Controls Jasmonate-Regulated Plant Defense. Mol. Cell 50, 504–515 (2013). [DOI] [PubMed] [Google Scholar]
- Wasternack C. & Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi S. et al. Isolation of jasmonate-induced sesquiterpene synthase of rice: Product of which has an antifungal activity against Magnaporthe oryzae. J. Plant Physiol. 171, 625–632 (2014). [DOI] [PubMed] [Google Scholar]
- Okada K., Abe H. & Arimura G. Jasmonates Induce Both Defense Responses and Communication in Monocotyledonous and Dicotyledonous Plants. Plant Cell Physiol. 56, 16–27 (2015). [DOI] [PubMed] [Google Scholar]
- Miyamoto K., Shimizu T. & Okada K. Transcriptional regulation of the biosynthesis of phytoalexin: A lesson from specialized metabolites in rice. Plant Biotechnol. 31, 377–388 (2014). [Google Scholar]
- Horie K. et al. Identification of UV-Induced Diterpenes Including a New Diterpene Phytoalexin, Phytocassane F, from Rice Leaves by Complementary GC/MS and LC/MS Approaches. J. Agric. Food. Chem. 63, 4050–4059 (2015). [DOI] [PubMed] [Google Scholar]
- Kodama O., Miyakawa J., Akatsuka T. & Kiyosawa S. Sakuranetin, a flavanone phytoalexin from ultraviolet-irradiated rice leaves. Phytochem. 31, 3807–3809 (1992). [Google Scholar]
- Hasegawa M. et al. Analysis on blast fungus-responsive characters of a flavonoid phytoalexin sakuranetin; accumulation in infected rice leaves, antifungal activity antd detoxification by fungus. Molecules 19, 11404–11418 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann M. et al. Identification of rice Allene Oxide Cyclase mutants and the function of jasmonate for defence against Magnaporthe oryzae. Plant J. 74, 226–238 (2013). [DOI] [PubMed] [Google Scholar]
- Shimizu T. et al. OsJAR1 Contributes Mainly to Biosynthesis of the Stress-Induced Jasmonoyl-Isoleucine Involved in Defense Responses in Rice. Biosci Biotechnol Biochem. 77, 1556–1564 (2013). [DOI] [PubMed] [Google Scholar]
- Shimizu T. et al. Purification and identification of naringenin 7-O-methyltransferase, a key enzyme in biosynthesis of flavonoid phytoalexin sakuranetin in rice. J. Biol. Chem. 287, 19315–19325 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K. et al. Jasmonoyl-L-isoleucine is required for the production of a flavonoid phytoalexin but not diterpenoid phytoalexins in ultraviolet-irradiated rice leaves. Biosci. Biotechnol. Biochem. 80, 1934–1938 (2016). [DOI] [PubMed] [Google Scholar]
- Shoji T., Kajikawa M. & Hashimoto T. Clustered Transcription Factor Genes Regulate Nicotine Biosynthesis in Tobacco. Plant Cell 22, 3390–3409 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga H. et al. Identification and Characterization of ANAC042, a Transcription Factor Family Gene Involved in the Regulation of Camalexin Biosynthesis in Arabidopsis. Mol. Plant Microbe Interact. 25, 684–696 (2012). [DOI] [PubMed] [Google Scholar]
- Chujo T. et al. Overexpression of Phosphomimic Mutated OsWRKY53 Leads to Enhanced Blast Resistance in Rice. PLoS ONE 9, e98737 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K. et al. Identification of Target Genes of the bZIP Transcription Factor OsTGAP1, Whose Overexpression Causes Elicitor-Induced Hyperaccumulation of Diterpenoid Phytoalexins in Rice Cells. PLoS ONE 9, e105823 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K. et al. Overexpression of the bZIP transcription factor OsbZIP79 suppresses the production of diterpenoid phytoalexin in rice cells. J. Plant Physiol. 173, 19–27 (2015). [DOI] [PubMed] [Google Scholar]
- Yamamura C. et al. Diterpenoid phytoalexin factor, a bHLH transcription factor, plays a central role in the biosynthesis of diterpenoid phytoalexins in rice. Plant J. 84, 1100–1113 (2015). [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P. et al. The Arabidopsis bHLH Transcription Factors MYC3 and MYC4 Are Targets of JAZ Repressors and Act Additively with MYC2 in the Activation of Jasmonate Responses. Plant Cell 23, 701–715 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. & Manners J. M. MYC2: the master in action. Mol. Plant 6, 686–703 (2013). [DOI] [PubMed] [Google Scholar]
- Goossens J., Swinnen G., Vanden Bossche R., Pauwels L. & Goossens A. Change of a conserved amino acid in the MYC2 and MYC3 transcription factors leads to release of JAZ repression and increased activity. New Phytol. 206, 1229–1237 (2015). [DOI] [PubMed] [Google Scholar]
- Cai Q. et al. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 5, 3476 (2014). [DOI] [PubMed] [Google Scholar]
- Uji Y. et al. Overexpression of OsMYC2 Results in the Up-Regulation of Early JA-Responsive Genes and Bacterial Blight Resistance in Rice. Plant Cell Physiol. 57, 1814–1827 (2016). [DOI] [PubMed] [Google Scholar]
- Li S. Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 9, e27522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoji T. & Hashimoto T. Tobacco MYC2 Regulates Jasmonate-Inducible Nicotine Biosynthesis Genes Directly and By Way of the NIC2-Locus ERF Genes. Plant Cell Physiol. 52, 1117–1130 (2011). [DOI] [PubMed] [Google Scholar]
- Chavali G. B., Vijayalakshmi C. & Salunke D. M. Analysis of sequence signature defining functional specificity and structural stability in helix-loop-helix proteins. Proteins 42, 471–480 (2001). [DOI] [PubMed] [Google Scholar]
- Sawasaki T. et al. Arabidopsis HY5 protein functions as a DNA-binding tag for purification and functional immobilization of proteins on agarose/DNA microplate. FEBS Lett. 582, 221–228 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Mitsuda N., Matsui K. & Ohme-Takagi M. Identification of the minimal repression domain of SUPERMAN shows that the DLELRL hexapeptide is both necessary and sufficient for repression of transcription in Arabidopsis. Biochem. Biophys. Res. Commun. 321, 172–178 (2004). [DOI] [PubMed] [Google Scholar]
- Niu Y., Figueroa P. & Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62, 2143–2154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y. et al. Basic Helix-Loop-Helix Transcription Factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 Are Negative Regulators of Jasmonate Responses in Arabidopsis. Plant Physiol. 163, 291–304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M. et al. A bHLH-Type Transcription Factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, Acts as a Repressor to Negatively Regulate Jasmonate Signaling in Arabidopsis. Plant Cell 25, 1641–1656 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D. & Shimamoto K. Simple RNAi Vectors for Stable and Transient Suppression of Gene Function in Rice. Plant Cell Physiol. 45, 490–495 (2004). [DOI] [PubMed] [Google Scholar]
- Toki S. et al. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 47, 969–976 (2006). [DOI] [PubMed] [Google Scholar]
- Hino T. et al. Two Sec13p Homologs, AtSec13A and AtSec13B, Redundantly Contribute to the Formation of COPII Transport Vesicles in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 75, 1848–1852 (2011). [DOI] [PubMed] [Google Scholar]
- Yano T. et al. AGIA Tag System Based on a High Affinity Rabbit Monoclonal Antibody against Human Dopamine Receptor D1 for Protein Analysis. PLoS ONE 11, e0156716 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M., Ohme-Takagi M. & Shinshi H. Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 22, 29–38 (2000). [DOI] [PubMed] [Google Scholar]
- Shimizu T. et al. Effects of a bile acid elicitor, cholic acid, on the biosynthesis of diterpenoid phytoalexins in suspension-cultured rice cells. Phytochem. 69, 973–981 (2008). [DOI] [PubMed] [Google Scholar]
- Madin K., Sawasaki T., Ogasawara T. & Endo Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc. Natl. Acad. Sci. USA 97, 559–564 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawasaki T. et al. A bilayer cell-free protein synthesis system for high-throughput screening of gene products. FEBS Lett. 514, 102–105 (2002). [DOI] [PubMed] [Google Scholar]
- Nemoto K., Takemori N., Seki M., Shinozaki K. & Sawasaki T. Members of the Plant CRK Superfamily Are Capable of Trans- and Autophosphorylation of Tyrosine Residues. J. Biol. Chem. 290, 16665–16677 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.