Abstract
Formins are large multidomain proteins required for assembly of actin cables that contribute to the polarity and division of animal and fungal cells. Formin homology-1 (FH1) domains bind profilin, and highly conserved FH2 domains nucleate actin filaments. We characterized the effects of two formins, budding yeast Bni1p and fission yeast Cdc12p, on actin assembly. We used evanescent wave fluorescence microscopy to observe assembly of actin filaments (i) nucleated by soluble formin FH1FH2 domains and (ii) associated with formin FH1FH2 domains immobilized on microscope slides. Bni1p(FH1FH2)p and Cdc12p(FH1FH2)p nucleated new actin filaments or captured the barbed ends of preformed actin filaments that grew by insertion of subunits between the immobilized formin and the barbed end of the filament. Both formins remained bound to growing actin filament barbed ends for >1,000 sec. Elongation of a filament between an immobilized formin and a second anchor point buckled filament segments as short as 0.7 μm, demonstrating that polymerization of single actin filaments produces forces of >1 piconewton, close to the theoretical maximum. After buckling, further growth produced long loops that did not supercoil, suggesting that formins do not stair step along the two subunits exposed on the growing barbed end. In agreement, Arp2/3 complex branched filaments did not rotate as they grew from formins attached to the slide surface. Formins are not mechanistically identical because barbed end elongation from Cdc12(FH1FH2)p, but not Bni1(FH1FH2)p, requires profilin. However, profilin increased the rate of Bni1(FH1FH2)p-mediated barbed end elongation from 75% to 100% of full-speed.
Many cellular structures composed of actin filaments depend on formins, so the discovery that the FH2 domain of budding yeast Bni1p stimulates spontaneous polymerization of pure actin was a major advance (1, 2). Biochemical experiments in those pioneering papers and subsequent work (3–5) suggest that Bni1(FH1FH2)p nucleates filaments and then allows both the fast growing barbed end and slowly growing pointed end of the filaments to elongate.
Several lines of evidence suggest that formins bind actin filament barbed ends and remain in place as filaments grow. Although Bni1(FH1FH2)p does not raise the critical concentration like a barbed end capping protein, it was observed to bind near barbed ends by immunoelectron microscopy (1) and inhibits association of capping protein with barbed ends (4, 5). The FH1FH2 domains of fission yeast formin Cdc12p tightly cap actin filament barbed ends and nucleate short filaments that grow slowly from their pointed ends (6). The actin–monomer binding protein profilin allows filaments nucleated by Cdc12(FH1FH2)p to grow at their barbed ends (6). Because Cdc12p inhibits end to end annealing of actin filaments even in the presence of profilin (6), profilin might not simply dissociate Cdc12p from barbed ends. Indeed, Higashida et al. (7) recently showed that aggregates of the mammalian formin mDia1 persistently associate with elongating barbed ends. Here, we used real-time evanescent wave fluorescence microscopy to observe the assembly of individual filaments attached to yeast formins. We measured the force produced by actin polymerization, addressed some of the unresolved kinetic and thermodynamic problems raised by the presence of a formin on a growing barbed end, and compared the mechanisms of budding yeast Bni1p and fission yeast Cdc12p.
Materials and Methods
Plasmid Construction and Protein Purification. Bacterial expression plasmids for Cdc12(FH1FH2)p and Bni1(FH1FH2)p were constructed by using standard methods (8). Proteins were purified as described (6) with minor modifications (Fig. 4, which is published as supporting information on the PNAS web site). For details, refer to Supporting Text, which is published as supporting information on the PNAS web site.
Total Internal Reflection Fluorescence (TIRF) Microscopy. We collected images of fluorescent actin filaments excited by total internal reflection at 15-sec intervals with a Hamamatsu C4742–95 CCD camera (Orca-ER) on an Olympus IX-70 microscope and processed them with imagej software (http://rsb.info.nih.gov/ij) as described in detail (6, 9).
Glass flow cells of 5 × 25 × 0.3 mm (width/length/height) (9) were incubated with either 100 nM Bni1(FH1FH2)p, 100 nM GST-Bni1(FH1FH2)p, 250 nM GST-Cdc12(FH1FH2)p, or 10 nM N-ethylmaleimide (NEM)-myosin and Bni1(FH1FH2)p or GST-Bni1(FH1FH2)p for 2–5 min, washed extensively with 1% BSA, and equilibrated with TIRF buffer {10 mM imidazole, pH 7.0/50 mM KCl/1 mM MgCl2/1 mM EGTA/50 mM DTT/0.2 mM ATP/50 μM CaCl2/15 mM glucose/20 μg/ml catalase/100 μg/ml glucose oxidase, 0.5% [4,000 cP (centipoise; 1 P = 0.1 Pa·sec)] methylcellulose}. Mixtures of unlabeled Mg-ATP actin and either Mg-ATP Oregon green (OG) actin or Mg-ATP Alexa green (AG) actin monomers were mixed with 2× TIRF buffer supplemented with water and formin, profilin, Arp2/3 complex, GST-WA, capping protein, or phalloidin to give the final concentrations indicated in the figure legends. These samples were transferred to a flow cell, and imaging began as soon as filaments appeared (2–5 min, depending on flow and polymerization rates). In some experiments, a second sample with twice the concentration of labeled actin replaced the initial sample during continuous imaging. Maximum intensity projections revealed movement-constricted NEM-myosin II and formin attachment points of individual filaments (6).
Calculation of Force. The force of actin assembly required to buckle actin filaments was calculated with a derivation of the beam equation (10)
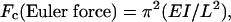 |
where EI is the flexural rigidity of an actin filament, and L is the distance between attachment points at the buckling moment. The flexural rigidity (EI = Lp/kBT) was calculated from measured persistence lengths (Lp): 9 μm(EI = 3.6 × 10–26 N·m2) for pure actin; and 18 μm (EI = 7.2 × 10–26 N·m2) for phalloidin-saturated actin (11, 12).
The theoretical stall force fs was calculated as follows (13):
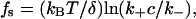 |
where kB is the Boltzmann constant, T is temperature (kBT = 4.116 × 10–21 J), δ is the added length per monomer (2.7 nm), k+ is the rate constant for subunit addition (11.6 μM–1·s–1), c is the actin concentration, and k– is the rate constant for subunit dissociation (1.4 s–1).
Results
Comparison of Budding Yeast Bni1p and Fission Yeast Cdc12p. We used time-lapse TIRF microscopy (6, 9) to observe the effects of fission yeast Cdc12(FH1FH2)p and budding yeast Bni1(FH1FH2)p on growing actin filaments attached to the surface of glass slides by NEM-myosin II (Fig. 1 and diagram 1 in Fig. 2a). Direct observation of individual actin filaments eliminated the need to make assumptions about the effects of formins on filament ends, as required with bulk assembly assays. We obtained similar results by using 1 μM unlabeled actin with a trace of fluorescent actin labeled on either cys-374 (OG-actin) or on lysines (AG-actin). Fluorescent subunits allow visualization of the filaments, but unlabeled actin accounts for most of the growth (9). NEM-myosin bound to the microscope slide anchored filaments at one or a few points along their length, leaving both ends free to elongate.
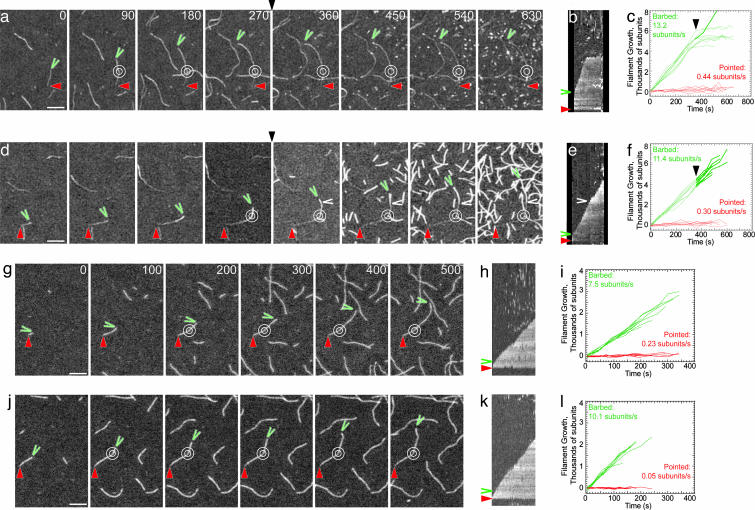
Fig. 1.
Time lapse evanescent wave fluorescence microscopy of the effect of formins on actin polymerization. (a, d, g, and j) Time lapse micrographs with the time in seconds indicated at the top. White double rings indicate points where filaments were attached to NEM-myosin on the slide surface; green wedges indicate barbed ends; red triangles indicate pointed ends; black triangles indicate time when the second solution replaced the initial solution; white wedges indicate filament lengths at the time the solution was changed. Movies of all time lapses and additional controls are published as supporting information on the PNAS web site. (b, e, h, and k) Kymographs of the length of the filament marked to the left (y axis) vs. time (x axis, 720 sec in b and e and 600 sec in h and k). (c, f, i, and l) Plots of the growth of individual filament barbed and pointed ends vs. time. (a–f) Effect of formins on growing actin filaments. (a–c) Cdc12(FH1FH2)p. The initial solution was 1.0 μM actin with 0.25 μM OG-actin. The second solution was 1.0 μM actin with 0.65 μM OG-actin with 10 nM Cdc12(FH1FH2)p. (d–f) Bni1(FH1FH2)p. The initial solution was 1.0 μM actin with 0.25 μM OG-actin. The second solution was 1.0 μM actin with 0.65 μM OG-actin and 25 nM Bni1(FH1FH2)p. (g–l) Effect of formins on the spontaneous assembly of 1.0 μM unlabeled Mg-ATP actin with 0.2 μM actin labeled with AG (AG-actin). (g–i) Actin with 20 nM Bni1(FH1FH2)p. (j–l) Actin with 20 nM Bni1(FH1FH2)p and 1.5 μM profilin. (Scale bar, 5 μm.)
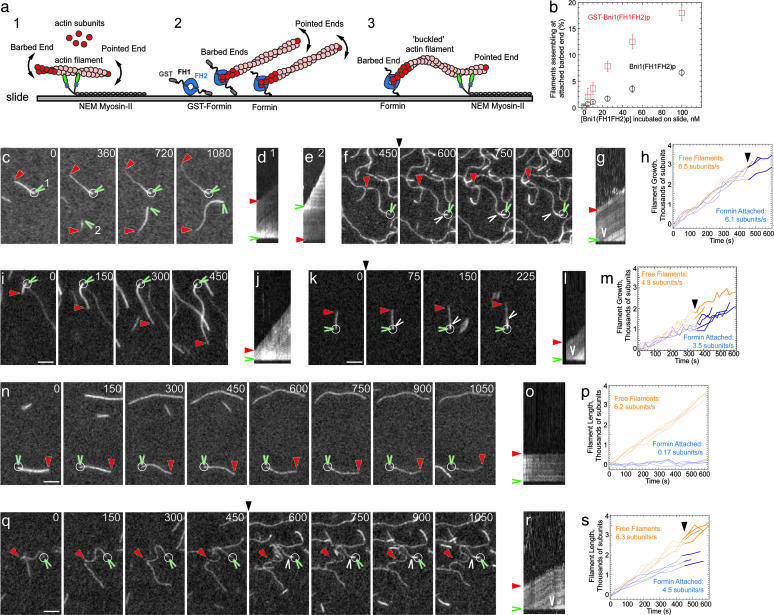
Fig. 2.
Time lapse evanescent wave fluorescence microscopy of actin filaments growing from formins attached to the slide surface. Conditions and symbols are as in Fig. 1. White circles indicate points where filaments were attached at their barbed end to formin or GST-formin on the slide surface. (a) Diagram of filament attachment and assembly on slides coated with NEM-myosin II (1), formin or GST-formin (2), and NEM-myosin II and formin (3). (b) Dependence of the fraction of filaments that were attached at their barbed ends on the concentration of GST-Bni1(FH1FH2)p or Bni1(FH1FH2)p incubated with slide. (c, f, i, k, n, and q) Time lapse micrographs with seconds indicated at the top. (d, e, g, j, l, o, and r) Kymographs of the length of filaments marked to the left (y axis) vs. time (x axis, 900 sec in d and e and 450 sec in g, j, l, o, and r). (h, m, p, and s) Plots of the growth of free filaments (orange) and filaments attached to formin immobilized on the slide (blue). (c–e) The slide was preincubated with 100 nM GST-Bni1(FH1FH2)p followed by 0.5 μM Mg-ATP actin and 0.2 μM OG-actin. Filament 1 grew away from its attached barbed end (indicated by the brighter, less bleached actin). The barbed end of filament 2 was free-elongating away from the pointed end. (f–h) The slide was preincubated with 100 nM GST-Bni1(FH1FH2)p followed by 0.75 μM Mg-ATP actin and 0.1 μM AG-actin, and subsequently, 0.75 μM Mg-ATP actin and 0.2 μM AG-actin to visualize new monomer addition. Filaments grew at their attached barbed ends. (i and j) The slide was preincubated with 100 nM Bni1(FH1FH2)p followed by 1.0 μM Mg-ATP actin and 0.5 μM OG-actin. (k–m) The slide was preincubated with 100 nM Bni1(FH1FH2)p followed by 0.5 μM Mg-ATP actin and 0.15 μM OG-actin, and subsequently, 0.5 μM Mg-ATP actin and 0.30 μM OG-actin to visualize new monomer addition. Filaments grew at their attached barbed ends. (n–p) The slide was preincubated with 250 nM GST-Cdc12(FH1FH2)p followed by 0.75 μM Mg-ATP actin and 0.1 μM AG-actin. Free filaments grew, but filaments attached to formin by their barbed end did not. (q–s) The slide was preincubated with 250 nM GST-Cdc12(FH1FH2)p followed by 0.75 μM Mg-ATP actin and 0.1 μM AG-actin with 1 μM profilin, and subsequently, 0.75 μM Mg-ATP actin and 0.2 μM AG-actin with 1 μM profilin. Filaments grew at their attached barbed ends. (Scale bar, 5 μm.)
First we compared the effects of formin on the elongation of preexisting actin filaments (Fig. 1 a–f and Fig. 5 a–c, which is published as supporting information on the PNAS web site, and Table 1). Movies 1–24, corresponding to all time lapse micrographs, are published as supporting information on the PNAS web site. Bni1(FH1FH2)p (residues 1227–1766) did not block barbed end growth, and nucleated many filaments that also grew at their barbed ends (Fig. 1 d–f). Bni1(FH1FH2)p had no effect on pointed end growth. In contrast, Cdc12(FH1FH2)p (residues 882–1390) completely blocked growth at the barbed ends of most (95%) filaments without affecting growth at pointed ends (Fig. 1 a–c), as we described previously for a Cdc(FH1FH2)p construct consisting of residues 882–1375 (6). Cdc12(FH1FH2)p also nucleated numerous filaments that grew only at their pointed ends.
Table 1. Comparison of actin assembly rates in the presence of formin.
| Conditions* | Barbed end rate, s1/frequency, % | Pointed end rate, s-1/frequency, % |
|---|---|---|
| Filaments + actin | 10.2 ± 0.2/100 | 0.38 ± 0.07/100 |
| Filaments + actin + 10 nM | 0.0 ± 0.0/95 | 0.44 ± 0.15/100 |
| Cdc12(FH1FH2)p | 13.2 ± 0.3/5 | 0.38 ± 0.03/100†‡ |
| Filaments + actin + 25 nM | 11.4 ± 0.1/100 | 0.30 ± 0.04/100 |
| Bni1(FH1FH2)p | 6.9 ± 0.2/100†§ | 0.39 ± 0.04/100† |
| Actin | 11.2 ± 0.8/100 | 0.35 ± 0.04/100 |
| Actin + profilin | 12.1 ± 0.5/100 | 0.03 ± 0.05/100¶ |
| Actin + 20 nM | 0.0 ± 0.0/> 99 | 0.39 ± 0.04/100‡ |
| Cdc12(FH1FH2)p | 10.6 ± 0.2/< 1 | |
| Actin + 20 nM | 7.4 ± 0.3/100∥ | 0.05 ± 0.05/100¶ |
| Cdc12(FH1FH2)p + profilin | ||
| Actin + 20 nM Bni1(FH1FH2)p | 7.5 ± 0.2/100§ | 0.23 ± 0.03/100 |
| Actin + 20 nM Bni1(FH1FH2)p + profilin | 10.1 ± 0.3/100** | 0.05 ± 0.06/100¶ |
Experiments where formin was not attached to the slide surface, as reported in Figs. 1 and 5. Twenty individual filaments were measured from two independent experiments.
Freshly nucleated filaments.
The rate that Cdc12(FH1FH2)p-nucleated filaments assembled was not statistically different from the rate of control pointed ends, by the two-tailed t test (P = 0.433-0.612).
Bni1(FH1FH2)p-nucleated filaments grew significantly slower at their barbed ends than control barbed ends (P < 0.002).
In the presence of profilin, Cdc12(FH1FH2)p-nucleated barbed ends grew significantly slower than control barbed ends (P = 0.0006).
Profilin forced pointed ends to grow significantly slower than control pointed ends (P < 0.0001).
In the presence of profilin, Bni1(FH1FH2)p-nucleated barbed ends grew significantly faster than in the absence of profilin (P = 0.0002), but not statistically different from control barbed ends (P = 0.752).
Second, we compared the effects of formins on spontaneous assembly of new filaments (Figs. 1 g–l and 5 d–o and Table 1). Bni1(FH1FH2)p nucleated filaments that grew at their barbed ends at 75% the normal rate, without affecting pointed end growth (Fig. 1 g–i). With Bni1(FH1FH2)p and profilin, barbed ends grew at the full rate, but pointed ends did not grow (Fig. 1 j–l). Cdc12(FH1FH2)p nucleated filaments that grew at their pointed ends only (Fig. 5 j–l). Rare barbed ends (<1%) in these samples grew normally, indicating that the monomer pool was not depleted. Profilin inhibited pointed end growth, but allowed filaments nucleated by Cdc12(FH1FH2)p to grow at their barbed ends at ≈75% the normal rate (Fig. 5 m–o).
Persistent Attachment of Yeast Formins to Growing Barbed Ends. Next, we used TIRF microscopy to observe actin filaments growing on the surface of glass slides coated with Cdc12(FH1FH2)p or Bni1(FH1FH2)p. Higashida et al. (7) used a similar strategy to observe polymerization of rhodamine phalloidin-labeled filaments relative to immobilized clusters of antibodies and the mammalian formin mDia1.
Incubation of actin monomers on glass slides coated with formin FH1FH2 domains under polymerizing conditions (diagram 2 in Fig. 2a) produced two types of actin filaments: filaments with free ends, and filaments growing with one end fixed to a point on the slide (Fig. 2 c–e). Actin alone was required for growth from immobilized Bni1(FH1FH2)p (Fig. 2 c–m), whereas growth from immobilized Cdc12(FH1FH2)p required actin and profilin (Fig. 2 n–s). Filaments with one fixed end grew at rates near those of filaments with free barbed ends (Fig. 2 c and e; filament 2) in the same samples. Increasing the fraction of fluorescent actin by replacing the contents of the chamber during an experiment showed that new subunits inserted at the attached barbed end (Fig. 2 f, g, k, l, q, and r). Some filaments rotated about their attachment points through angles up to 90° in static preparations and up to 180° when buffer flowed through the chamber. Growing filaments rarely dissociated from either formin: 8.0 × 10–4 sec–1 for Bni1(FH1FH2)p and <1.0 × 10–5 sec–1 for Cdc12(FH1FH2)p. GST-tagged Bni1(FH1FH2)p (Fig. 2b) and GST-Cdc12(FH1FH2)p were more active than the untagged constructs, but GST was not required for insertional assembly (Figs. 2 i–m and 3 b–f).
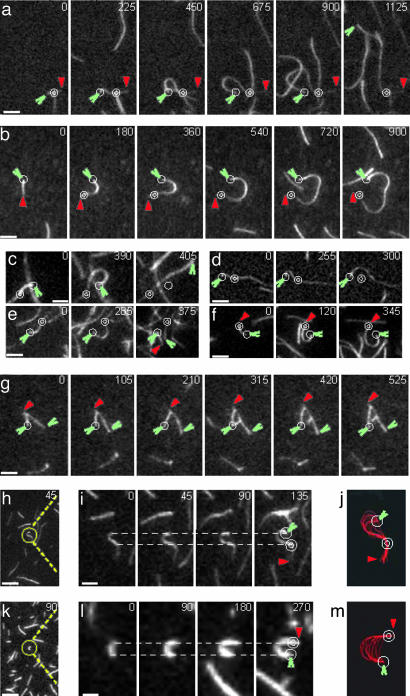
Fig. 3.
Measurement of actin polymerization force by buckling of filaments growing at their barbed end attached to a formin immobilized on the slide and their side attached to an NEM-myosin also immobilized on the slide surface. Conditions and symbols as in Figs. 1 and 2. Slides were preincubated with 100 nM NEM-myosin (a–f and h–m) and 100 mM GST-Bni1(FH1FH2)p (a and h–m) or 100 nM Bni1(FH1FH2)p (b–f). (a–g, i, and l) Time lapse evanescent wave fluorescence micrographs with seconds indicated at the top (i and l, magnified ×5). (h and k) Unmagnified micrographs of a single time-point (in seconds indicated at top) from the corresponding time lapse to the right. (j and m) Projection of Gaussian line fits showing the position of the filament to the left over time. (a-f) Mg-ATP actin (1.0 μM) with 0.4 μM OG-actin polymerized on the slide. Buckling between Bni1(FH1FH2)p and NEM-myosin (n = 123) was followed by detachment from Bni1(FH1FH2)p and springing away from this attachment point (42%) (c), detachment from the NEM-myosin and springing away from this attachment point (8%) (d), breaking between the attachment points (5%) (e), or remaining buckled throughout the time course (45%) (f). (g) Mg-ATP actin (1.0 μM) with 0.4 μM OG-actin and 1 nM Arp2/3 complex, 3 nM GST-WA, and 5 nM capping protein, assembled on a slide incubated with 100 nM GST-Bni1(FH1FH2)p. The branched filaments did not rotate, with respect to the formin, as they grew at their attached barbed ends. (h–j) Mg-ATP actin (0.25 μM) with 0.1 μM OG-actin polymerized on the slide. Elongation at the barbed end buckled the 1.0-μm segment of the filament between the NEM-myosin and formin attachment points, which required 0.4 pN of force. (k–m) Mg-ATP actin (0.5 μM) with 0.2 μM OG-actin was polymerized on the slide with 20 μM phalloidin. Elongation at the barbed end buckled the 0.75-μm segment of the filament between the NEM-myosin and formin attachment points, which required 1.3 pN of force. (Scale bar, 5 μmin a–h and k and 1 μmin i and l.)
Although the number of formins immobilized on glass greatly exceeded the number of captured barbed ends, the active formins were fully functional. We measured bound formins by SDS/PAGE and immunoblotting for GST. Approximately 20% of 100 nM GST-Bni1(FH1FH2)p remained bound to the surfaces of flow chambers and was eluted with SDS. The bound formin corresponded to 40 formin dimers per μm2 of glass surface. Over 30 min, an average of 100 filaments per field grew from such a surface, corresponding to one filament per 6,500 attached Bni1p molecules (0.015%). This calculation underestimates the number of active immobilized formins, because the surface of the slide continued to nucleate (≈25%) and capture (≈75%) filaments at the same rate of two filaments per min throughout the 30-min observation. In solution, the yield from 100 nM Bni1(FH1FH2)p is about one filament per 10 Bni1(FH1FH2)p molecules, so nucleation is less favorable on the surface than in solution. The active immobilized formins were indistinguishable from soluble formins: both supported persistent barbed end elongation at rates in the range of 50–100% (Fig. 2 d, h, m, and s) of the rate of free barbed ends, and profilin allowed immobilized Cdc12(FH1FH2)p to nucleate and support barbed end growth of actin filaments (Fig. 2 q and r).
The following observations are consistent with a single formin dimer anchoring each growing filament to the slide. Both tagged and untagged formins supported barbed end growth of single filaments from points on the slide. We never observed two filaments growing from the same point on the slide among the ≈1,000 that we observed. Gel filtration of Bni1(FH1FH2)p removed any aggregates, but did not change its properties when immobilized on a slide (Figs. 2 i and j and 3 b–f). The Stokes' radius of this homogeneous Bni1(FH1FH2)p was consistent with an asymmetric dimer. The fraction of filaments originating from or captured by an immobilized formin depended on the concentration of formin used to coat the slide, with GST-Bni1(FH1FH2)p capturing ≈3-fold more filaments than Bni1(FH1FH2)p at all concentrations tested (Fig. 2b).
Force Produced by Polymerization of Individual Actin Filaments. To measure the force produced by polymerization of individual actin filaments, we polymerized actin on slides preincubated with both a formin and NEM-myosin II to provide a second attachment point along the length of the filament (diagram 3 in Fig. 2a). When tethered by NEM-myosin, growth of a filament from immobilized Bni1(FH1FH2)p (Fig. 3 b–f) or GST-Bni1(FH1FH2)p (Fig. 3 a and h–m) caused the filament to buckle between the formin and the NEM-myosin. As buckled filaments grew, they looped out randomly to the right or left between the two attachment points (Fig. 3 a–f and h–m). As these loops grew, the maximum rotation around the point of barbed end attachment was 180°. Some doubly tethered filaments dissociated from the formin (Fig. 3c) or NEM-myosin (Fig. 3d) or broke between attachment points (Fig. 3e).
The force required to buckle a rod is proportional to its stiffness and inversely proportional to its length squared (10). Thus, the length of the shortest filament that buckled revealed the maximum observed force produced by actin polymerization. Polymerization of 0.25 μM Mg-ATP actin with 0.1 μM OG-actin buckled filaments between GST-Bni1(FH1FH2)p and NEM-myosin attachment points separated by as little as 0.8–1.2 μm (Fig. 3 h–j), corresponding to forces of 0.25–0.56 pN. Polymerization of 0.5 μM Mg-ATP actin with 0.2 μM OG-actin and 20 μM phalloidin buckled filaments between attachments separated by as little as 0.75 μm (Fig. 3 k–m). Phalloidin increases the stiffness of actin filaments 2-fold, so polymerization produced a force of at least 1.3 pN, close to the theoretical maximum (≈2 pN) under the experimental conditions. The flexibility of actin filaments and the resolution of the microscope limited the detection of buckling filaments shorter than 0.7 μm and measurement of forces >1.3 pN. We expect that force transiently slows the rate of assembly as the filament buckles, but the temporal and spatial resolution (≈100 subunits) were too small to detect this change.
Mechanistic Details of Formin-Mediated Insertional Assembly at Actin Filament Barbed Ends. Buckling experiments provided further evidence that a single formin dimer is associated with an elongating barbed end. Gaussian line fits of buckled filaments showed that growing barbed ends remained fixed to a space representing a single pixel (Fig. 3 j and m). Thus, barbed end attachment points of buckled filaments deviated <0.2 μm for times as long as 500 sec. Buckled filaments growing in association with a formin or GST-formin remained attached to a single point during the addition of >7,000 new subunits (Fig. 3 a and b). Upon dissociation from an immobilized formin, the elastic restoring force in a bent filament caused it to “snap out” from the attachment point (Fig. 3 a and c), almost never rebinding to another fixed point on the slide. The low probability of recapture during elastic recoil means that a filament growing under tension from a single point over hundreds of seconds must associate continuously with a single active formin dimer rather than jumping locally between formins in an aggregate. Additionally, we note that the barbed end elongation rate in the presence of formins is 30-fold higher than the ATP-hydrolysis rate, measured without formin (14), suggesting that ATP hydrolysis does not regulate formin-mediated insertional assembly.
Elongating filaments did not rotate relative to the immobilized formin. First, branches formed by Arp2/3 complex did not rotate as they grew from an immobilized GST-Bni1(FH1FH2)p (Fig. 3g). Capping protein included in these experiments limited the growth of branches formed by Arp2/3 complex, but not elongation of barbed ends anchored to the slide by formins. This finding confirms that formins protect barbed ends from capping protein (4, 5, 15). Second, although loops between a formin and a myosin grew to ≈20 μm (284 turns of the actin helix) (Fig. 3 a and b), none of >50 of these long loops attached to Bni1(FH1FH2) or GST-Bni1(FH1FH2)p supercoiled or twisted like tethered filaments translocated by myosin (16). This finding provides strong evidence that filaments growing from immobilized formins do not rotate.
Discussion
By direct comparison, we show that budding yeast Bni1p and fission yeast Cdc12p are not mechanistically identical. Cdc12p, but not Bni1p, requires profilin to allow barbed end assembly. However, we found that profilin does increase the barbed end elongation rate of individual Bni1p nucleated filaments, from 75% to 100% of full-speed, which explains why bulk assembly rates in the presence of Bni1p are increased by profilin (2, 4) even though profilin reduces Bni1p-dependent actin nucleation (3). Additionally, we provide evidence that the barbed end of an individual growing filament remains bound to a single formin molecule for >1,000 sec, 50 times longer than predicted from indirect experiments (5). Independently, Higashida et al. (7) observed barbed end growth of numerous actin filaments from “large aggregates” of mDia1 and antibodies. They also made spectacular movies of “single or a few EGFP molecules coupled to multimeric mDia1” surfing on actin filaments in cells. Their use of fluorescent formins is ideal for observing surfing of formins in cells. The single, immobilized formins used here have advantages for biophysical analysis, because the formin is between the surface and the growing barbed end of the filament.
Our observations of formin-mediated barbed end insertional polymerization are completely consistent with experiments on several formins (1–6, 15, 17). However, few mechanistic details were known. A popular idea is that one FH2 domain of a formin dimer maintains contact with the barbed end of one strand of the actin helix, whereas the other FH2 domain interacts with the end of the other long pitch helix (4, 5, 18). Flexible linkers between the two formin FH2 domains (18) might accommodate the 2.7-nm offset of the two actin subunits. This mechanism postulates that growth of a barbed end involves dissociation of the trailing formin FH2 domain from its strand, addition of an actin subunit to that strand, and reassociation of the formin FH2 domain with the new terminus of the filament. Because barbed ends grow at >10 subunits per second in vitro and >100 subunits per second in vivo (19), FH2 domains would have to dissociate rapidly from the penultimate actin subunit, but very slowly (half time of >1,000 sec according to our measurements) from the terminal subunit.
Two experiments show that filaments elongating while anchored by a formin do not twist as expected if the formin were immobilized and tracked along the growing actin helix: loops between a myosin anchor and an immobilized formin do not supercoil, and branches formed by Arp2/3 complex do not rotate. If a filament is prevented from rotating by a myosin anchor, growth at its barbed end will rotate the orientation of subunits 360° for every 26 subunits added. Thus, the formin must reorient rapidly on the end of the growing filament or the tether between the formin and the glass must include a rotatable bond. We have never observed rotation of a filament about its attachment point by >180°, even under fluid flow. Thus, alternatives to simple stair-stepping mechanisms should be considered, such as rotation of a growing actin helix relative to the FH2 homodimer like a shaft in a bearing. It has also been proposed that formins might bind to the side at the barbed end of the actin filament (15). Rigorous experimentation will be required to differentiate between these and other possible mechanisms.
It has long been appreciated that actin polymerization produces force for cell motility (20, 21), but analysis of this force has been restricted to theoretical studies and investigations of samples of numerous filaments. Biomimetic systems allow measurements of the force produced by populations of actin filaments pushing on a bacterium, bead, or vesicle (22–25). However, these systems are subject to both propulsive forces and opposing friction, and do not reveal the force produced by individual actin filaments.
Polymerization of an individual actin filament tethered between a formin and a myosin molecule in the presence of phalloidin produced a force of at least 1.3 pN, close to the theoretical maximum (≈2 pN) under the experimental conditions. Thus, the polymerization motor is very efficient. Microtubules are much stiffer, and their elongation produces buckling forces up to 10 pN (13, 26).
Because only short filaments are sufficiently stiff to transmit the polymerization force without buckling, capping protein is useful to limit the length of the force-producing filaments at the leading edge of motile cells. The capacity of actin polymerization to produce force in association with a formin supports the idea that formins mediate some protrusive forces in cells (7, 27).
In the presence of profilin, looped filaments attached along their length by NEM-myosin and growing from immobilized GST-Bni1(FH1FH2) have been observed to rotate up to 360°.
Supplementary Material
Acknowledgments
We thank Ikuko Fujiwara, Jeff Kuhn, Matt Lord, Rachel Mahaffy, and Brad Nolen for assistance and discussions. This work was supported by National Institutes of Health Grants GM-26132 and GM-26338 (to T.D.P.) and a National Institutes of Health Postdoctoral Fellowship (to D.R.K.).
Abbreviations: FH, formin homology; AG, Alexa green; OG, Oregon green; TIRF, total internal reflection fluorescence; NEM, N-ethylmaleimide.
See Commentary on page 14685.
References
- 1.Pruyne, D., Evangelista, M., Yang, C., Bi, E., Zigmond, S., Bretscher, A. & Boone, C. (2002) Science 297, 612–615. [DOI] [PubMed] [Google Scholar]
- 2.Sagot, I., Rodal, A. A., Moseley, J., Goode, B. L. & Pellman, D. (2002) Nat. Cell Biol. 4, 626–631. [DOI] [PubMed] [Google Scholar]
- 3.Pring, M., Evangelista, M., Boone, C., Yang, C. & Zigmond, S. H. (2003) Biochemistry 42, 486–496. [DOI] [PubMed] [Google Scholar]
- 4.Moseley, J. B., Sagot, I., Manning, A. L., Xu, Y., Eck, M. J., Pellman, D. & Goode, B. L. (2004) Mol. Biol. Cell 15, 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zigmond, S. H., Evangelista, M., Boone, C., Yang, C., Dar, A. C., Sicheri, F., Forkey, J. & Pring, M. (2003) Curr. Biol. 13, 1820–1823. [DOI] [PubMed] [Google Scholar]
- 6.Kovar, D. R., Kuhn, J. R., Tichy, A. L. & Pollard, T. D. (2003) J. Cell Biol. 161, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higashida, C., Miyoshi, T., Fujita, A., Oceguera-Yanez, F., Monypenny, J., Andou, Y., Narumiya, S. & Watanabe, N. (2004) Science 303, 2007–2010. [DOI] [PubMed] [Google Scholar]
- 8.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 9.Amann, K. J. & Pollard, T. D. (2001) Proc. Natl. Acad. Sci. USA 98, 15009–15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howard, J. (2001) Mechanics of Motor Proteins and the Cytoskeleton (Sinauer, Sunderland, MA).
- 11.Gittes, F., Mickey, B., Nettleton, J. & Howard, J. (1993) J. Cell Biol. 120, 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Isambert, H., Venier, P., Maggs, A. C., Fattoum, A., Kassab, R., Pantaloni, D. & Carlier, M. F. (1995) J. Biol. Chem. 270, 11437–11444. [DOI] [PubMed] [Google Scholar]
- 13.Dogterom, M. & Yurke, B. (1997) Science 278, 856–860. [DOI] [PubMed] [Google Scholar]
- 14.Blanchoin, L. & Pollard, T. D. (2002) Biochemistry 41, 597–602. [DOI] [PubMed] [Google Scholar]
- 15.Harris, E. S., Li, F. & Higgs, H. N. (2004) J. Biol. Chem. 279, 20076–20087. [DOI] [PubMed] [Google Scholar]
- 16.Nishizaka, T., Yagi, T., Tanaka, Y. & Ishiwata, S. (1993) Nature 361, 269–271. [DOI] [PubMed] [Google Scholar]
- 17.Li, F. & Higgs, H. N. (2003) Curr. Biol. 13, 1335–1340. [DOI] [PubMed] [Google Scholar]
- 18.Xu, Y., Moseley, J. B., Sagot, I., Poy, F., Pellman, D., Goode, B. L. & Eck, M. J. (2004) Cell 116, 711–723. [DOI] [PubMed] [Google Scholar]
- 19.Yang, H. C. & Pon, L. A. (2002) Proc. Natl. Acad. Sci. USA 99, 751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mogilner, A. & Oster, G. (2003) Biophys. J. 84, 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlier, M. F., Le Clainche, C., Wiesner, S. & Pantaloni, D. (2003) BioEssays 25, 336–345. [DOI] [PubMed] [Google Scholar]
- 22.Giardini, P. A., Fletcher, D. A. & Theriot, J. A. (2003) Proc. Natl. Acad. Sci. USA 100, 6493–6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Upadhyaya, A. & van Oudenaarden, A. (2003) Curr. Biol. 13, R734–R744. [DOI] [PubMed] [Google Scholar]
- 24.Marcy, Y., Prost, J., Carlier, M. F. & Sykes, C. (2004) Proc. Natl. Acad. Sci. USA 101, 5992–5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz, I. M., Ehrenberg, M., Bindschadler, M. & McGrath, J. L. (2004) Curr. Biol. 14, 1094–1098. [DOI] [PubMed] [Google Scholar]
- 26.Janson, M. E., de Dood, M. E. & Dogterom, M. (2003) J. Cell Biol. 161, 1029–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wallar, B. J. & Alberts, A. S. (2003) Trends Cell Biol. 13, 435–446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.