Telomerase maintains telomere length by adding G-rich telomeric repeats to the ends of linear eukaryotic chromosomes. The core telomerase enzyme consists of two components: an essential RNA component and a catalytic protein component. The catalytic component, telomerase reverse transcriptase (TERT), contains sequence motifs homologous to those in the catalytic domain of reverse transcriptase enzymes. Although the TERT component of telomerase is fairly conserved among eukaryotes, the RNA component varies dramatically in sequence composition and in size. The structure of the telomerase RNA has been well established in ciliates and vertebrates (1–4). These structural models have led to experiments that provided fundamental insight into telomerase RNA function. However, it has been difficult to determine the global secondary structure of the very large yeast RNA. In the past month, there have been four new reports proposing a secondary structure for the yeast telomerase RNA (5–8). In this issue of PNAS, Blackburn and colleagues (7) propose a core structure for the yeast telomerase RNA that resembles the structure present in ciliate and vertebrate telomerase RNAs.
The Ciliate and Vertebrate Telomerase RNAs Share a Common Core Structure
Telomerase RNA is very divergent across eukaryotic species. The size ranges from ≈150 nt in ciliates to >1,300 nt in fungi. Although there is some sequence similarity within each group of closely related eukaryotes, there is no sequence similarity between groups. Thus, the telomerase RNA sequences from vertebrates cannot be aligned with the RNA sequences from ciliates. The rapid divergence in RNA size and sequence has made it difficult to identify common secondary structures for telomerase RNAs.
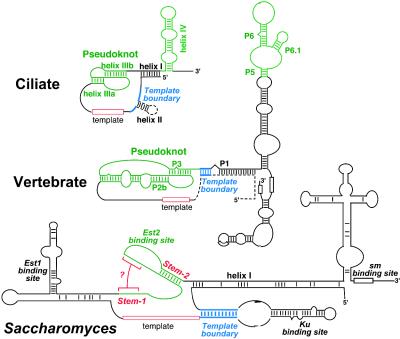
Phylogenetic comparative analysis has proven to be the most powerful approach for inferring higher-order RNA structures. In this method, base paired helices in RNA secondary structure are determined by nucleotide covariation between species (9). For example, an A:U base pair in one species may change to a G:C, C:G, or U:A Watson–Crick base pair in a different species. Secondary structure models of both the ciliate and vertebrate RNAs were established by using phylogenetic analysis (2, 4, 10). Remarkably, despite differences in length and sequence, there is a similar core secondary structure in both groups of organisms that includes a long-range base pairing interaction that encloses the template and an adjacent pseudoknot structure in a larger loop (Fig. 1). In most vertebrates, this long-range base-pairing, called the P1 helix, plays a role in defining the template boundary (11). In the ciliate RNAs, a highly conserved sequence motif that is located between the helix I and the template defines the template boundary (12, 13).
Fig. 1.
Secondary structures of ciliate, vertebrate, and yeast (Saccharomyces) telomerase RNAs. Structural elements in green represent conserved regions that bind to TERT. The structures that are present in some but not all species within a group are shown by dashed lines. The structures that define template region (red) and the template boundary (blue) are indicated. The RNA structural elements bound by protein components are indicated. In the Saccharomyces RNA, the putative stem-1 is indicated by red brackets. The yeast helices are named according to Dandjinou et al. (6) and Lin et al. (7).
Determining the Yeast Telomerase RNA Structure
Given the conservation of telomerase RNA structure in ciliates and vertebrates, elucidation of the structure of the yeast Saccharomyces cerevisiae is of great interest. With the availability of genome sequences from eight Saccharomyces species, four groups have now proposed structures for the yeast telomerase RNA.
Zappulla and Cech (8) and Dandjinou et al. (6) both took a similar approach of using the RNA structure prediction programs mfold or alifold (14, 15) together with phylogenetic analysis to propose a model for the entire 1,300-nt RNA (Fig. 1). This proposed structure encompases several known helical regions that were previously known binding sites for the telomerase Est1 protein and the Ku70/80 heterodimer (16). Chappell and Lundblad (5) and Lin et al. (7) took a somewhat different approach to study the structure and function of the yeast telomerase RNA. They also combined both phylogenetic and computer prediction to examine specific structural elements of the RNA core, and then tested the potential base pairings experimentally.
Lin et al., Zappulla and Cech, and Dandjinou et al. propose a long-range base-pairing, termed helix I (Fig. 1), that brings the template in close proximity to the Est2-binding site (6–8). Such spatial arrangement of RNA elements generate a core structure that is analogous (and potentially homologous) to the ciliate and vertebrate structures (1, 2, 4). Compared to ciliate and vertebrate RNA, the yeast helix I is unusually long, 86 bp, with several internal loops and bulges (6, 8). Although the proposed base pairings need refinement or confirmation by experimental data, the presence of this helix is supported by both phylogenetic evidence (6, 8) and mutagenesis (7).
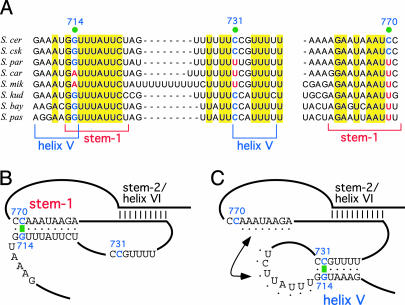
Another interesting structure in the model proposed by Lin et al. is a pseudoknot that resembles those in ciliate and vertebrate telomerase RNAs (Figs. 1 and 2B). This region of the Saccharomyces telomerase RNA binds the telomerase catalytic subunit Est2 (the yeast TERT protein) and is essential for telomerase function (17). In the vertebrate pseudoknot, one of the loops contains a stretch of U residues that are invariant and essential for activity (4). Lin et al. also point out a similar conserved U-rich sequence in the yeast pseudoknot. The proposed pseudoknot structure suggests a conserved core for ciliate, yeast, and vertebrate telomerase RNAs. However, an alternative solution was proposed by the other three groups. In their models, there is an additional small stem loop, called the helix V, that forms immediately adjacent to stem-2 of the proposed pseudoknot (Fig. 2C). Interestingly, the covariation evidence from the available sequences supports either structure. The proposed stem 1 pairing is supported by covariation at 714G:770C (Fig. 2 A and B), whereas the proposed helix V includes the same 714G nucleotide, which in this case is paired with 731C, and this pairing is also supported by covariation data (Fig. 2 A and C). Some of these papers cite a single change from a G:C to a G:U wobble pair (i.e., 713G:771C changes to G:U) as positive evidence for phylogenetic covariation. However, covariation is defined as two independent changes that maintain Watson–Crick base pairing (9); thus, single-event changes should be considered as neither positive nor negative evidence for a helix (Fig. 2A).
Fig. 2.
Two possible structures in yeast telomerase RNA core. (A) Sequence alignment of regions of the proposed pseudoknot and helix V. The sequence alignment and nucleotide numbering of telomerase RNAs from eight Saccharomyces species are adapted from Dandjinou et al. (6). Invariant residues are highlighted in yellow. Residues that show covariation are indicated by green dots. (B) A possible secondary structure of Est2 binding domain, proposed by Lin et al. (7), consists of the stem-1, supported by one nucleotide covariation at the base pair 714G:770C. (C) An alternative structure, proposed by Dandjinou et al. (6), consists of the helix V, supported by one nucleotide covariation at the base pair 714G:731C. Potential base pairings between the loops of helix V and helix VI, as proposed by Dandjinou et al. (6), are indicated by black dots. Both structures consist of a helix called stem-2 (7) or helix VI (6).
Dandjinou et al. proposed a hybrid between the two-stem model and the pseudoknot model. They propose a loop–loop pseudoknot between the loop of helix V and the loop of helix VI (Fig. 2C). However, the structural feasibility of such interaction remains to be tested, and this base-pairing interaction lacks covariation supports (Fig. 2A). To resolve the issue of which helix is most likely to be present, more distantly related yeast sequences are needed to provide more sequence variations.
A Conserved RNA-Binding Protein Implies a Conserved RNA Structure
In contrast to the RNA component, the catalytic protein component of telomerase, TERT, is highly conserved throughout eukaryotes (18). Most TERT proteins contain a telomerase-specific domain, called the motif T, that is important for RNA binding (19, 20). If the RNA-binding domain of TERT is conserved among ciliates, yeasts, and vertebrates, the RNA element that is recognized ought to be conserved as well. All four papers propose the pairing of stem-2/helix VI. Both Chappell and Lundblad (5) and Lin et al. (7) provide strong experimental evidence that this pairing is needed for the Est2 binding. Both groups also propose an additional stem-1 that pulls the stem-2 into a pseudoknot structure. Surprisingly, they also both report that the stem-1 needed for this pseudoknot formation can be deleted with no apparent effect on telomere length or Est2 binding. Although these results argue against the pseudoknot structure, Chappell et al. (5) went on to show that this region of the yeast RNA can be functionally replaced by the equivalent pseudoknot element from the ciliate Oxytricha nova, implying that a pseudoknot is indeed recognized by the yeast Est2 protein.
The identification of a possible pseudoknot and a conserved core structure in yeast telomerase is a significant advance in understanding telomerase. The structures provided by these four papers are readily testable by using nucleotide substitutions to probe the importance of potential base pairings. They provide useful working models that will stimulate new experimental questions.
See companion article on page 14713.
References
- 1.Romero, D. P. & Blackburn, E. H. (1991) Cell 67, 343–353. [DOI] [PubMed] [Google Scholar]
- 2.ten Dam, E., van Belkum, A. & Pleij, K. (1991) Nucleic Acids Res. 19, 6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lingner, J., Hendrick, L. L. & Cech, T. R. (1994) Genes Dev. 8, 1984–1998. [DOI] [PubMed] [Google Scholar]
- 4.Chen, J.-L., Blasco, M. A. & Greider, C. W. (2000) Cell 100, 503–514. [DOI] [PubMed] [Google Scholar]
- 5.Chappell, A. S. & Lundblad, V. (2004) Mol. Cell. Biol. 24, 7720–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dandjinou, A. T., Levesque, N., Larose, S., Lucier, J. F., Abou Elela, S. & Wellinger, R. J. (2004) Curr. Biol. 14, 1148–1158. [DOI] [PubMed] [Google Scholar]
- 7.Lin, J., Ly, H., Hussain, A., Abraham, M., Pearl, S., Tzfati, Y., Parslow, T. G. & Blackburn, E. H. (2004) Proc. Natl. Acad. Sci. USA 101, 14713–14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zappulla, D. C. & Cech, T. R. (2004) Proc. Natl. Acad. Sci. USA 101, 10024–10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woese, C. R. & Pace, N. R. (1993) in The RNA World, eds. Gesteland, R. F. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 91–117.
- 10.Tzfati, Y., Knight, Z., Roy, J. & Blackburn, E. H. (2003) Genes Dev. 17, 1779–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J.-L. & Greider, C. W. (2003) Genes Dev. 17, 2747–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Autexier, C. & Greider, C. W. (1995) Genes Dev. 9, 2227–2239. [DOI] [PubMed] [Google Scholar]
- 13.Lai, C. K., Miller, M. C. & Collins, K. (2002) Genes Dev. 16, 415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofacker, I. L., Fekete, M. & Stadler, P. F. (2002) J. Mol. Biol. 319, 1059–1066. [DOI] [PubMed] [Google Scholar]
- 15.Zuker, M. (2003) Nucleic Acids Res. 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson, S. E., Stellwagen, A. E., Diede, S. J., Singer, M. S., Haimberger, Z. W., Johnson, C. O., Tzoneva, M. & Gottschling, D. E. (2001) Nat. Genet. 27, 64–67. [DOI] [PubMed] [Google Scholar]
- 17.Livengood, A. J., Zaug, A. J. & Cech, T. R. (2002) Mol. Cell. Biol. 22, 2366–2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamura, T. M. & Cech, T. R. (1998) Cell 92, 587–590. [DOI] [PubMed] [Google Scholar]
- 19.Friedman, K. L. & Cech, T. R. (1999) Genes Dev. 13, 2863–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelleher, C., Teixeira, M. T., Forstemann, K. & Lingner, J. (2002) Trends Biochem. Sci. 27, 572–579. [DOI] [PubMed] [Google Scholar]