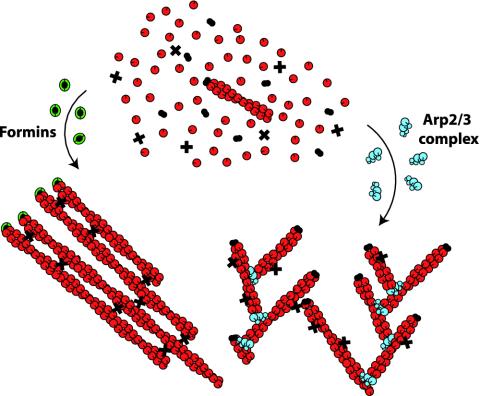
While cellular conditions strongly favor the growth of actin filaments, growth occurs only when actin monomers have access to a filament's fast-growing barbed end. This is the key to understanding how the only known de novo generators of actin filaments in cells, the Arp2/3 complex and members of the formin family, can give rise to such structurally different networks (Fig. 1). The Arp2/3 complex binds to the sides of an actin filament where it nucleates a daughter filament with a free barbed end at an acute angle to the mother (1). The new filament grows for only a short period before being blocked by barbed-end capping proteins, and so networks generated by the actions of many Arp2/3 complexes consist of short filaments arranged in a dendritic brush-like structure. By contrast, some formins generate filaments that can grow at their barbed ends while protected from the activity of capping proteins (2). Assemblies of these formins create elongated parallel bundles of filaments like those comprising the actin cables of budding yeast (2).
Fig. 1.
Actin network geometry depends on how long the barbed end is free during filament nucleation. Starting with the same building blocks, the formins and Arp2/3 complex will create very different networks because of their opposite treatment of barbed ends. Arp2/3 complex (blue) nucleates filaments with free barbed ends from the sides of preexisting filaments. Because these filaments are quickly blocked by capping proteins (black), the resulting network is composed of short filaments arranged in a brush-like structure. Such networks are seen, for example, at the leading edges of crawling cells. By contrast, formins (green) protect barbed ends from capping as they grow to create long filaments that can be arranged into bundles by cross-linkers (x). Formin-induced bundles are found, for example, in the actin cables of budding yeast.
Armed with crystal structures and numerous inferences from actin assembly kinetics in the presence of formins and capping protein, investigators have imagined that formins stay affixed to the barbed ends of growing filaments even while they permit actin monomers to assemble onto the filament barbed end (3). In this issue of PNAS, Kovar and Pollard (4) confirm this mechanism by directly visualizing single filaments growing from isolated and immobilized domains of two yeast formins. An exciting by-product of this effort is the first measurement of the pushing force generated by a growing actin filament.
A New Look at Formins
To watch formins at work, Kovar and Pollard coated a microscope slide with highly diluted solutions of the FH1-FH2 domains of Bni1p or Cdc12p. After isolated formins adsorbed to the slide, they were exposed to a solution of fluorescent actin monomers under polymerizing conditions. Using total internal reflection microscopy to create clear images of filaments on the surface of the slide, Kovar and Pollard saw single actin filaments growing while attached at one end. To confirm that filaments were growing at the points of attachment, they replaced the original monomer solution with a solution containing brighter actin. The newer actin clearly assembled at the attachment point at rates only feasible for the barbed end of actin. Because seeing is believing, these observations provide the most definitive evidence imaginable that individual formins remain associated with barbed ends of actin filaments while permitting the insertion of new monomers at those same ends.
Formins do not rotate as they track a filament's barbed end.
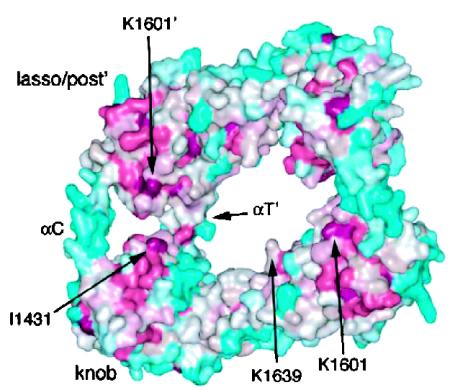
Even as Kovar and Pollard confirm key ideas about how formins operate, they offer a major challenge. The crystal structure of FH2 (5) indicates that this actin-binding domain dimerizes to form a flexible, doughnut-shaped structure with head-to-tail connections (Fig. 2). In the prevailing “stair-stepping” model for processive capping by formins, one FH2 domain is bound to the terminal subunit in each strand of actin's double helix (3). The domain bound to the lagging strand releases to allow addition of one monomer and then advances to bind the new monomer. This process repeats for the second strand to complete a simple processive cycle. The challenge that Kovar and Pollard present is the finding that formins do not rotate as they track a filament's barbed end. They establish this idea by using modified myosins to anchor filaments at a point distant from their barbed ends. In several cases, filaments grow 10 or more micrometers between the formin and myosin anchors. Because the formins appear unable to freely spin while attached to the glass, the filament should rotate 14 times for each micrometer of growth between the anchored proteins if the stair-stepping mechanism is correct. Such an extreme rotation should cause obvious supercoiling, but by using Arp2/3 complex to mark the sides of filaments with branches, Kovar and Pollard observe that the filaments don't even twist.
Fig. 2.
The structure of an FH2 domain dimer of the yeast formin Bni1p. Color indicates the degree of evolutionary conservation with deep magenta representing highly conserved regions and dark teal representing the most variable regions. The four magenta patches are putative actin binding sites. [Reprinted with permission from ref. 5 (Copyright 2004, with permission from Elsevier).]
Pushing by Single Filaments
Although formin-nucleated filaments do not appear to rotate as they grow between anchored points, they clearly buckle from the compressive forces generated by polymerization. This allowed Kovar and Pollard to make the first measures of the forces exerted by a single polymerizing filament. Because the buckling force scales inversely with the square of the length between attachment points, the shortest filament observed to buckle provides a lower bound on the force a polymerizing filament must be able to exert. Kovar and Pollard observed a 0.75-μm filament buckle and calculated that this would require 1.3 pN of force, only ≈0.7 pN shy of the force that should stall polymerization under the conditions in the article. Future experiments should reveal how polymerization rates slow with compression forces.
More to Come
The demonstration that a growing filament can push against the formin that nucleates it should also keep the biophysics community busy. Thermodynamics may have no objections, but the mechanism could be difficult to decipher. Faced with a similar puzzle when it became clear that Listeria monocytogenes bind to the Arp2/3 complex-generated networks that propel them through cytoplasm (6, 7), some investigators proposed separate populations of anchored and growing filaments (8). Clearly this option does not exist for a single formin/filament complex. Another group proposed that a barbed-end clamp permitted anchored filaments to grow (9). This “actoclampin” model may be a good starting point for thinking about the biophysics of formin movements.
Many exciting measurements are sure to come. Repeating the observations of Kovar and Pollard with formins bound to surfaces through two or more engineered attachment points would confirm the lack of formin/filament rotation during filament growth. Single-molecule fluorescence anisotropy imaging (10) might also be used to analyze rotational motions. Single formin movement seems amenable to tracking and force measurements with optical traps (11). Such data will be key to a physical and energetic explanation of the formin machinery. The results of Kovar and Pollard may mean that filaments fit through the lumen of the FH2 doughnut and that there are many available binding configurations. In this way, small twisting strains could be relieved because the formin slips into nearby configurations without sliding off the filament. Cross-linking actin to formins should reveal whether binding is limited to the four conserved sites in the crystal (magenta in Fig. 2).
Acknowledgments
We thank attendees of a recent Institute for Complex Adaptive Matter workshop on the Biophysics of Actin-Based Motility for tremendous discussions about the mysteries of formins. This work was supported by Whitaker Research Grant RG-01-0338.
See companion article on page 14725.
References
- 1.Welch, M. D. & Mullins, R. D. (2002) Annu. Rev. Cell. Dev. Biol. 18, 247–288. [DOI] [PubMed] [Google Scholar]
- 2.Sagot, I., Klee, S. K. & Pellman, D. (2002) Nat. Cell. Biol. 4, 42–50. [DOI] [PubMed] [Google Scholar]
- 3.Zigmond, S. H., Evangelista, M., Boone, C., Yang, C., Dar, A. C., Sicheri, F., Forkey, J. & Pring, M. (2003) Curr. Biol. 13, 1820–1823. [DOI] [PubMed] [Google Scholar]
- 4.Kovar, D. R. & Pollard, T. D. (2004) Proc. Natl. Acad. Sci. USA 101, 14725–14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu, Y., Moseley, J. B., Sagot, I., Poy, F., Pellman, D., Goode, B. L. & Eck, M. J. (2004) Cell 116, 711–723. [DOI] [PubMed] [Google Scholar]
- 6.Gerbal, F., Laurent, V., Ott, A., Carlier, M. F., Chaikin, P. & Prost, J. (2000) Eur. Biophys. J. 29, 134–140. [DOI] [PubMed] [Google Scholar]
- 7.Kuo, S. C. & McGrath, J. L. (2000) Nature 407, 1026–1029. [DOI] [PubMed] [Google Scholar]
- 8.Mogilner, A. & Oster, G. (2003) Biophys. J. 84, 1591–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickinson, R. B. & Purich, D. L. (2002) Biophys. J. 82, 605–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adachi, K., Yasuda, R., Noji, H., Itoh, H., Harada, Y., Yoshida, M. & Kinosita, K., Jr. (2000) Proc. Natl. Acad. Sci. USA 97, 7243–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Block, S. M., Goldstein, L. S. & Schnapp, B. J. (1990) Nature 348, 348–352. [DOI] [PubMed] [Google Scholar]