Abstract
A critical early event in the HIV type 1 (HIV-1) particle assembly pathway is the targeting of the Gag protein to the site of virus assembly. In many cell types, assembly takes place predominantly at the plasma membrane. Cellular factors that regulate Gag targeting remain undefined. The phosphoinositide phosphatidylinositol (4,5) bisphosphate [PI(4,5)P2] controls the plasma membrane localization of a number of cellular proteins. To explore the possibility that this lipid may be involved in Gag targeting and virus particle production, we overexpressed phosphoinositide 5-phosphatase IV, an enzyme that depletes cellular PI(4,5)P2, or overexpressed a constitutively active form of Arf6 (Arf6/Q67L), which induces the formation of PI(4,5)P2-enriched endosomal structures. Both approaches severely reduced virus production. Upon 5-phosphatase IV overexpression, Gag was no longer localized on the plasma membrane but instead was retargeted to late endosomes. Strikingly, in cells expressing Arf6/Q67L, Gag was redirected to the PI(4,5)P2-enriched vesicles and HIV-1 virions budded into these vesicles. These results demonstrate that PI(4,5)P2 plays a key role in Gag targeting to the plasma membrane and thus serves as a cellular determinant of HIV-1 particle production.
Retrovirus particle production is a multistep process promoted by the viral Gag precursor protein (1, 2). The HIV type 1 (HIV-1) Gag polyprotein, Pr55Gag, is composed of four domains that are cleaved by the viral protease (PR) concomitantly with virus release to generate the mature Gag proteins: matrix (MA or p17), capsid (CA or p24), nucleocapsid (NC or p7) and p6, and two small spacer peptides, SP1 and SP2. The N-terminal portion of MA, which is myristylated, facilitates the binding of Gag to membrane, whereas CA and NC promote Gag multimerization. p6 plays a central role in the release of virus particles from the cell by interacting with the class E vacuolar protein sorting protein Tsg101 (3, 4).
Mutational analyses have established that, in addition to directing Gag membrane binding, the HIV-1 MA domain regulates the targeting of Gag to the site of virus particle assembly (5-14). Recent reports suggest that wild-type HIV-1 assembly occurs either on the plasma membrane or in a late endosome/multivesicular body (MVB) compartment. In cell types such as HeLa and T cells, the majority of virus assembly takes place at the plasma membrane. In contrast, in primary macrophages, assembly occurs predominantly in late endosomes/MVBs (15-18). Some cell types appear to support assembly both at the plasma membrane and in late endosomes/MVBs (19, 20). Mutations in a highly basic domain of MA (amino acids 17-31), or between MA amino acids 84 and 88, shift Gag localization from the plasma membrane to intracellular vesicles (10-13), a compartment we recently identified as the MVB (18). In addition to its role in Gag targeting, the MA basic domain has been suggested to contribute to the membrane binding of Gag by interacting with acidic phospholipids on the cytoplasmic leaflet of cellular membranes (21). Mutations in the MA basic domain can thus affect both Gag targeting and overall membrane binding efficiency (12, 21-23). The above-mentioned studies on the cell type-dependent nature of Gag targeting strongly suggest that not only the MA domain of Gag but also host cell factors play an active role in determining the subcellular location of HIV-1 particle assembly. However, the identity of cellular cofactors for HIV-1 Gag targeting remains to be defined.
Many cellular proteins are known to contain membrane targeting domains in which basic amino acids play a key role (24-26). These structural determinants, which include pleckstrin homology (PH) domains, engage in interactions with negatively charged lipid molecules collectively known as phosphoinositides (24-26). Members of the phosphoinositide family of lipids differ from each other in the position and number of phosphate moieties on the inositol ring. Phosphoinositides can thus be converted from one type to another via the action of specific lipid kinases and phosphatases (27, 28). Importantly, different phosphoinositides localize preferentially to distinct subcellular membranes, thereby influencing the targeting of proteins to which they bind (29, 30). For example, phosphatidylinositol (4,5) bisphosphate [PI(4,5)P2] is concentrated primarily on the cytoplasmic leaflet of the plasma membrane. As a result, the interactions between PI(4,5)P2 and various basic domains (e.g., the PH domain of phospholipase Cδ1) can direct a wide variety of proteins with such domains specifically to the plasma membrane (27-33). Intracellular levels of PI(4,5)P2 can be regulated by phosphatases, including polyphosphoinositide 5-phosphatases, which reduce PI(4,5)P2 levels by hydrolyzing the phosphate at the D5 position of PI(4,5)P2. In addition, the activity of phosphatidylinositol-4-phosphate 5-kinase, an enzyme that generates the majority of PI(4,5)P2, is regulated by a number of factors including the small G protein ADP-ribosylation factor 6 (Arf6) (ref. 34, reviewed in ref. 35). Expression of a constitutively active form of Arf6 defective for GTP hydrolysis, Arf6/Q67L, alters cellular PI(4,5)P2 localization by inducing the formation of PI(4,5)P2-enriched endosomal structures (36, 37).
Because the basic amino acid sequence in HIV-1 MA is critically involved in Gag targeting to the plasma membrane, we hypothesized that the subcellular localization of Gag may be determined by PI(4,5)P2 in a manner analogous to proteins containing phosphoinositide-binding domains. Consistent with this hypothesis, inositol phosphates, which are structurally related to phosphoinositides, bind Gag and modulate particle assembly in vitro (38).
In the current study, we examined whether cellular PI(4,5)P2 is involved in HIV-1 Gag targeting and virus particle production. To address this question, we perturbed PI(4,5)P2 levels in virus-producing cells by using two different approaches: (i) overexpressing polyphosphoinositide 5-phosphatase IV (5ptaseIV), and (ii) overexpressing the constitutively active Arf6 mutant Arf6/Q67L. The results of this study strongly suggest that PI(4,5)P2 plays a key role in localizing Gag to the plasma membrane and promoting efficient virus particle production. These findings shed light on a critically important step in HIV-1 replication and reveal a previously undescribed function of PI(4,5)P2.
Materials and Methods
Cells, Plasmids, Transfections, and Virus Release Assays. HeLa cells were maintained as described (39). The HIV-1 molecular clone pNL4-3 (40) and its derivatives, pNL4-3/PR- (41) and pNL4-3/55FLAG (12), were reported. The 5ptaseIV expression plasmid, pcDNA4TO/Myc5ptaseIV, was a kind gift from P. Majerus (Washington University School of Medicine, St. Louis) (42). The Δ1 5ptaseIV mutant lacking the phosphatase signature domain was constructed by removing a 1.1-kb fragment between the NarI and BstBI restriction sites in pcDNA4TO/Myc5ptaseIV. Plasmids expressing the AP180 C-terminal fragment and PHGFP were generous gifts from L. Greene [National Heart, Lung, and Blood Institute, National Institutes of Health (NIH), Bethesda] (43) and T. Balla (National Institute of Child Health and Human Development, NIH) (44), respectively. In the experiment described in Fig. 4, both Gag and PHGFP were expressed from a pNL4-3 derivative, pNL4-3/KFS/398/PHGFP, that expresses PHGFP from the nef ORF. This plasmid was constructed by using a transfer vector p398-6, a kind gift from K.-T. Jeang (National Institute of Allergy and Infectious Diseases, NIH) (45). A plasmid expressing HA-tagged Arf6/Q67L, pXS/Arf6Q67L-HA, was a generous gift from J. Donaldson (National Heart, Lung, and Blood Institute, NIH). Transfection and metabolic labeling of HeLa cells and immunoprecipitation of viral proteins were performed as described (39). Virus release efficiency was calculated as the amount of virion-associated Gag as a fraction of total (cell plus virion) Gag synthesized during a 2-h metabolic labeling period.
Fig. 4.
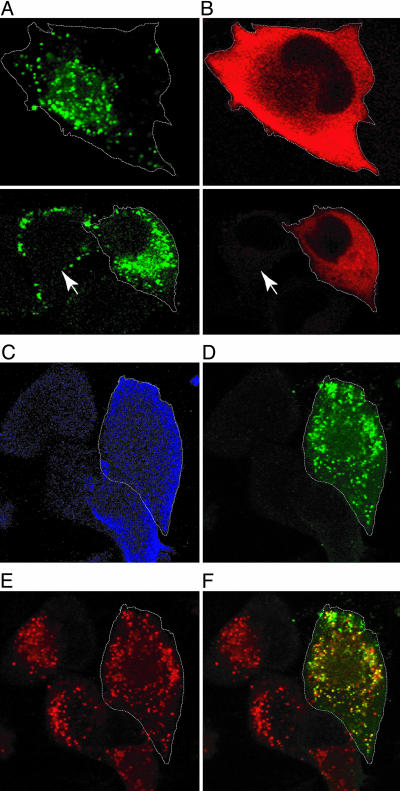
Gag localizes to PI(4,5)P2-enriched vesicles induced by Arf6/Q67L. HeLa cells were cotransfected with the HA-tagged Arf6/Q67L expression plasmid and either pNL4-3/KFS/398/PHGFP (A-H) or pNL4-3 (I-L). Arf6/Q67L (blue in A, E, and I) and Gag (red in C and G and green in K) were detected by anti-HA and anti-p17 (MA) Abs, respectively. PHGFP localization is shown in green in B and F. In I-L, cells were also immunostained with anti-CD63 Ab (red in J). Merged images of Gag and PHGFP or CD63 signals are shown in D, H, and L.
Abs, Fluorescent Reagents, and Immunostaining. Abs and fluorescent reagents were obtained from the sources listed in Supporting Text, which is published as supporting information on the PNAS web site. Immunostaining and confocal microscopy were performed as detailed (12, 18). Quantitative analyses of colocalization between Gag and CD63 were performed by using the colocalization measurement function (46) of mipav (Medical Image Processing, Analysis, and Visualization) software (Division of Computational Bioscience, Center for Information Technology, National Institutes of Health).
Electron Microscopy (EM). Fixation and processing of transfected HeLa cells were carried out as described (47). Cells were examined by a Hitachi H7000 electron microscope.
Results
Overexpression of 5ptaseIV Inhibits HIV-1 Particle Production. To determine whether PI(4,5)P2 is involved in HIV-1 particle assembly and release, we reduced cellular PI(4,5)P2 levels by overexpressing 5ptaseIV (48). To monitor plasma membrane PI(4,5)P2 levels, we used a GFP-tagged PH domain from phospholipase Cδ1 (PHGFP; ref. 44). As reported (44), when singly expressed, PHGFP was localized primarily on the plasma membrane (Fig. 6 A and D, which is published as supporting information on the PNAS web site). In contrast, when PHGFP was coexpressed with Myc-epitope-tagged 5ptaseIV, the GFP signal was detected throughout the cytoplasm and nucleus but not on the plasma membrane (Fig. 6 B and E). These observations demonstrate that 5ptaseIV overexpression indeed reduced the levels of plasma membrane PI(4,5)P2 as reported in 293 cells (42). As a control, we constructed a mutant form of 5ptaseIV (Δ1) lacking the 5-phosphatase signature domain that contains two motifs highly conserved among different classes of 5-phosphatases. As anticipated, the Δ1 mutant had no effect on the localization of PHGFP (Fig. 6 C and F).
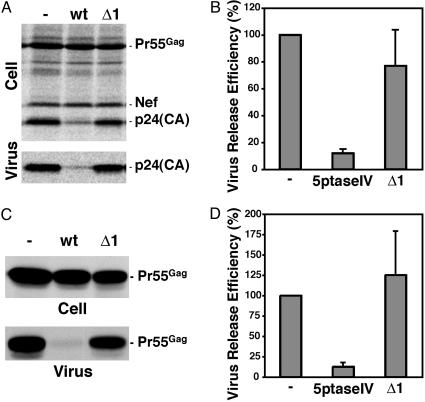
Having established that 5ptaseIV overexpression reduces the level of cell surface PI(4,5)P2 in HeLa cells, we examined the impact of this depletion on virus particle production (Fig. 1). HeLa cells were transfected singly with the full-length, infectious HIV-1 molecular clone pNL4-3 or were cotransfected with pNL4-3 and the 5ptaseIV expression vector. Cell- and virus-associated Gag proteins were analyzed by immunoprecipitation after metabolic labeling of the transfected cells (Fig. 1 A). In the culture overexpressing 5ptaseIV, virus release efficiency was reduced ≈8-fold compared with the control (Fig. 1B). In contrast, in cells expressing the Δ1 mutant, virus release efficiency was unaffected (Fig. 1 A and B). Marked inhibition of virus release by 5ptaseIV overexpression was also observed in Jurkat T cells (data not shown). Significantly, the observed impact of 5ptaseIV on WT HIV-1 release is not caused by global cytotoxicity, as particle release mediated by an NC-deleted Gag derivative in which the MA domain is replaced by the N terminus of Fyn kinase [Fyn(10)ΔMA/delNC] was markedly less affected by 5ptaseIV overexpression (Fig. 7, which is published as supporting information on the PNAS web site). These results strongly suggest that PI(4,5)P2 is involved in virus particle production.
Fig. 1.
HIV-1 particle production is inhibited by 5ptaseIV overexpression. (A) HeLa cells were transfected with pNL4-3 alone (-) or were cotransfected with pNL4-3 and expression plasmids encoding 5ptaseIV (wt) or the Δ1 5ptaseIV mutant (Δ1). One day after transfection, cells were metabolically labeled for 2 h with [35S]Met/Cys, and labeled viral proteins in cell and virion lysates were immunoprecipitated with anti-HIV Ig and analyzed by SDS/PAGE followed by fluorography. Viral proteins Pr55Gag, p24 (CA), and Nef are indicated. (B) Virus release efficiency from pNL4-3-expressing cells was calculated as described in Materials and Methods. Data from three independent experiments were quantified by PhosphorImager analysis and are shown as means ± SD. Note that the reduction in virus release efficiency measured upon 5ptaseIV overexpression may be an underestimate, because a small number of cells in the cotransfected culture express pNL4-3 but not 5ptaseIV (see Fig. 2). (C) HeLa cells transfected with pNL4-3/55FLAG alone (-) or cotransfected with pNL4-3/55FLAG and plasmids encoding 5ptaseIV (wt) or the Δ1 5ptaseIV mutant (Δ1) were analyzed as in A. (D) Virus release efficiency of cells expressing the FLAG-tagged Gag was calculated based on the data from seven independent experiments as described above and in Materials and Methods.
We noted that 5ptaseIV overexpression caused an increase in the ratio of cell-associated Pr55Gag to p24, indicating that PI(4,5)P2 depletion was accompanied by a defect in PR-mediated Gag processing (Fig. 1 A). To determine whether the effect of PI(4,5)P2 depletion on Gag processing was linked to the defect in virus particle production, we examined the impact of 5ptaseIV overexpression in the absence of a functional PR. For this purpose, we used two PR-defective pNL4-3 derivatives: one containing a mutation in the PR active site (pNL4-3/PR-; ref. 41), the other encoding a C-terminally FLAG-tagged Pr55Gag and lacking an intact PR gene (pNL4-3/55FLAG; ref. 12). In both cases, virus production was markedly reduced (8-fold) by 5ptaseIV overexpression but not by the Δ1 mutant (Fig. 1 C and D and data not shown). These results indicate that depletion of plasma membrane PI(4,5)P2 impairs HIV-1 particle production in a manner independent of PR-mediated Gag processing.
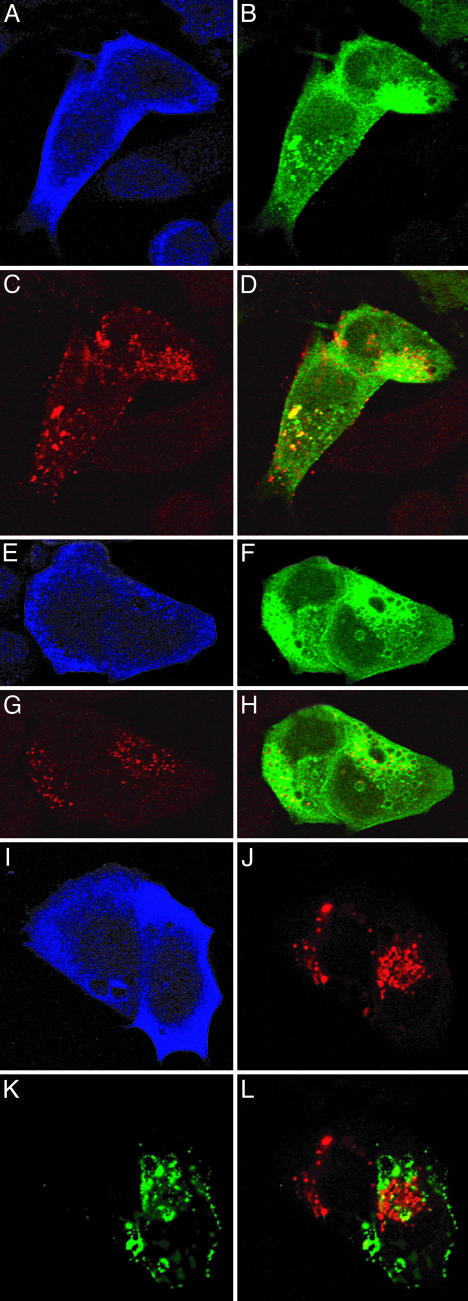
Overexpression of 5ptaseIV Inhibits Gag Localization to the Plasma Membrane and Retargets Gag to Late Endosomes. We next examined whether localization of Gag to the plasma membrane is affected by PI(4,5)P2 depletion. HeLa cells were cotransfected with pNL4-3 and the 5ptaseIV expression vector and were analyzed by confocal microscopy (Fig. 2). When singly expressed, the majority of Gag visualized with an anti-p17 (MA) antibody was present in a punctate pattern on the plasma membrane (Fig. 2A). In contrast, Gag-positive cells overexpressing 5ptaseIV (Fig. 2 A-D, outlined by dotted line) showed little or no Gag signal at the cell surface, but instead typically displayed an accumulation of Gag in intracellular vesicles (see also Fig. 8 A and B, which is published as supporting information on the PNAS web site). These Gag-containing vesicles were also positive for CD63, a marker for late endosomes/MVBs (49) (Fig. 2 C-F). Quantitative image analyses showed that 78% of Gag colocalized with CD63 in 5ptaseIV overexpressing cells, whereas only 6% of Gag did so in the absence of 5ptaseIV overexpression. These results, which were confirmed in >10 independent experiments, suggest that depletion of plasma membrane PI(4,5)P2 blocks the localization of Gag at the cell surface and causes mistargeting of virus assembly to a late endosomal compartment. This mistargeting of virus assembly was confirmed by electron microscopy (EM) (Fig. 9, which is published as supporting information on the PNAS web site).
Fig. 2.
Overexpression of 5ptaseIV inhibits Gag localization to the plasma membrane and retargets Gag to a late endosomal compartment. HeLa cells were cotransfected with pNL4-3 and the Myc-tagged 5ptaseIV expression plasmid. Gag (green in A and D) and 5ptaseIV (red in B and blue in C) were detected by mouse monoclonal anti-p17 (MA) and rabbit polyclonal anti-Myc Abs, respectively. The boundary of cells coexpressing Gag and 5ptaseIV is indicated by white dotted lines. Note that, in cells not overexpressing 5ptaseIV (arrow), Gag is localized on the cell surface, whereas, in the 5ptaseIV-overexpressing cells, Gag is predominantly detected in the perinuclear region. In C-F, cells were also immunostained with anti-CD63 Ab (red in E). (F) A merged image of Gag and CD63 signals, with colocalization indicated in yellow, is shown.
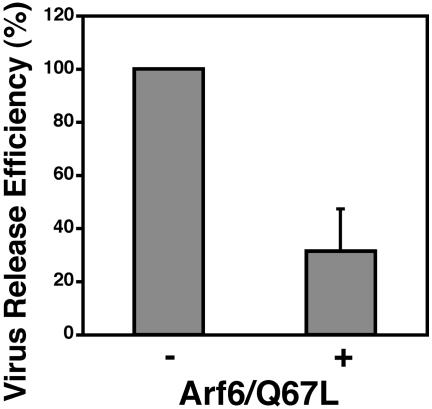
Expression of the Constitutively Active Arf6 Mutant Arf6/Q67L Decreases Virus Particle Production and Induces Accumulation of Gag in PI(4,5)P2-Enriched Intracellular Vesicles. Because 5ptaseIV dephosphorylates not only PI(4,5)P2 but also PI(3,4,5)P3 (42), we sought to validate the results presented above by employing an independent approach to perturb cellular PI(4,5)P2 levels. As mentioned in the Introduction, Arf6 regulates the activity of the PI(4,5)P2-generating kinase phosphatidylinositol-4-phosphate 5-kinase (ref. 34, reviewed in ref. 35). When we cotransfected HeLa cells with pNL4-3 and a plasmid expressing HA-tagged Arf6/Q67L, we observed a 3-fold reduction in virus release (Fig. 3), confirming that perturbation of cellular PI(4,5)P2 inhibits virus particle production. Significantly, particle release mediated by the Fyn(10)ΔMA/delNC mutant Gag was unaffected by Arf6/Q67L expression (Fig. 7), demonstrating that the impact of Arf6/Q67L on WT virus release is not caused by global cytotoxicity.
Fig. 3.
Expression of the constitutively active Arf6 mutant Arf6/Q67L inhibits virus particle production. HeLa cells were transfected with pNL4-3 alone (-) or were cotransfected with pNL4-3 and an expression plasmid encoding HA-tagged Arf6/Q67L (+) and were analyzed as in Fig. 1. Virus release efficiency was calculated based on data from four independent experiments as described in Materials and Methods. Data are shown as means ± SD.
In contrast to the effect of 5ptaseIV overexpression (Fig. 6), constitutively active Arf6/Q67L stimulates PI(4,5)P2 production (34, 37, 50) and induces the accumulation of PI(4,5)P2-enriched endosomal structures (36, 37). If PI(4,5)P2 influences Gag localization by directly or indirectly interacting with Gag, one might expect to see a relocalization of Gag to these Arf6/Q67L-induced structures. To address this possibility, we examined the distribution of Gag and PI(4,5)P2 in Arf6/Q67L-expressing cells (Fig. 4). Confirming previous reports (36, 37), in cells expressing Arf6/Q67L, PHGFP localized to intracellular vesicular structures (Fig. 4B). Strikingly, Gag partially relocalized to the intracellular region containing these PI(4,5)P2-enriched vesicles (Fig. 4 C and D). We also observed that, in a number of cells, the PHGFP-positive membranes formed large vacuole-like structures (Fig. 4F). In these cells, the p17 (MA) signal colocalized with these vacuoles (Fig. 4 G and H). These results suggest that PI(4,5)P2 accumulation in intracellular vesicles shifts Gag localization from the plasma membrane to these endosomal structures thereby inhibiting virus particle production from the plasma membrane. We found little colocalization between Gag accumulated in Arf6/Q67L-induced vesicles and CD63 (Fig. 4 I-L). Together, the results shown in Fig. 4 suggest that the altered Gag localization in Arf6/Q67L-expressing cells is caused by a different mechanism from that observed in cells overexpressing 5ptaseIV.
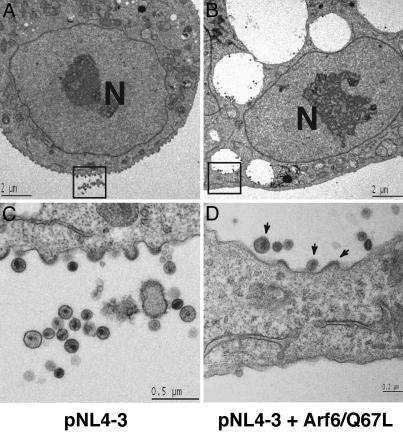
To examine further the localization of Gag in PI(4,5)P2-enriched endosomal structures, we performed electron microscopy (EM) analysis of HeLa cells cotransfected with pNL4-3 and the Arf6/Q67L expression vector (Fig. 5). In contrast to HeLa cells transfected with pNL4-3 alone (Fig. 5 A and C), cells cotransfected with pNL4-3 and the Arf6/Q67L vector contained large vesicles with an electron-lucent luminal space (Figs. 5 B and D and 9). We believe that these vesicles correspond to the PI(4,5)P2-enriched vesicles shown in Fig. 5. In cells singly expressing NL4-3, virus particles were seen budding predominantly from the plasma membrane (Fig. 5 A and C). In Arf6/Q67L-expressing cells, virus particles in the process of budding were observed on the membrane of the enlarged intracellular vesicles and virions with condensed cores were found in the lumen of these vesicles (Figs. 5 B and D and 9). Taken together, the confocal and EM data indicate that Arf6/Q67L expression leads to the formation of PI(4,5)P2-enriched vesicles to which virus particle assembly is directed.
Fig. 5.
HIV-1 particles assemble in Arf6/Q67L-induced vesicles. HeLa cells transfected with pNL4-3 alone (A and C) or cotransfected with pNL4-3 and the HA-tagged Arf6/Q67L expression plasmid (B and D) were observed by transmission electron microscopy (EM). Higher magnification of boxed areas in A and B is shown in C and D, respectively. Note that virus particles as well as budding structures are detected on the membrane of Arf6/Q67L-induced vesicles (arrows in D). In the cultures cotransfected with pNL4-3 and the Arf6/Q67L expression plasmid, 42% of the virus particles were detected in association with these vesicles, whereas only 6% of virions in cultures transfected with pNL4-3 alone was associated with intracellular vesicles. Scale bars are shown in each panel. Nuclei are indicated (N).
Discussion
In this study, we examined the role of the phosphoinositide PI(4,5)P2 in HIV-1 Gag targeting and virus assembly. To this end, we perturbed cellular PI(4,5)P2 levels by exogenous overexpression of the phosphatase 5ptaseIV and the constitutively active Arf6 mutant Arf6/Q67L. Overexpression of 5ptaseIV severely reduced levels of plasma membrane PI(4,5)P2 (Fig. 1 and ref. 42) and strongly inhibited virus production (Fig. 1). The latter defect is likely caused by the lack of Gag localization to the plasma membrane and the accompanying targeting of Gag to CD63-positive late endosomes (Fig. 2). Arf6/Q67L expression also markedly decreased virus production (Fig. 3), but most likely by a mechanism distinct from that induced by 5ptaseIV. Arf6/Q67L expression induced PI(4,5)P2-enriched endosomal vesicles (Fig. 5 and refs. 36 and 37) to which Gag was localized, thereby reducing virus particle production and release from the cell surface (Figs. 4, 5, and 9). As two independent methods of PI(4,5)P2 perturbation altered Gag localization, the results presented here strongly suggest that PI(4,5)P2 plays an important role in Gag targeting and virus production. To our knowledge, this is the first report demonstrating an involvement of PI(4,5)P2 in virus assembly and release.
There are several nonmutually exclusive models that could account for the role of PI(4,5)P2 in Gag targeting. PI(4,5)P2 could promote or stabilize the interaction of Gag with the plasma membrane by interacting with Gag directly, or indirectly through an unknown adaptor molecule, or by creating a lipid microenvironment favorable for Gag binding and virus assembly. This model is consistent with the data obtained with the Arf6/Q67L mutant; upon induction of PI(4,5)P2-enriched vesicles by Arf6/Q67L, Gag accumulated in these newly formed structures (Fig. 4). In addition, virus production by a Gag deletion mutant [Fyn(10)ΔMA/delNC] containing the N terminus of Fyn but lacking MA and NC domains (and hence the Gag highly basic sequences) was significantly less sensitive to PI(4,5)P2 perturbation (Fig. 7). Therefore, an interaction between PI(4,5)P2 and Gag may at least partially determine the subcellular location of Gag targeting and virus assembly. Alternatively, PI(4,5)P2 may stabilize the interaction between Gag and the plasma membrane by affecting Gag conformation or enhancing its multimerization. This latter possibility is consistent with the observation that Gag interaction with inositol derivatives affects the stability and size of virus-like particles assembled in vitro from purified Gag (38). Although assays widely used to measure protein-phosphoinositide binding (e.g., protein-lipid overlay and liposome binding) require careful interpretation (see Supporting Text), such analyses aimed at probing possible Gag-PI(4,5)P2 interactions should be performed. When plasma membrane PI(4,5)P2 is not available, Gag may bind other membranes, e.g., those of the late endosomes/MVBs. In this regard, it is noteworthy that steady-state binding of Gag to total membrane is unaffected by 5ptaseIV overexpression (A.O. and E.O.F., unpublished data). The fact that the Gag proteins of many retroviruses in addition to HIV-1 use a basic cluster of amino acids in MA to direct the binding of Gag to membrane (reviewed in ref. 51) suggests that a role for PI(4,5)P2 or other phosphoinositides in Gag targeting may be a general phenomenon among retroviruses. Indeed, preliminary results suggest that the release of equine infectious anemia virus and Mason-Pfizer monkey virus is inhibited by 5ptaseIV overexpression (A.O. and E.O.F., unpublished data).
An alternative model to explain the effects of perturbing cellular PI(4,5)P2 on Gag targeting and virus release postulates that cellular functions regulated by PI(4,5)P2 could play an important role in promoting Gag trafficking to the plasma membrane. PI(4,5)P2 has been implicated in several cellular processes through direct interactions with key regulatory proteins. PI(4,5)P2 involvement in clathrin-mediated endocytosis and actin cytoskeleton rearrangement is well documented (27-33). However, inhibition of clathrin-mediated endocytosis or disruption of the actin cytoskeleton by PI(4,5)P2-independent methods did not reduce virus production (Fig. 8). Therefore, the effects we observe on Gag targeting and virus release are unlikely to be caused by disruption of clathrin-mediated endocytosis or rearrangement of the actin cytoskeleton. We do not exclude the possibility, however, that other cellular functions regulated by PI(4,5)P2 may impact Gag targeting and virus production. Several reports have suggested that retroviral Gag can be transported to the plasma membrane through the retrograde trafficking of endosomes (19, 20, 52, 53). Although we did not detect PI(4,5)P2 on late endosomal membranes in HeLa cells, if PI(4,5)P2 is critically involved in membrane trafficking from the late endosome to the cell surface, the accumulation of Gag in late endosomes upon 5ptaseIV overexpression could be explained by this trafficking block. Similarly, the impact of Arf6/Q67L on Gag localization could be influenced by a block in the endosomal pathway as Arf6 has been shown to regulate endosomal trafficking (37, 54-57). Monitoring Gag trafficking in living cells in real time will help elucidate whether PI(4,5)P2 influences Gag localization to the plasma membrane by direct or indirect interactions, by regulating endosomal transport pathways, or by a combination of both mechanisms.
In cell types such as HeLa and T cells, the majority of Gag is targeted to the plasma membrane, whereas in primary human macrophages, Gag assembles predominantly in late endosomes (15-18). Cellular determinants of Gag localization to late endosomes in macrophages have not been defined. p6 is dispensable for this localization (18), suggesting that an interaction between Gag and Tsg101 is not essential. Our preliminary data show that, in human monocyte-derived macrophages, PI(4,5)P2 colocalizes with HIV-1 Gag on late endosomes (A.O. and E.O.F., unpublished data). Therefore, PI(4,5)P2 may play a role in Gag targeting to late endosomes in macrophages. Given that PI(4,5)P2 is also abundant on the macrophage plasma membrane (A.O. and E.O.F., unpublished data), other factors are likely to be involved in regulating Gag localization in this cell type.
In summary, we used two independent approaches to perturb cellular PI(4,5)P2 levels to show that this phosphoinositide is critically involved in determining the subcellular location of HIV-1 assembly. The data presented in this report not only suggest a previously undescribed role for this important lipid, but also provide significant insights into the molecular mechanism of Gag trafficking. Further studies will need to explore the extent to which Gag targeting of divergent retroviruses is regulated by PI(4,5)P2 or other phosphoinositides. Elucidating the viral and cellular determinants of Gag targeting is essential, because the localization of assembly in virus-producing cells likely affects retrovirus replication and pathogenesis by enhancing viral transmission through cell-cell contact and by creating intracellular viral reservoirs that are inaccessible to the host immune system.
Supplementary Material
Acknowledgments
We thank J. Donaldson, A. Rein, V. KewalRamani, B. Luttge, U. Munshi, C. Adamson, and A. Waheed for critical review of the manuscript and helpful discussions; O. Schwartz, J. Kabat, and E. Cho for assistance with confocal microscopy; M. McAuliffe for the use of the mipav software; and T. Balla, J. Donaldson, L. Greene, K.-T. Jeang, P. Majerus, and D. Ott (SAIC-Frederick, National Cancer Institute) for providing plasmids. HIV Ig was obtained through the National Institutes of Health AIDS Research Reference and Reagent Program.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HIV-1, HIV type 1; PR, protease; MA, matrix; NC, nucleocapsid; MVB, multivesicular body; PH, pleckstrin homology; PI(4,5)P2, phosphatidylinositol (4,5) biphosphate; 5ptaseIV, polyphosphoinositide 5-phosphatase IV; Arf6, ADP-ribosylation factor 6.
References
- 1.Swanstrom, R. & Wills, J. W. (1997) in Retroviruses, eds. Coffin, J. M., Hughes, S. H. & Varmus, H. E. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 263-334.
- 2.Freed, E. O. (1998) Virology 251, 1-15. [DOI] [PubMed] [Google Scholar]
- 3.Freed, E. O. (2002) J. Virol. 76, 4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pornillos, O., Garrus, J. E. & Sundquist, W. I. (2002) Trends Cell Biol. 12, 569-579. [DOI] [PubMed] [Google Scholar]
- 5.Facke, M., Janetzko, A., Shoeman, R. L. & Krausslich, H. G. (1993) J. Virol. 67, 4972-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gallina, A., Mantoan, G., Rindi, G. & Milanesi, G. (1994) Biochem. Biophys. Res. Commun. 204, 1031-1038. [DOI] [PubMed] [Google Scholar]
- 7.Lee, P. P. & Linial, M. L. (1994) J. Virol. 68, 6644-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reil, H., Bukovsky, A. A., Gelderblom, H. R. & Gottlinger, H. G. (1998) EMBO J. 17, 2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang, C. T., Lai, H. Y. & Li, J. J. (1998) J. Virol. 72, 7950-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan, X., Yu, X., Lee, T. H. & Essex, M. (1993) J. Virol. 67, 6387-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed, E. O., Orenstein, J. M., Buckler-White, A. J. & Martin, M. A. (1994) J. Virol. 68, 5311-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ono, A., Orenstein, J. M. & Freed, E. O. (2000) J. Virol. 74, 2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermida-Matsumoto, L. & Resh, M. D. (2000) J. Virol. 74, 8670-8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannon, P. M., Matthews, S., Clark, N., Byles, E. D., Iourin, O., Hockley, D. J., Kingsman, S. M. & Kingsman, A. J. (1997) J. Virol. 71, 3474-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raposo, G., Moore, M., Innes, D., Leijendekker, R., Leigh-Brown, A., Benaroch, P. & Geuze, H. (2002) Traffic 3, 718-729. [DOI] [PubMed] [Google Scholar]
- 16.Pelchen-Matthews, A., Kramer, B. & Marsh, M. (2003) J. Cell Biol. 162, 443-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen, D. G., Booth, A., Gould, S. J. & Hildreth, J. E. (2003) J. Biol. Chem. 278, 52347-52354. [DOI] [PubMed] [Google Scholar]
- 18.Ono, A. & Freed, E. O. (2004) J. Virol. 78, 1552-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherer, N. M., Lehmann, M. J., Jimenez-Soto, L. F., Ingmundson, A., Horner, S. M., Cicchetti, G., Allen, P. G., Pypaert, M., Cunningham, J. M. & Mothes, W. (2003) Traffic 4, 785-801. [DOI] [PubMed] [Google Scholar]
- 20.Nydegger, S., Foti, M., Derdowski, A., Spearman, P. & Thali, M. (2003) Traffic 4, 902-910. [DOI] [PubMed] [Google Scholar]
- 21.Zhou, W., Parent, L. J., Wills, J. W. & Resh, M. D. (1994) J. Virol. 68, 2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiernan, R. E., Ono, A., Englund, G. & Freed, E. O. (1998) J. Virol. 72, 4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kiernan, R. E., Ono, A. & Freed, E. O. (1999) J. Virol. 73, 4728-4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurley, J. H. & Meyer, T. (2001) Curr. Opin. Cell Biol. 13, 146-152. [DOI] [PubMed] [Google Scholar]
- 25.Lemmon, M. A. (2003) Traffic 4, 201-213. [DOI] [PubMed] [Google Scholar]
- 26.DiNitto, J. P., Cronin, T. C. & Lambright, D. G. (2003) Sci. STKE 2003, re16. [DOI] [PubMed] [Google Scholar]
- 27.Takenawa, T. & Itoh, T. (2001) Biochim. Biophys. Acta 1533, 190-206. [DOI] [PubMed] [Google Scholar]
- 28.Toker, A. (2002) Cell Mol. Life Sci. 59, 761-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simonsen, A., Wurmser, A. E., Emr, S. D. & Stenmark, H. (2001) Curr. Opin. Cell Biol. 13, 485-492. [DOI] [PubMed] [Google Scholar]
- 30.Wenk, M. R. & De Camilli, P. (2004) Proc. Natl. Acad. Sci. USA 101, 8262-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin, T. F. (2001) Curr. Opin. Cell Biol. 13, 493-499. [DOI] [PubMed] [Google Scholar]
- 32.Yin, H. L. & Janmey, P. A. (2003) Annu. Rev. Physiol. 65, 761-789. [DOI] [PubMed] [Google Scholar]
- 33.Czech, M. P. (2003) Annu. Rev. Physiol. 65, 791-815. [DOI] [PubMed] [Google Scholar]
- 34.Honda, A., Nogami, M., Yokozeki, T., Yamazaki, M., Nakamura, H., Watanabe, H., Kawamoto, K., Nakayama, K., Morris, A. J., Frohman, M. A. & Kanaho, Y. (1999) Cell 99, 521-532. [DOI] [PubMed] [Google Scholar]
- 35.Donaldson, J. G. (2003) J. Biol. Chem. 278, 41573-41576. [DOI] [PubMed] [Google Scholar]
- 36.Aikawa, Y. & Martin, T. F. (2003) J. Cell Biol. 162, 647-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brown, F. D., Rozelle, A. L., Yin, H. L., Balla, T. & Donaldson, J. G. (2001) J. Cell Biol. 154, 1007-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campbell, S., Fisher, R. J., Towler, E. M., Fox, S., Issaq, H. J., Wolfe, T., Phillips, L. R. & Rein, A. (2001) Proc. Natl. Acad. Sci. USA 98, 10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freed, E. O. & Martin, M. A. (1994) J. Virol. 68, 2503-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adachi, A., Gendelman, H. E., Koenig, S., Folks, T., Willey, R., Rabson, A. & Martin, M. A. (1986) J. Virol. 59, 284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang, M., Orenstein, J. M., Martin, M. A. & Freed, E. O. (1995) J. Virol. 69, 6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kisseleva, M. V., Cao, L. & Majerus, P. W. (2002) J. Biol. Chem. 277, 6266-6272. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, X., Greener, T., Al-Hasani, H., Cushman, S. W., Eisenberg, E. & Greene, L. E. (2001) J. Cell Sci. 114, 353-365. [DOI] [PubMed] [Google Scholar]
- 44.Varnai, P. & Balla, T. (1998) J. Cell Biol. 143, 501-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang, L. M., Joshi, A., Willey, R., Orenstein, J. & Jeang, K. T. (1994) EMBO J. 13, 2886-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Costes, S. V., Daelemans, D., Cho, E. H., Dobbin, Z., Pavlakis, G. & Lockett, S. (2004) Biophys. J. 86, 3993-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonda, M. A., Aaronson, S. A., Ellmore, N., Zeve, V. H. & Nagashima, K. (1976) J. Natl. Cancer Inst. 56, 245-263. [DOI] [PubMed] [Google Scholar]
- 48.Kisseleva, M. V., Wilson, M. P. & Majerus, P. W. (2000) J. Biol. Chem. 275, 20110-20116. [DOI] [PubMed] [Google Scholar]
- 49.Escola, J. M., Kleijmeer, M. J., Stoorvogel, W., Griffith, J. M., Yoshie, O. & Geuze, H. J. (1998) J. Biol. Chem. 273, 20121-20127. [DOI] [PubMed] [Google Scholar]
- 50.Krauss, M., Kinuta, M., Wenk, M. R., De Camilli, P., Takei, K. & Haucke, V. (2003) J. Cell Biol. 162, 113-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conte, M. R. & Matthews, S. (1998) Virology 246, 191-198. [DOI] [PubMed] [Google Scholar]
- 52.Basyuk, E., Galli, T., Mougel, M., Blanchard, J. M., Sitbon, M. & Bertrand, E. (2003) Dev. Cell 5, 161-174. [DOI] [PubMed] [Google Scholar]
- 53.Lindwasser, O. W. & Resh, M. D. (2004) J. Virol. 78, 6013-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D'Souza-Schorey, C., Li, G., Colombo, M. I. & Stahl, P. D. (1995) Science 267, 1175-1178. [DOI] [PubMed] [Google Scholar]
- 55.Peters, P. J., Hsu, V. W., Ooi, C. E., Finazzi, D., Teal, S. B., Oorschot, V., Donaldson, J. G. & Klausner, R. D. (1995) J. Cell Biol. 128, 1003-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radhakrishna, H. & Donaldson, J. G. (1997) J. Cell Biol. 139, 49-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Naslavsky, N., Weigert, R. & Donaldson, J. G. (2003) Mol. Biol. Cell 14, 417-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.