Abstract
Fusion of biological membranes is mediated by distinct integral membrane proteins, e.g., soluble N-ethylmaleimide-sensitive factor attachment protein receptors and viral fusion proteins. Previous work has indicated that the transmembrane segments (TMSs) of such integral membrane proteins play an important role in fusion. Furthermore, peptide mimics of the transmembrane part can drive the fusion of liposomes, and evidence had been obtained that fusogenicity depends on their conformational flexibility. To test this hypothesis, we present a series of unnatural TMSs that were designed de novo based on the structural properties of hydrophobic residues. We find that the fusogenicity of these peptides depends on the ratio of α-helix-promoting Leu and β-sheet-promoting Val residues and is enhanced by helix-destabilizing Pro and Gly residues within their hydrophobic cores. The ability of these peptides to refold from an α-helical state to a β-sheet conformation and backwards was determined under different conditions. Membrane fusogenic peptides with mixed Leu/Val sequences tend to switch more readily between different conformations than a nonfusogenic peptide with an oligo-Leu core. We propose that structural flexibility of these TMSs is a prerequisite of fusogenicity.
Biological membrane fusion requires drastic restructuring of lipid bilayers mediated by fusogenic membrane proteins whose structure/function relationships are intensely investigated. Intracellular fusion of eukaryotic membranes is driven by the single membrane-span soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins. A widely accepted model posits that SNARE complex formation establishes membrane proximity leading to complete fusion, albeit SNARE action is precisely regulated by a multitude of accessory proteins (1, 2). In support of this model, recombinant SNARE proteins mediate liposome–liposome fusion in vitro (3–5). When enveloped viruses enter their host cells, fusion is mediated by envelope proteins. In the case of influenza hemagglutinin, a pH-driven global conformational change of the ectodomain ejects an amphipathic fusion peptide toward the target bilayer to establish initial contact between both membranes followed by bilayer merger (1, 2). Thus, establishment of membrane proximity mediated by soluble fusion protein domains is an early step of fusion.
Fusion protein function critically depends on proteinaceous transmembrane segments (TMSs). For example, replacement of the TMSs of yeast exocytotic SNAREs by prenyl anchors blocks exocytosis at hemifusion; i.e., proximal monolayers mix, but distal monolayers remain intact (6). Similarly, a prenyl moiety in place of a yeast vacuolar SNARE TMS abolishes vacuole–vacuole fusion (7). Replacing TMSs of influenza hemagglutinin or of the vesicular stomatitis virus (VSV) G protein with a glycosylphosphatidylinositol membrane anchor (8) allows for hemifusion but blocks full fusion. Similar observations were made upon mutating the TMS sequences of influenza hemagglutinin (9), VSV G protein (10), or other viral fusion proteins (11–13). The TMSs of fusogenic proteins therefore appear to constitute functional domains and may promote the transition from hemifusion to full fusion at a late step.
A role of TMSs in fusion is also supported by recent in vitro studies. In these studies, synthetic peptides representing the TMSs of SNAREs or VSV G protein drive liposome–liposome fusion (14–16) that was enhanced by calcium-induced liposome aggregation (14, 15) or by osmotic stress (16). In case of the VSV G protein, mutations that affected fusogenicity of the full-length protein in vivo (10) also affected fusogenicity of the TMS peptide in vitro (15, 16). CD spectroscopy showed these TMS peptides to exist in an equilibrium of α-helical and β-sheet structures in solution. As the fusogenic activity of mutant peptides decreased with increasing stability of their α-helical conformations, it was proposed that structural plasticity of TMSs may support fusion protein function (14, 16). These data are consistent with an unusual amino acid composition of fusion protein TMSs as β-sheet-promoting β-branched amino acids and/or helix-destabilizing Gly residues are overrepresented in TMSs of SNAREs (14) and viral fusogens (10).
Here, we took a direct approach to test this hypothesis. We designed a number of hydrophobic model TMS sequences that were analyzed with respect to their membrane fusogenicity and secondary structures. We find that hydrophobic sequences that are made up of mixtures of helix- and sheet-promoting residues plus helix-destabilizing residues are most fusogenic and that this functional property is paralleled by a large degree of structural flexibility.
Methods
Peptide Synthesis. Peptides were synthesized by tert-butoxycarbonyl (Peptide Specialty Laboratories, Heidelberg) or fluorenylmethoxycarbonyl chemistry and purified by HPLC to purities >90% as determined by MS or used without purification when synthesis yielded products that were >90% pure. Concentrations were determined by means of tryptophan absorbance.
Preparation and Fusion of Small Unilamellar Liposomes. Liposomes were prepared by sonication as described in ref. 14 from mixtures of egg phosphatidylcholine/brain phosphatidylethanolamine (PE)/brain phosphatidylserine (phosphatidylcholine was purified from fresh egg yolk; PE and phosphatidylserine were from Avanti Polar Lipids) at a ratio of 6:2:2 (wt/wt/wt) with or without 0.8% (wt/wt) N-(7-nitro-2,1,3-benzoxadiazol-4-yl)hexadecyl-PE (NBD-PE) and N-(lissamine rhodamine B sulfonyl)hexadecyl-PE (Molecular Probes). Peptide/lipid (P/L) ratios were determined as described in ref. 15, except that liposomes were lysed with 1% (wt/vol) SDS before determining peptide concentrations by tryptophan fluorescence. Fusion assays were done by using a fluorescence dequenching assay (17) as described in refs. 14 and 15. Initial rates of fusion were calculated by fitting the first 10 min of the kinetics by a polynomial function and determining its first derivative. All values were corrected for the peptide-independent, spontaneous fusion of pure liposomes and for detergent quenching of NBD fluorescence (<4% of the total values).
CD Spectroscopy. For polarity titrations, peptides dissolved at 1 mg/ml in either 10 mM Tris·HCl buffer (pH 7.5) or in 100% trifluoroethanol (TFE) were brought to the respective buffer/TFE ratios while maintaining a peptide concentration of 100 μg/ml. CD spectra were obtained by using a J-710 automatic recording spectral polarimeter (Jasco, Easton, MD) calibrated with 0.05% β-androsterone in spectral grade dioxane as a standard. Spectra were routinely measured from 190 nm to 240 nm in a 1-mm dichroically neutral quartz cuvette at 20°C by using a time constant of 4 sec per scan speed of 5 nm/min and a sensitivity of 100 millidegrees per cm. Spectra were the signal-averaged accumulation of four scans with the baseline subtracted. For measurements in inverse micelles, sodium bis(2-ethylhexyl)sulfosuccinate/isooctane/water inverse micelles were prepared according to Grandi et al. (18). Aqueous peptide solutions were injected into a solution of 50 mM sodium bis(2-ethylhexyl)sulfosuccinate in isooctane (effective peptide concentrations were 40 μM), shaken for 2 min, and measured immediately. Signals were averaged eight times. Micelles were measured with 0.3%, 1%, 1.4%, 2%, or 3% aqueous peptide solution. All spectra were converted to mean residue ellipticity (θmr), and secondary structures were calculated from the CD spectra normalized to θmr by using the program pepfit (19).
Fourier Transform Infrared (FTIR) Spectroscopy. FTIR spectra were recorded on a Tensor 27 spectrometer (Bruker, Billerica, MA) equipped with a liquid nitrogen-cooled mercury–cadmium–tellurium detector and a manually operated flowcell (Micro Biolytics, Freiburg, Germany) that was thermostated at 10°C to avoid TFE evaporation and that had a 6-μm pathlength. Then 64 interferograms of a spectral range of 2,000–1,000 cm–1 with 2 cm–1 instrumental resolution were averaged before Fourier transformation. Peptides were diluted from a stock in TFE to a concentration of 1.5 mg/ml into mixtures of 10%, 20%, 40%, 60%, 80%, or 100% TFE in 2H2O. Three spectra were recorded of each titration step, and averages are shown. Spectra were smoothed with a Fourier self-deconvolution algorithm. The secondary structure was determined by deconvolution of the amide-I band (1,700–1,600 cm–1) as described in ref. 20. The number of bands, band position, band intensity, full width at one-half height, curvature shape (Cauchy curves were used with variable G/Lorentz contents), and an overall baseline for the whole amide-I band were varied. All start parameters (except band intensity and baseline) were identical for all spectra of the same peptide. A minimum number of bands was used to deconvolute the measured spectra.
Cross Polarization Magic Angle Spinning (CP-MAS) NMR Experiments. Peptides (5 mg) in 50% acetic acid (vol/vol) were frozen in liquid nitrogen, lyophilized, and put in a 4-mm MAS-NMR rotor. The 1D 13C-natural abundance NMR spectra were recorded on a 13C-radio frequency of 188 MHz by using CP-MAS NMR spectroscopy on a Bruker AV-750 spectrometer equipped with a double-channel CP-MAS probe head. All data were recorded at room temperature by using a MAS spin frequency ωR/2π of 12.5 kHz in a magnetic field strength of 17.6 T. The 13CO resonance of U-[13C,15N]-Tyr·HCl at 172.1 ppm was used as an external reference for the calibration of the isotropic 13C-chemical shifts.
Results
Design of Fusogenic Peptides. The peptides characterized in this study, designated “LV peptides,” contained hydrophobic core sequences whose design was based on hydrophobic residues with different secondary structure propensities, i.e., helix-promoting Leu and sheet-promoting Val residues (21, 22) at different ratios. In some peptides, the core sequences also contained Gly and/or Pro residues that are known to destabilize helices (23, 24). All sequences were flanked by Lys for better solubility and membrane anchoring (25). A Trp residue was included for quantitation (Fig. 1).
Fig. 1.
Design of fusogenic peptides. Peptides contain hydrophobic core sequences based on different Leu/Val (L/V) ratios and helix-destabilizing Pro and Gly residues (in bold).
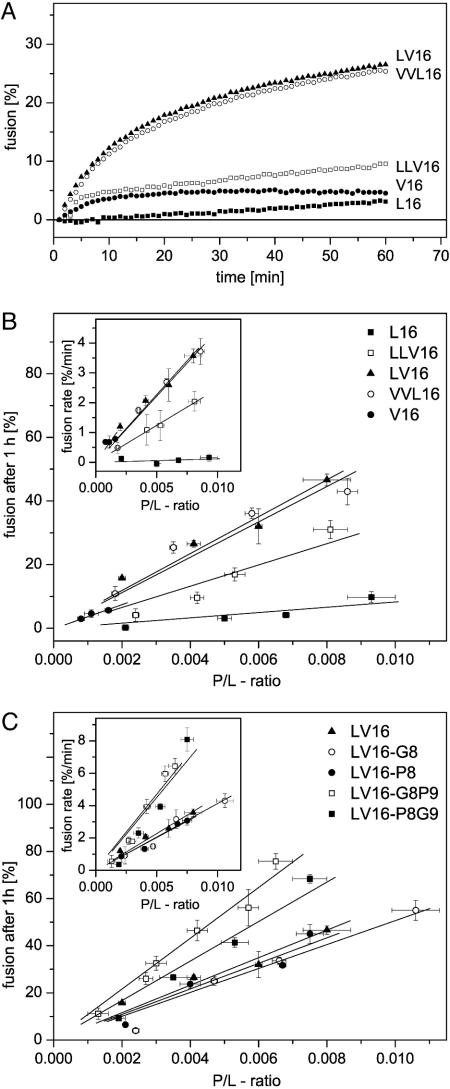
Fusogenic Activity of LV Peptides. The peptides were incorporated into liposomal membranes by sonication at different P/L ratios. Liposome–liposome fusion was examined by a standard fluorescence dequenching assay (17) upon rapidly shifting the temperature to 37°C. This assay is based on fluorescence resonance energy transfer between the membrane-bound fluorophores NBD-PE and N-(lissamine rhodamine B sulfonyl)hexadecyl-PE being present at quenching concentrations in liposomal membranes. Upon fusion of labeled membranes with peptide-containing unlabeled membranes, the average distance between the fluorophores and, thus, NBD fluorescence increases over time; this result is taken as a measure of lipid mixing. A mixture of egg phosphatidylcholine, brain PE, and brain phosphatidylserine (at a 6:2:2 weight ratio) was used (14). Fig. 2 compares membrane fusogenicity of different LV peptides. Initial fusion rates reflect the probability by which a random liposome–liposome collision turns into a productive fusion event and were obtained from the initial steepness of the kinetics. The extents of fusion, as seen after 1 h of incubation, reflect the sum of all fusion events within that time period. Both parameters were plotted against the experimentally determined P/L ratios. The resulting dose/response relationships revealed that fusion rates and fusion extents increased linearly with the P/L ratios. Interestingly, fusogenicities were strongly dependent on the Leu/Val ratios. Whereas a Leu-based peptide (L16) was virtually nonfusogenic, as seen in refs. 14 and 15, mixtures of both residue types exhibited intermediate (LLV16) to strong (LV16 and VVL16) fusogenicities (Fig. 2 A and B). The Val-based peptide (V16) was a special case because it led to massive lipid/peptide aggregates upon sonication. Upon removal of these aggregates by centrifugation, the liposomes in the supernatant exhibited only P/L ratios <0.002 such that the fusogenicity of V16 could not be determined with certainty.
Fig. 2.
Fusogenic activities of TMS peptides. (A) Typical fusion kinetics reveal that fusogenicity depends on the Leu/Val ratio of the hydrophobic peptide core sequences. P/L ratios were from 0.0035 to 0.005, except for peptide V16, which was incorporated up to a P/L ratio of only 0.0011. (B) Quantitative comparison of fusogenicities of the different aliphatic core sequences by plotting fusion extents, as seen after 1 h, or initial rates (Insets) as a function of P/L ratio. (C) Fusogenicities of peptides containing Gly and/or Pro relative to the parental LV16. Data points represent means ± SE (n = 4–18 independent experiments).
We next examined the influence of helix-destabilizing residue types within the context of our most fusogenic aliphatic sequence LV16. Placing either a Gly or a Pro residue at the center of the peptide (LV16-G8 and LV16-P8) had little effect on fusogenicity. However, a Gly/Pro (LV16-G8P9) or a Pro/Gly (LV16-P8G9) pair significantly enhanced fusion (Fig. 2C).
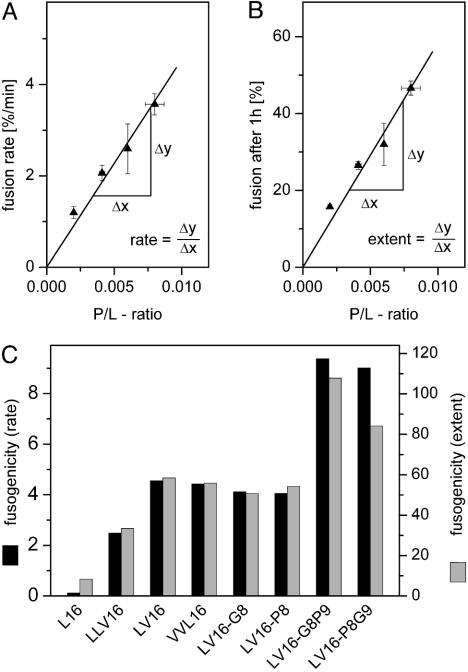
Fig. 3 summarizes all fusogenicities. To obtain a parameter that is independent of P/L ratio, we calculated the slopes of the dose/response relationships shown in Fig. 2; i.e., we obtained a general dimensionless measure for initial rates and extents of fusion for each peptide. Accordingly, fusion extents and rates increase up to ≈10-fold and ≈80-fold, respectively, in the following rank order: L16 < LLV16 < VVL16 ≈ LV16 ≈ LV16-G8 ≈ LV16-P8 < LV16-G8P9 ≈ LV16-P8G9.
Fig. 3.
Comparison of fusogenicities. (A) Taking LV16 as an example, the schematic depicts how the slopes of the relationships between P/L ratio and initial fusion rate (Left) or fusion extent (Right) were derived for the comparison shown in B. V16 was omitted here because of its inefficient membrane incorporation.
We conclude that we designed fusogenic peptides by mixing helix-promoting Leu and sheet-promoting Val residues and that fusogenicity is enhanced by helix-destabilizing residues.
To show that fusion involved both membrane leaflets rather than being arrested at hemifusion, we extinguished the fluorescence of the NBD moiety in the outer leaflet by dithionite treatment (15). By using LV16 and LV16-P8G9 as examples, we found that NBD fluorescence dequenching was indistinguishable with or without previous outer-leaflet bleaching. This indicates that inner- and outer-membrane leaflets fused with similar efficiencies (data not shown).
Structural Flexibility of LV Peptides. To assess whether the different fusogenicities of our peptides are related to their structural properties, we determined their secondary structures under different conditions.
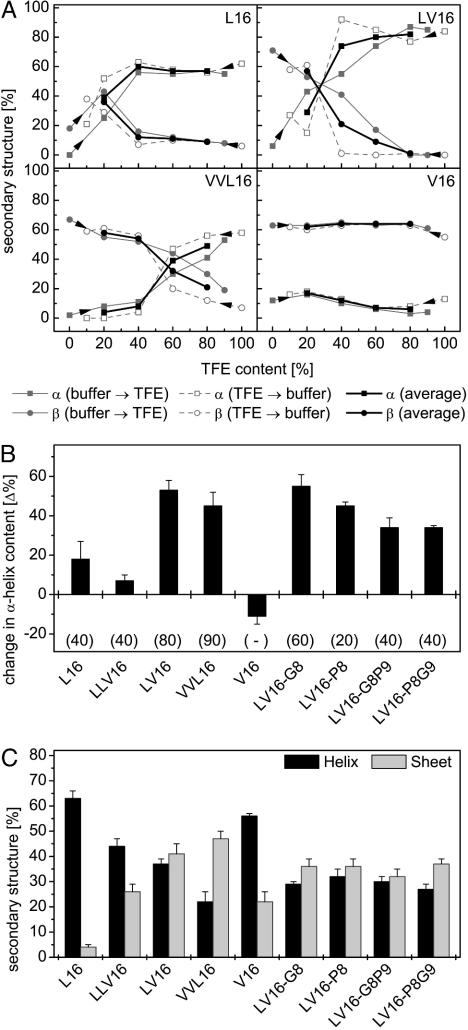
First, we determined secondary structures by CD spectroscopy in various mixtures of aqueous buffer and TFE. TFE stabilizes helical structures indirectly by partial desolvation, i.e., destabilization, of the unfolded state (26), which is similar to the situation in a membrane. Changes in secondary structure upon stepwise addition of aqueous buffer to peptides dissolved in TFE or vice versa are therefore taken as rough measures of conformational flexibility of the helices. At peptide concentrations of 0.1 mg/ml, all peptides, except V16, were highly helical. Increasing solvent polarity decreased α-helix contents and favored β-sheet formation. The changes of turn plus random coil structures followed no systematic order; it should be noted that they dropped by ≈50% for L16 and LLV16 but only by <20% for all other peptides when titrating from buffer to 20% TFE. In general, secondary structure changes were rather similar irrespective of the direction (buffer → TFE or TFE → buffer) of the titrations indicating their largely reversible nature. Fig. 4A shows some of the titration curves for which the values (from 20% to 80% TFE) of the forward and backward titrations were averaged (Fig. 4A, bold lines). Fig. 4B compares the differences between the average α-helicities at 20% and 80% TFE. Interestingly, within the series L16, LLV16, LV16, VVL16, and V16, the largest secondary structure changes were seen for the most fusogenic peptides (LV16 and VVL16). Furthermore, the crossover points of the helix/sheet transitions are shifted to higher TFE concentrations with VVL16, compared with LV16 (Fig. 4A). V16 displayed mostly β-sheet structure with little α-helicity under all conditions. Those peptides with Gly and/or a Pro residues changed secondary structure similar to LV16, although over narrower polarity ranges (in terms of percentage TFE).
Fig. 4.
Structural plasticity of LV peptides determined by CD spectroscopy. (A) Contents of α-helical and β-sheet structure are shown for some peptides (0.1 mg/ml) upon titrating from either aqueous buffer or from TFE solution as indicated by arrowheads. Bold lines represent the averaged values of both types of titrations. (B) Conformational change of peptides as revealed by differences in average helix content seen at 20% and 80% TFE, respectively. Gains in secondary structure upon decreasing polarity result in positive values. Note that the change in helix content tended to be larger with the more fusogenic peptides. Data are shown as means ± SE (n = 4–5). (C) Contents of α-helical and β-sheet structure in inverse sodium bis(2-ethylhexyl)sulfosuccinate/water/iso-octane micelles. Data are shown as means ± SE (n = 8–10).
Second, CD spectroscopy was done with peptides dissolved in inverse micelles where small water droplets are dispersed by sodium bis(2-ethylhexyl)sulfosuccinate in a bulk of iso octane (27). In this solvent, the charged termini of the peptides are thought to be solvated by water, whereas their hydrophobic cores are exposed to hydrocarbon mimicking the hydrophobic core of a lipid membrane. Overall regular secondary structure contents of our LV peptides were between 60% and 80% (Fig. 4C). Interestingly, almost equal α-helix and β-sheet contents were seen with the most fusogenic peptides (LV16, LV16-G8, LV16-P8, LV16-G8P9, and LV16-P8G9), except VVL16 that contained more sheet than helix. Those peptides with lower fusogenicity (L16 and LLV16) displayed more helix (63% and 44%) than sheet (4% and 26%). A surprising observation is that V16 also formed more helix than sheet under these conditions.
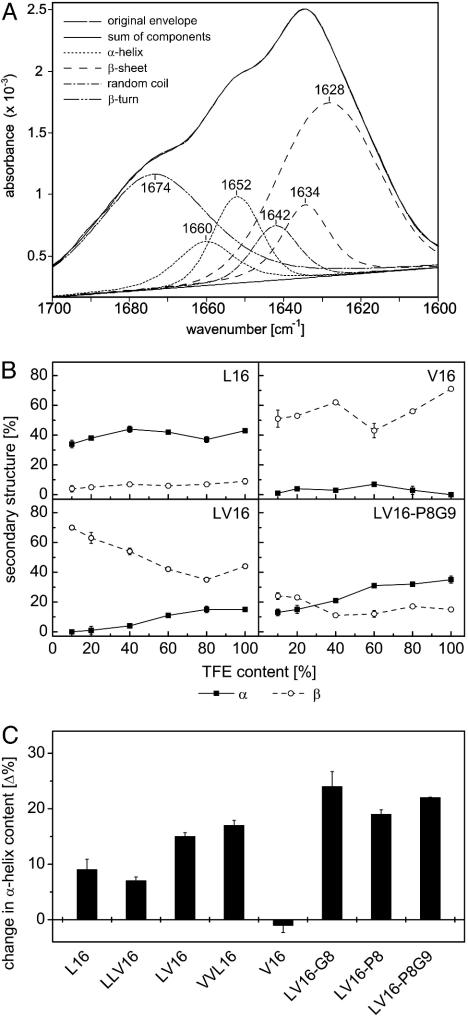
Third, we performed the polarity titrations at a 15-fold higher peptide concentration to cover a broad concentration range. Because peptide solutions of 1.5 mg/ml are difficult to examine by CD spectroscopy, these concentrations were analyzed by FTIR spectroscopy, a further tool to analyze secondary structures of proteins by deconvoluting the amide-I band (28). We started from pure TFE solutions and stepwisely added buffer before taking spectra. L16 and LLV16 were mostly α-helical (≈40%) under all conditions. The fusogenic LV16 and VVL16 peptides displayed only ≈20% α-helix in TFE and polarity increases reduced α-helix contents, albeit to a smaller degree than at 0.1 mg/ml (see Fig. 4) V16 displayed mostly β-sheet (Fig. 5B). Thus, the notion that the more fusogenic peptides exhibit higher structural flexibility is confirmed at the higher peptide concentration. Here, crossover of helical to sheet structure upon polarity increase was only seen for LV16-P8G9 (Fig. 5B).
Fig. 5.
Structural plasticity of LV peptides determined by FTIR spectroscopy. (A) Original spectrum of LV16 in 100% TFE and decomposition of the amide-I band into individual bands according to ref. 27; the superposition of the original envelope with the sum of components demonstrates the quality of the fit. (B) α-Helix and β-sheet contents of some peptides (1.5 mg/ml) upon titration from TFE solution. (C) Conformational change of peptides as revealed by differences in helix content seen at 10% and 100% TFE, respectively. Gains in secondary structure upon decreasing polarity result in positive values. Note that the changes in helix contents tended to be larger with the more fusogenic peptides. Data are shown as means ± SE (n = 3); error bars were omitted when they were smaller than the symbols. Results obtained with peptide LV16-G8P9 are not shown because it adhered to the cuvette surface with unknown effects on its secondary structure.
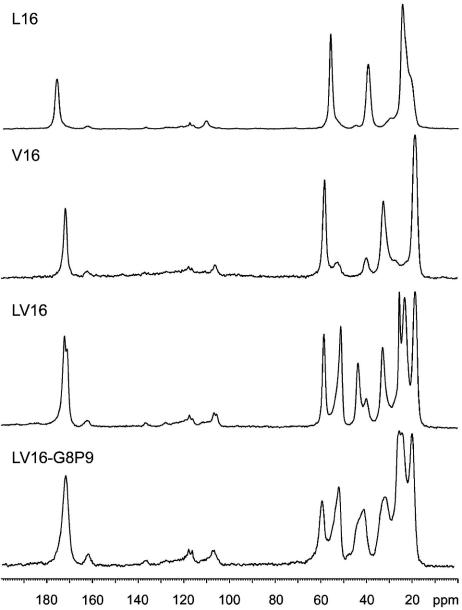
Fourth, to determine how the peptides fold when all solvent is removed, high-resolution 13C-1D solid-state NMR spectra were obtained in the lyophilized state (Fig. 6) that is the starting material for the fusion assays. The 13C-carbonyl shift reflects that conformation as a response at ≈175 ppm is characteristic for an α-helix, whereas a response at ≈170 ppm is in general attributed to a β-sheet (29). Hence, L16 (175.8 ppm) forms an α-helix, whereas V16 (170.3 ppm) forms a β-sheet. LV16 shows two signals at 172.5 ppm and 171.5 ppm. This splitting indicates slightly different signals for the carbonyl 13C of the alternating Leu and Val residues in a unique overall conformation. Because the chemical shift is in the upfield region, the data indicate a β-sheet conformation. The resonance observed for LV16-G8P9 (171.3 ppm) also is indicative of predominantly β-sheet. This carbonyl line is asymmetric with a downfield extension at the base into the shift range for α-helix. Furthermore, all peptides showed relatively broad signals indicating considerable structural disorder at the microscopic scale, relative to a fully crystalline peptide preparation. The structural disorder appears most pronounced for LV16 and LV16-G8P9, which is in line with their fusion-promoting activity.
Fig. 6.
One-dimensional 13C-natural abundance CP-MAS solid-state NMR spectra of lyophilized peptides. Please note the different 13C-carbonyl resonances for L16 and V16. The resonances from 20 ppm to 60 ppm correspond to Cα, Cβ, and side-chain carbons.
Discussion
We have examined whether the membrane-fusogenic activity of simplified hydrophobic model peptides is related to their conformational flexibility. Our results show that mixed Leu/Val sequences exhibit high fusogenicities that are further increased by including Gly/Pro or Pro/Gly pairs. At the same time, the more fusogenic peptides were distinguished by a generally lower stability of their helical conformation. When comparing secondary structures at low (0.1 mg/ml) and high (1.5 mg/ml) peptide concentrations, β-sheet contents of LV16 and VVL16 are increased at 1.5 mg/ml. It should be borne in mind that absolute secondary structure contents determined by CD or FTIR spectroscopy may differ because of different underlying physical principles. Nevertheless, both techniques agree in reporting polarity-dependent refolding of our most fusogenic peptides. Differences may also be due to stabilization of β-sheet through aggregate formation at the higher peptide concentration, in line with the solid-state NMR data. LV16 and VVL16 might be particularly prone to aggregation due to their high Val contents, whereas insertion of Gly and/or Pro decrease sheet contents. In the solid-state, all fusogenic peptides analyzed predominantly fold as β-sheet, although the asymmetry of the carbonyl line is evidence for a mixture of α-helical and β-sheet conformation for LV16 and LV16-G8P9.
Membrane fusion can be divided into early and late steps. At the early step, membranes are brought into close proximity by the action of membrane-extrinsic fusion protein domains (1, 2). TMSs may support this early step by enhancing the stability and/or multimerization of viral (30, 31) or SNARE-based (32–34) fusion complexes. Actual lipid mixing is a late step and may be facilitated by structurally flexible TMSs. Although structural flexibility of our fusogenic LV variants manifested itself as helix to sheet transition in isotropic solution, we do not necessarily consider that this transition takes place in the low-dielectric environment of a membrane. Rather, the corresponding α-helical structures may be conformationally flexible in a sense that they locally unwind in the bilayer and thus facilitate lipid mixing. We cannot rule out that peptide aggregation upon β-sheet formation contributes to lipid mixing; however, the lack of cooperativity seen when examining fusion at different P/L ratios argues against this possibility.
That structurally flexible TMSs are required for fusion is supported by observations with natural fusion proteins. β-Branched residues and/or Gly are overrepresented in the TMSs of SNAREs (14) and viral fusion proteins (10), and a Gly/Pro pair is conserved in the fusion protein TMS from type C retroviruses (35). Functional studies demonstrated that mutating Gly residues within the TMSs of influenza hemagglutinin (9), the VSV G protein (10), or the reovirus fusion associated small transmembrane protein (13) or mutating a Pro within the Moloney virus fusion protein TMS (12) drastically reduced the fusogenicities of these full-length proteins in cellular assays. Thus, helix-destabilizing residues may enhance fusion protein function by means of destabilization of TMS helices known from previous model studies (36, 37). Our polarity titration curves show that LV peptides with Gly and/or Pro tend to unfold over a more narrow range of TFE concentration, suggesting a more cooperative helix/sheet transition compared with the parental LV16. We note, however, that the increased fusogenicities of peptides with Gly/Pro or Pro/Gly pairs relative to those with single Gly or Pro are not reflected by significantly different polarity titration curves. We may speculate that the pairs may speed up conformational dynamics during lipid mixing that is known to proceed on a millisecond time scale (38).
Conformational flexibility appears to be one important structural property of membrane-active fusion protein domains. The interplay of flexibility and other structural features with lipids will have to be studied in the future for a more complete understanding of membrane fusion.
Acknowledgments
We thank Dr. M. Gütlich for critical comments on the manuscript and Bettina Brosig for initial experiments. This work was supported by the Volkswagenstiftung Conformational Control of Biomolecular Function Funding Initiative.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TFE, trifluoroethanol; CP-MAS, cross polarization magic angle spinning; FTIR, Fourier transform infrared; PE, phosphatidylethanolamine; NBD-PE, N-(7-nitro-2,1,3-benzoxadiazol-4-yl)hexadecyl-PE; P/L, peptide/lipid; SNARE, soluble N-ethylmaleimide-sensitive factor attachment protein receptor; TMS, transmembrane segment; VSV, vesicular stomatitis virus.
References
- 1.Jahn, R., Lang, T. & Südhof, T. C. (2003) Cell 112, 519–533. [DOI] [PubMed] [Google Scholar]
- 2.Tamm, L. K., Crane, J. & Kiessling, V. (2003) Curr. Opin. Struct. Biol. 13, 453–466. [DOI] [PubMed] [Google Scholar]
- 3.Weber, T., Zemelman, B. V., McNew, J. A., Westermann, B., Gmachl, M., Parlati, F., Söllner, T. H. & Rothman, J. E. (1998) Cell 92, 759–772. [DOI] [PubMed] [Google Scholar]
- 4.Schuette, C. G., Hatsuzawa, K., Margittai, M., Stein, A., Riedel, D., Küster, P., König, M., Seidel, C. & Jahn, R. (2004) Proc. Natl. Acad. Sci. USA 101, 2858–2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tucker, W. C., Weber, T. & Chapman, E. R. (2004) Science 304, 435–438. [DOI] [PubMed] [Google Scholar]
- 6.Grote, E., Baba, M., Ohsumi, Y. & Novick, P. J. (2000) J. Cell Biol. 151, 453–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rohde, J., Dietrich, L., Langosch, D. & Ungermann, C. (2003) J. Biol. Chem. 278, 1656–1662. [DOI] [PubMed] [Google Scholar]
- 8.Kemble, G. W., Danieli, T. & White, J. M. (1994) Cell 76, 383–391. [DOI] [PubMed] [Google Scholar]
- 9.Melikyan, G. B., Markosyan, R. M., Roth, M. G. & Cohen, F. S. (2000) Mol. Biol. Cell 11, 3765–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleverley, D. Z. & Lenard, J. (1998) Proc. Natl. Acad. Sci. USA 95, 3425–3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Owens, R. J., Burke, C. & Rose, J. K. (1994) J. Virol. 68, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor, G. M. & Sanders, D. A. (1999) Mol. Biol. Cell 10, 2803–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shmulevitz, M., Salsman, J. & Duncan, R. (2003) J. Virol. 77, 9769–9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langosch, D., Crane, J. M., Brosig, B., Hellwig, A., Tamm, L. K. & Reed, J. (2001) J. Mol. Biol. 311, 709–721. [DOI] [PubMed] [Google Scholar]
- 15.Langosch, D., Brosig, B. & Pipkorn, R. (2001) J. Biol. Chem. 276, 32016–32021. [DOI] [PubMed] [Google Scholar]
- 16.Dennison, S. M., Greenfield, N., Lenard, J. & Lentz, B. R. (2002) Biochemistry 41, 14925–14934. [DOI] [PubMed] [Google Scholar]
- 17.Struck, D. K., Hoekstra, D. & Pagano, R. E. (1981) Biochemistry 20, 4093–4099. [DOI] [PubMed] [Google Scholar]
- 18.Grandi, C., Smith, R. E. & Luisi, P. L. (1981) J. Biol. Chem. 256, 837–843. [PubMed] [Google Scholar]
- 19.Reed, J. & Reed, T. A. (1997) Anal. Biochem. 254, 36–40. [DOI] [PubMed] [Google Scholar]
- 20.Goormaghtigh, E., Raussens, R. & Ruyschaert, J.-M. (1999) Biochim. Biophys. Acta 1422, 105–185. [DOI] [PubMed] [Google Scholar]
- 21.Minor, D. L. & Kim, P. S. (1994) Nature 367, 660–663. [DOI] [PubMed] [Google Scholar]
- 22.Street, A. G. & Mayo, S. L. (1999) Proc. Natl. Acad. Sci. USA 96, 9074–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordes, F. S., Bright, J. N. & Sansom, M. S. P. (2002) J. Mol. Biol. 323, 951–960. [DOI] [PubMed] [Google Scholar]
- 24.Monne, M., Hermansson, M. & von Heijne, G. (1999) J. Mol. Biol. 288, 141–145. [DOI] [PubMed] [Google Scholar]
- 25.de Planque, M. R., Kruijtzer, J. A., Liskamp, R. M., Marsh, D., Greathouse, D. V., Koeppe, R. E., II, de Kruijff, B. & Killian, J. A. (1999) J. Biol. Chem. 274, 20839–20846. [DOI] [PubMed] [Google Scholar]
- 26.Kentsis, A. & Sosnick, T. R. (1998) Biochemistry 37, 14613–14622. [DOI] [PubMed] [Google Scholar]
- 27.Waks, M. (1986) Proteins Struct. Funct. Genet. 1, 4–15.2453052 [Google Scholar]
- 28.Goormaghtigh, E., Cabiaux, V. & Ruyschaert, J.-M. (1994) Subcell. Biochem. 23, 329–450. [DOI] [PubMed] [Google Scholar]
- 29.Kameda, T., Takeda, N., Kuroki, S., Kurosu, H., Ando, S., Ando, I., Shoji, A. & Ozaki, T. (1996) J. Mol. Struct. 384, 17–23. [Google Scholar]
- 30.Doms, R. W. & Helenius, A. (1986) J. Virol. 60, 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tatulian, S. A. & Tamm, L. K. (2000) Biochemistry 39, 496–507. [DOI] [PubMed] [Google Scholar]
- 32.Margittai, M., Otto, H. & Jahn, R. (1999) FEBS Lett. 446, 40–44. [DOI] [PubMed] [Google Scholar]
- 33.Laage, R., Rohde, J., Brosig, B. & Langosch, D. (2000) J. Biol. Chem. 275, 17481–17487. [DOI] [PubMed] [Google Scholar]
- 34.Roy, R., Laage, R. & Langosch, D. (2004) Biochemistry 43, 4964–4970. [DOI] [PubMed] [Google Scholar]
- 35.Schroth-Diez, B., Ludwig, K., Baljinnyam, B., Kozerski, C., Huang, Q. & Herrmann, A. (2000) Biosci. Rep. 20, 571–595. [DOI] [PubMed] [Google Scholar]
- 36.Li, S. C. & Deber, C. M. (1992) FEBS Lett. 311, 217–220. [DOI] [PubMed] [Google Scholar]
- 37.Liu, L.-P. & Deber, C. M. (1998) J. Biol. Chem. 273, 23645–23648. [DOI] [PubMed] [Google Scholar]
- 38.Nomura, F., Inaba, T., Ishikawa, S., Nagata, M., Takahashi, S., Hotani, H. & Takiguchi, K. (2004) Proc. Natl. Acad. Sci. USA 101, 3420–3425. [DOI] [PMC free article] [PubMed] [Google Scholar]