Abstract
The U1 small nuclear ribonucleoprotein particle U1C protein has a zinc finger-like structure (C2H2 motif) at its N terminus, which is conserved from yeast to humans. Mutations of amino acid L13 within this domain rescue the essential function of the helicase protein Prp28p. Prp28p has been implicated in unwinding the 5′ splice site (5′ss)–U1 small nuclear RNA (snRNA) base-pairing, to allow replacement of U1 snRNA with U6 snRNA during spliceosome assembly. The L13 phenotype has therefore been interpreted to indicate that WT U1C contributes to 5′ss–U1 snRNA stabilization by binding to the RNA duplex. We show here that an L13 mutant extract cannot form stable base-pairing at room temperature but is permissive for U1–5′ss base-pairing at low temperature. This phenotype is similar to that of a U1C-depleted extract, indicating that the U1C L13 mutation is a strong loss-of-function mutation. The two mutant extracts are unlike a WT extract, which undergoes stable pairing at room temperature but little or no pairing at low temperature. Taken together with previous results and the failure to observe a direct interaction of U1C with the U1–5′ss duplex, the data suggest that U1C contributes indirectly to stable U1–5′ss base-pairing under permissive conditions. A model is proposed to account for the L13 results.
Eukaryotic genes are usually interrupted by introns, which must be precisely excised. Intron removal or pre-mRNA splicing takes place within a large RNA–protein machine termed the spliceosome. The five splicing small nuclear ribonucleoprotein particles (snRNPs) and many additional non-snRNP spliceosomal proteins play specific roles in the splicing process (1–8). In both yeast and mammals, pre-mRNA recognition by the spliceosome relies on a set of consensus sequences within the pre-mRNA intronic regions. These are the 5′ splice site (5′ss), branchpoint, and 3′ splice site regions, which associate with protein and RNA components of the spliceosome (2, 3, 9, 10).
During in vitro splicing, U1 snRNP recognizes the 5′ss in an ATP-independent fashion and joins the pre-mRNA to form commitment complex in yeast or the E complex in mammals (11, 12). Because spliceosome assembly is cotranscriptional and the 5′ss is synthesized before the other two cis-acting regions, it presumably associates with U1 snRNP before other splicing components join during subsequent steps of spliceosome assembly (13, 14). An alternative view is that a preassembled penta-snRNP recognizes the 5′ss, and the subsequent steps of “spliceosome assembly” occur as conformational changes in vivo (15). In either case, the 5′ss sequence functions not only in initial intron recognition but also in subsequent splicing steps, such as splice site partner assignment and even catalysis (16).
Commitment complex and E complex formation involve base-pairing between the highly conserved pre-mRNA 5′ss and the single-stranded 5′ end of U1 small nuclear RNA (snRNA). This base-pairing is critical for the U1 snRNP–5′ss interaction and contributes significantly to 5′ss selection, in both mammals and yeast (17–19). Nonetheless, U1 snRNP that is incapable of forming canonical RNA–RNA base-pairing can undergo surprisingly normal interactions with pre-mRNA in vitro (20). Although these mutant complexes are much less stable than WT complexes, they still maintain considerable sequence specificity for a proper 5′ss. The canonical RNA–RNA base-pairing is also not essential for either 5′ss selection or splicing in the HeLa in vitro system (21). All of these data indicate that other factors, probably U1 snRNP proteins, make significant contributions to 5′ss recognition and complex formation in mammals and yeast.
Early results in the human system had focused on the U1C protein as an important factor for efficient complex formation between U1 snRNP and the pre-mRNA 5′ss (22, 23). Biochemical depletion of U1C from U1 snRNP in HeLa nuclear extracts dramatically decreased U1 snRNP binding to a 5′ss (22). The Saccharomyces cerevisiae U1C gene was subsequently identified in our laboratory and shown to be essential (24). Genetic depletion of the protein inhibits splicing in vivo and commitment complex formation in vitro, indicating that yeast U1C is critical for U1 snRNP function (24). Our subsequent study suggested that yeast U1C plays a major role in 5′ss recognition, as the recombinant protein binds single-stranded RNA and has sequence specificity for 5′ss-like sequences (25).
U1C has a zinc finger-like structure (C2H2 motif) at its N terminus, which is conserved from yeast to humans (24, 26). A recent genetic study showed that mutations of amino acid L13 within this conserved domain could bypass the essential function of the DExH/D box helicase protein Prp28p (26). Prp28p had been previously implicated in unwinding the 5′ss–U1 snRNA base-pairing to allow replacement of U1 snRNA with U6 snRNA during progression of the spliceosome assembly pathway (26, 27). The L13 phenotype was therefore interpreted to indicate that WT U1C and specifically its zinc finger region normally contributes to U1 snRNA–5′ss duplex stabilization. Because the L13 mutant has low levels of U1C activity, the result is an unstable duplex that no longer requires Prp28p activity. All of this fit a model in which the U1C Zn finger region binds to the U1 snRNA–5′ss duplex.
Because our own yeast studies had suggested that U1C binds to the single-stranded 5′ss rather than to an RNA duplex within the context of U1 snRNP (25), we investigated in more detail the contribution of U1C to yeast U1 snRNP–pre-mRNA complex formation and stabilization. Our results provide direct biochemical evidence for the proposed Prp28p bypass model (26, 27), as in vitro complex formation with L13 U1C-containing U1 snRNP gives rise to unstable commitment complexes. However, our data show no evidence for a direct association of U1C with the U1 snRNA–5′ss duplex. Rather, they suggest that yeast U1C contributes to a temperature-dependent conformational change that takes place during commitment complex formation and helps to convert unstable complexes into stable complexes.
Materials and Methods
Materials. Methylene blue (MB) and ascorbic acid were purchased from Sigma. 4-Thiouridine (4-thio)-UTP and an in vitro transcription kit were purchased from Ambion, Austin, TX. 4′-Aminomethyl-4,5′,8-trimethyl psoralen was purchased from HRI Associates (Emeryville, CA).
Yeast Strains and Extracts. Y59 (WT) (MATa ade2 arg4 leu2–3,112 trp1–289 ura3–52), U1–70KHA (MAT ade2–101 his3–200 leu2–1 lys2–208 trp1–63 ura3–53 snp1::pJT-70KHA), U1CHA (MATα ade2 trp1 leu2 his3 ura3 canR yhc1::LEU2 pGAL-YU1CHA-URA3-CEN4), and U1C L13S (α prp28::HIS3 YHC1–2 ura3–53 lys2–801 ade2–101 trp1-Δ1 hisΔ200 leu2-Δ1) (a gift from T. H. Chang, Ohio State University, Columbus) were used in this work. Splicing extracts were prepared from the WT strain Y59 and tagged strains by using a modified miniextract protocol (28).
Constructs and RNA Substrates. WT pre-mRNA substrate containing the first 72 nt of RP51A pre-mRNA (WT-72) was generated by in vitro transcription of plasmid BT81. The 5′ss mutant RNA substrate was generated by in vitro transcription of plasmid pHD43, in which GTATGT was mutated to ATTTGT in the 5′ss region. The RNAs used for psoralen UV cross-linking, MB cross-linking, and immunoprecipitation assays were uniformly labeled with [α-32P]UTP by standard protocols. 4-Thio-UTP 32P-labeled RNAs were synthesized according to the previously published procedure from this laboratory (9). All RNAs were gel-purified.
MB-Mediated Cross-Linking. MB cross-linking was carried out as reported with minor modifications (29). Radiolabeled RNA was incubated with splicing extract to form commitment complex for 20 min at 25°C. After incubation, 0.5 μg/μl yeast total RNA was added. The samples (10 μl) were irradiated for 25 min with 4 ng of MB, 2 mM ascorbic acid, and 4 mM Tris·HCl (pH 7.5) on a precooled parafilmed block. The light source for the MB crosslinking was a 40-W fluorescent tube mounted 2–3 cm above the samples. After irradiation, the samples were subjected to RNase A/T1 digestion (RNase mixture, Ambion) and analyzed by SDS/PAGE and autoradiography.
Psoralen and 4-Thio-UTP Cross-Linking Assays. Psoralen crosslinking was performed as described (20). Commitment complexes were formed under splicing conditions in 10 μl and placed on ice. Psoralen was added to 10 μg/ml, and samples were irradiated with 365-nm UV light for 10 min. Samples were then deproteinized, extracted with phenol/chloroform, and precipitated with ethanol, and samples were analyzed on a 6% denaturing gel. 4-Thio-UTP UV cross-linking and immunoprecipitation were performed as described (9).
Chase Experiments. 32P-labeled RNA substrate was incubated with splicing extract in a standard commitment complex reaction for 20 min. A 400-fold molar excess of unlabeled WT-72 was then added and the incubation continued for the specified time periods. Psoralen crosslinking and 4-thio-UTP cross-linking were then performed as described above. Additionally, chased commitment complexes were immunoprecipitated with anti-Prp40 antibody and quantified by liquid scintillation counting.
Results
To investigate the role of U1C in canonical U1 snRNA–5′ss duplex formation and specifically address U1 snRNA–5′ss base-pairing, we assayed commitment complex formation in vitro with psoralen cross-linking (20). Splicing extracts were from a U1C-L13 mutant strain (26), a strain genetically depleted of U1C (24), and a control WT strain.
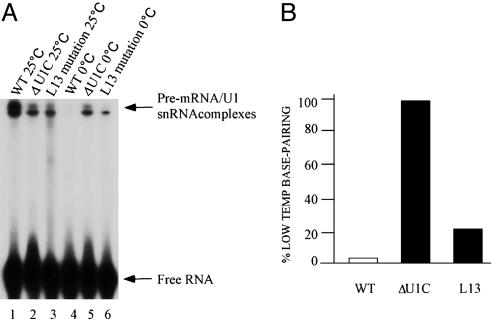
At room temperature, we observed a modest reduction in cross-linking with the two mutant extracts, compared with the WT extract, suggesting some contribution of U1C to in vitro commitment complex formation under these standard conditions (Fig. 1A, lanes 1–3). This finding fits with the observation that the L13 mutant strain is viable and grows well. At 0°C, however, there was much more cross-linking in the two mutant extracts than in the WT extract (Fig. 1 A, lanes 4–6, and B). For the WT and U1C-depleted extracts, the results were consistent with our previous report, i.e., there was no detectable crosslinking in the WT extract and substantial cross-linking in the U1C-depleted extract (25). In fact, cross-linking in the depleted extract showed little difference as a function of temperature, in striking contrast to the WT extract (Fig. 1B). An intermediate phenotype was observed for the L13 mutant extract, which still had much more cross-linking at 0°C than the WT extract despite having less cross-linking at room temperature (Fig. 1). The results were consistent with an intermediate loss-of-function etiology for the L13 mutation (Fig. 1A, lane 6 vs. lanes 5 and 4).
Fig. 1.
The effect of U1C on U1 snRNA–5′ss base-pairing at normal and low temperatures. (A) RNA–RNA base-pairing assayed by psoralen cross-linking at both high and low temperatures. Standard commitment complex formation assays were performed in WT, U1C L13 mutant, and U1C depletion extracts with 32P-radiolabeled WT-72 at the two temperatures. Psoralen cross-linking was carried out after 20 min of incubation as described (20). U1 snRNA–5′ss interactions and free RNA substrates are indicated by arrows. (B) Percentage of low-temperature base-pairing. The relative intensity of lanes 4–6 of A was measured, and the base-pairing in the U1C-depleted extract at low temperature was arbitrarily set to 100%.
The low-temperature data are paradoxical, i.e., a loss-of-function mutation that is supposed to destabilize the U1 snRNA–5′ss duplex is not expected to give rise to more psoralen-dependent cross-linking than WT U1C-containing snRNPs at any temperature. The apparent greater level of duplex formation in the L13 and depletion extracts suggests that WT U1C contributes to an inhibition of duplex formation at low temperature. This interpretation is also consistent with our previous observations indicating that WT U1C contacts the single-stranded 5′ss, the same sequence that base-pairs with the 5′ end of U1 snRNA (25). It would appear that U1C-binding competes with and inhibits base-pair formation, at least at low temperature (see below).
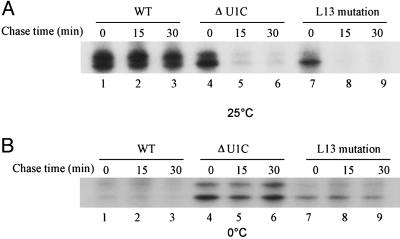
We also used a chase protocol to assess the stability of complexes formed at different temperatures in the three extracts. Incubation of radioactive pre-mRNA in extract was continued after addition of a 400-fold excess of cold pre-mRNA (Fig. 2). Psoralen cross-linking indicated that the U1 snRNA–pre-mRNA duplexes formed at 25°C in a WT extract are very stable, i.e., there was almost no change in signal after the 30-min incubation (Fig. 2 A, lanes 1–3). However, the signal had almost completely disappeared by 15 min of incubation in the L13 and U1C-depleted extracts (Fig. 2 A, lanes 4–9). These data provide direct biochemical evidence in support of a Prp28p bypass suppressor model: because the U1 snRNA–pre-mRNA duplex is inherently unstable when U1 snRNP contains the U1C L13 mutation, the Prp28p helicase is not necessary to unwind the duplex.
Fig. 2.
Stability of the U1 snRNA–5′ss base-pairing at two different temperatures. Chase assays were used to measure the stability of RNA–RNA interactions formed in the three different extracts. 32P-labeled transcripts were incubated with splicing extracts in standard commitment complex reactions for 20 min at either 25°C (A) or 0°C (B). Aliquots were removed, and a 400-fold molar excess of unlabeled RNAs was added. Psoralen was then added at the times indicated. The samples were UV-irradiated as described (20).
At low temperatures, however, the duplexes formed in U1C-depleted and L13 mutant extracts were stable under chase conditions (Fig. 2B, lanes 13–15 and 16–18). This finding is consistent with the steady-state data shown in Fig. 1 and might be relevant to the cold-sensitive phenotype of the Prp28p bypass suppressor strains (26), i.e., Prp28p might be more necessary at low temperature to help unwind the more stable L13-containing U1 snRNP–5′ss duplexes. At higher temperatures, however, Prp28p activity is unnecessary because of the less stable nature of these base pairs.
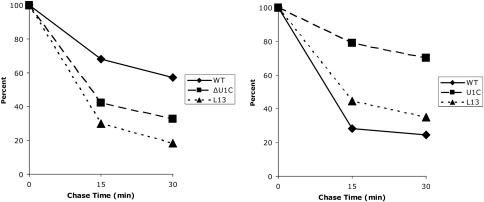
To assess commitment complex stability without focusing on U1 snRNA–5′ss base-pairing, an antibody against the U1 snRNP protein Prp40p was used for immunoprecipitations, and the amount of radioactive pre-mRNA in the pellet was determined after the same incubation and chase conditions (Fig. 3). Consistent with the psoralen results, WT complexes were more stable than those formed in the L13 mutant and U1C-depleted extracts after incubation at room temperature (Fig. 3). Also qualitatively consistent with the psoralen assay were the low-temperature data, which indicate that the mutant complexes are more stable than those formed in a WT extract at low temperature. There is, however, a substantial stability difference between the U1C-depleted and the L13 extracts at low temperature. Taken together with what appear to be complex stability curves (a mixture of more stable and less stable complexes), we interpret the data to indicate that the relatively stable component reflects the low-temperature base-paired complexes measured by psoralen (U1C > L13> WT), whereas the less stable component reflects complexes that lack normal base-pairing. This finding suggests further that the base-paired complexes in the mutant extracts are considerably more stable than the predominantly nonbased complexes formed in the WT extract at low temperature. Indeed, we attribute quantitative differences between the immunoprecipitation and the psoralen assays to the fact that the immunoprecipitation assay measures total complex formation (base-paired and nonbase-paired), whereas the psoralen assay only scores complexes containing canonical U1 snRNA–pre-mRNA base-pairing.
Fig. 3.
Stability of the RNA–protein complexes formed in three different extracts at 25°C (Left) and 0°C (Right). Commitment complex formation and chase assays were carried out as described in Materials and Methods. Complex stability was measured by liquid scintillation counter after immunoprecipitation with an antibody against Prp40. The value obtained after a 20-min incubation without unlabeled RNA was arbitrarily set to 100. Values were averaged from at least four experiments.
To address more generally protein–pre-mRNA contacts within commitment complex (i.e., not only protein contacts in complexes that also contain RNA–RNA duplexes), we used a standard 4-thio RNA–protein cross-linking assay at both high and low temperatures. We also subjected these complexes to the chase protocol as described above.
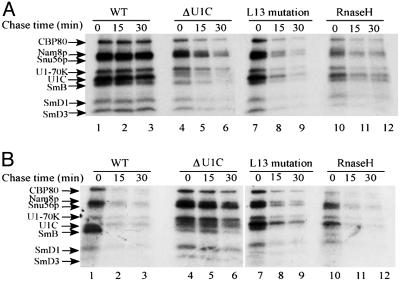
The cross-linking patterns observed were identical to what has been previously described for commitment complexes (9), i.e., all eight commitment complex components were detectable in all extracts and at both temperatures; the only exception is the U1C band, which is very faint in the U1C-depleted extract (Fig. 4). At room temperature (Fig. 4A), the relative band intensities between the three extracts (WT, U1C-depleted, and L13) were similar to what was observed with psoralen cross-linking (Fig. 2): strong signals in the WT extract and intermediate signals in the two mutant extracts. The RNase H extract (U1 snRNP treated with an oligonucleotide and RNase H to remove the 5′ end) served a control and indicated that most of the signal in the WT extract (Fig. 4, lanes 1–3 vs. lanes 10–12) reflected base-paired complexes as expected. The WT complexes also were highly stable, as evidenced by the failure to chase during incubation in the presence of competitor RNA, similar to what was observed in the psoralen chase experiment (Fig. 2). In contrast, the intermediate level signals in the mutant extracts were highly unstable (Fig. 4A, lanes 4–9), presumably reflecting the unstable nature of both the base-paired complexes and nonbase-paired complexes formed at room temperature in these extracts (Fig. 2).
Fig. 4.
Stability comparisons of RNA–protein interactions in the three extracts at two different incubation temperatures: 25°C(A) and 0°C(B). 4-Thio-UTP-labeled 32P WT-72 RNA (9) was used as a substrate for protein UV cross-linking. Results for the WT and RNase-H-treated extract at 25°C were as described (9, 20, 25). Previously identified proteins are indicated on the left.
At low temperature, lower-band intensities were detected in the WT extract (Fig. 4B). Comparable intensities were detected in the truncated-U1 snRNA extract (RNase H; Fig. 4B, compare lanes 1 and 10). This finding is consistent with the results shown above, namely, the absence of detectable psoralen cross-linking (base-pairing) in WT extracts at low temperature (Fig. 2), and previous results showing that the predominant interactions at low temperature are between U1 snRNP proteins and the single-stranded 5′ss region (25). These interactions are also unstable, as they were chased by incubation in the presence of cold competitor RNA (Fig. 4B, lanes 1–3 and 10–12), consistent with the chase experiment shown in Fig. 3. More prominent low-temperature bands were observed with the L13 and U1C-depleted extracts (Fig. 4B, lanes 4 and 7), consistent with the psoralen cross-linking and total complex measurements (Fig. 3). The U1C-depleted bands were also quite stable, compared with the L13 bands and especially compared with the WT bands (Fig. 4B). This finding also fits with the greater amount of signal in the psoralen cross-linking assay with the U1C-depleted extract (Figs. 1 and 2), suggesting that much of the low-temperature protein–RNA (4-thio-UTP) cross-linking signal in this extract was from base-paired complexes. These interactions were somewhat less stable in the L13 mutant extract (Fig. 4B, compare lanes 4–6 and 7–9), probably reflecting residual U1C activity and a lower fraction of base-paired complexes relative to the U1C-depleted extract.
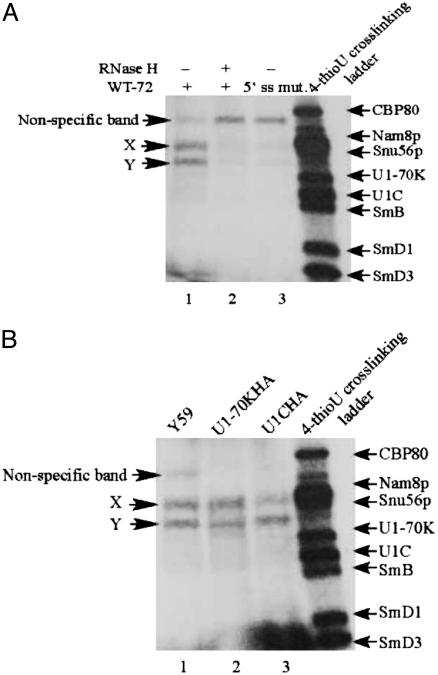
The dramatic stability difference between the WT and mutant complexes formed at room temperature indicates that U1C makes an important contribution to RNA duplex stabilization under these standard conditions. However, our published data are best explained by an interaction between U1C and single-stranded RNA (25), suggesting that U1C does not stabilize the duplex by contacting the U1 snRNA–pre-mRNA base pairs. To address more directly a U1C–duplex interaction, we performed a MB-mediated cross-linking assay (see Materials and Methods). MB can bind to double-stranded RNA, is highly photoreactive to visible light, and has been used to visualize proteins that contact directly double-stranded RNA (29).
With this assay, we observed three labeled proteins (Fig. 5A, lane 1). Two passed stringent specificity tests: they were both eliminated by truncation of the 5′ end of U1 snRNA with oligo-directed RNase H digestion (Fig. 5A, compare lanes 1 and 2), and they were prominently reduced when a mutated 5′ss instead of a WT 5′ss was used as a radioactive substrate (Fig. 5A, lane 3). This finding indicates that these two proteins bind to a bona fide U1 snRNA–5′ss duplex. Their apparent molecular weights indicate that neither of them is U1C or U1–70K (also implicated in 5′ss stabilization; ref. 30). To confirm this conclusion, we repeated the cross-linking with extracts containing hemagglutinin (HA)-tagged versions of U1C (U1CHA) or U1-70K (U1-70KHA). If a band was caused by one of these two proteins, it should disappear from its WT position and migrate with a lower mobility (9). There was no mobility change in either extract, consistent with the conclusion that neither band is U1C (or U1–70K). Although these data are negative, the absence of a band with the molecular weight of U1C suggests that it does not contact directly the U1 snRNA–5′ss duplex.
Fig. 5.
MB-mediated cross-linking to detect protein–double-stranded RNA interactions. (A) WT extract. (B) Comparison of WT and tagged extracts.32P-labeled transcripts were incubated with splicing extracts in standard commitment complex reactions for 20 min at 25°C. MB cross-linking was then carried out as described in Materials and Methods. Protein size ladder was the profile of WT-72 4-thio-UTP UV cross-linking. The intensity of a large nonspecific band was increased when that of the two specific bands was decreased (compare lane 1 with lanes 2 and 3 in A).
There is one additional complication that limits the strength of this conclusion: we found the identical pattern at low temperature, even with a WT extract (data not shown). This complication may indicate that MB is not entirely specific for protein contacts with double-stranded RNA. A simpler explanation reflects the fact that the samples were somewhat warmed by the fluorescent light bulb (data not shown). A third possibility is that canonical base-pairing exists at low temperature, despite the absence of a signal by psoralen cross-linking. MB but not psoralen may capture a small fraction of RNA–RNA base-pairing, which even at low temperatures is in dynamic equilibrium with protein–single-stranded RNA.
Discussion
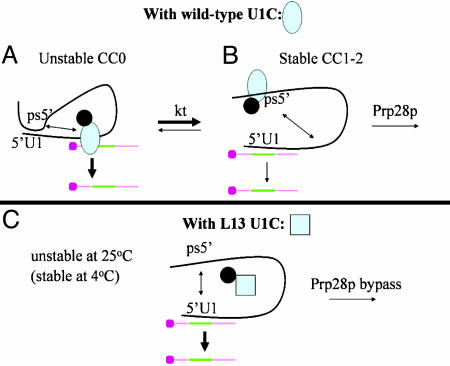
Based on these data and all of our previous reports (20, 25), we propose the following scheme: The first interaction between a single-stranded 5′ss and U1 snRNP is with U1C, which prevents base-pairing between the 5′ ss and the 5′ end of U1 snRNA (Fig. 6A, CC0). We suggest that U1C then aids a temperature-dependent conformational change, from this unstable nonbase-paired CC0 conformation to the canonical stable base-paired configuration (Fig. 6B, CC1-2). The splicing factor Prp28p then acts as an RNA helix to unwind the U1 snRNA–5′ss duplex or functions as an RNP remodeling enzyme (31) with an indirect destabilizing effect on this duplex. In either case, Prp28p activity allows subsequent steps of spliceosome assembly and the formation of the U6 snRNA–5′ss base-pairing interaction. These proposed events could equally well occur within a tetra-snRNP or penta-snRNP, if they turn out to reflect conformational changes rather than bona fide spliceosome assembly steps in vivo (15, 32).
Fig. 6.
Working model for the role of yeast U1C during in vitro commitment complex formation. Protein–protein interactions contribute to retaining WT U1C (blue oval, A and B; blue square, C) within U1 snRNP. The black ball is a putative U1 snRNP protein, which helps retain U1C within the snRNP when U1C interactions with U1 snRNA are weak or negligible (e.g., in A because of an RNA conformational change or in C because of a U1C protein conformational change). The colored bar is the pre-mRNA, with the intron in green. (A) There is little U1C binding to U1 RNA at low temperature, because of a favored RNA–RNA interaction, depicted as an intramolecular base-pairing interaction between a putative ps5′ and the 5′ss. Under these conditions, the 5′ substrate interaction is of low stability (indicated by a thick arrow in A) and predominantly with U1 snRNP proteins including U1C. (B) At higher temperatures (indicated by kt), there is a conformational change (indicated by a double arrow in A) that includes U1C binding to the ps5′. This process liberates the 5′ss and allows a base-pairing interaction with the pre-mRNA 5′ss (indicated by thin lines), which gives rise to a more stable interaction. (C) The L13 mutation causes a U1C conformational change (blue square rather than blue oval), which binds poorly to RNA. U1 RNA therefore has a tendency to adopt the low-temperature conformation as in A, which allows the ps5′ to interact with the 5′ end of U1 (double arrow) and compete with pre-mRNA base-pairing. This process gives rise to a lower stability base-pairing interaction (thick arrow).
The conformational change (CC0 > CC1-2) is proposed in part because almost no U1 snRNA–5′ss base-pairing is detectable at low temperature in a WT extract. Therefore, CC0 or something similar must be the predominant conformation at low temperature, whereas CC1-2 is the predominant conformation at high temperature. Although the absence of base-pairing at low temperature could be explained by the U1C–5′ss interaction and an energy requirement for the conformational change, it is tempting to suggest that some RNA sequence or structure within the large yeast U1 snRNA (33, 34) also contributes to the inhibition of base-pairing at low temperature. This inhibitory sequence or structure could be quite complex and/or the effect quite indirect, but for the sake of simplicity we have depicted it as a pseudo-5′ss sequence (ps5′) (Fig. 6) that can form intramolecular base pairs with the 5′ end of U1 snRNA (Fig. 6, 5′U1). This putative autoinhibitory interaction is mutually exclusive with canonical base-pairing between the 5′ end of U1 snRNA and the 5′ss. The data suggest that the intramolecular inhibitory interaction predominates at low temperature, which restricts the substrate 5′ss to a low stability interaction with U1C (CC0, Fig. 6A). At higher temperatures, the alternative conformation then allows standard 5′ss–U1 snRNA base-pairing (CC1-2; Fig. 6B).
A reason for the explicit suggestion of a ps5′ is the preference of U1C for single-stranded RNA with a 5′ss-like sequence (25). As a consequence, U1C could bind directly to a ps5′-like sequence and inhibit its action at normal temperatures. The notion is then that U1C switches RNAs during early splicing complex formation, from the substrate 5′ss to the U1 ps5′, and thereby contributes to the conformational change from CC0 to CC1-2 (Fig. 6). The intramolecular base-pairing would predominate at low temperature, whereas the U1C-U1 ps5′ interaction would win at higher temperatures. We note that there is an excellent candidate pseudo-5′ss at positions 420–427 with perfect complementarity to the 5′ end of U1 snRNA; the last six of these eight nucleotides are in a proposed single-stranded loop (loop Xc; ref. 35). Preliminary experiments indicate that there is enhanced low-temperature base-pairing when this complementary sequence is altered (data not shown).
A second reason for postulating the existence of a pseudo-5′ss sequence or structure is the low stability of complexes formed at room temperature in extracts containing the L13 mutation of U1C. Although a blocking model can explain the effects of the L13 mutation at low temperature (the potent 5′ss–U1C interaction normally inhibits the canonical 5′ss–U1snRNA interaction; Fig. 6, CC0; see below), this idea does not explain why the room temperature L13 base-paired complexes are of low stability, similar to those formed in the U1C-depleted extract and much different from WT complexes formed under these standard conditions (Figs. 2, 3, 4). Although psoralen does not provide a high-resolution picture of the base-pairing, we presume that it is quasi-normal in the L13 extracts (see below). If the L13 mutant U1C binds RNA less well than WT U1C, this weakened interaction would liberate the ps5′ or structure and allow it to compete with the 5′ss for base-pairing to the 5′ end of U1 snRNA. The low stability would then result from this cis competition (Fig. 6C). In any case, we suggest that the low stability of the L13 base-paired complex is the reason that this mutation can bypass the need for Prp28p, consistent with the original suggestion of Chen et al. (26).
In contrast, the L13 mutant extracts manifest stable and permissive base-pairing phenotypes at low temperatures (Figs. 2, 3, 4). The stability can explain why the bypass strains are cold sensitive (26). The permissive feature of the phenotype presumably reflects a failure to sequester the 5′ss (poor substrate RNA binding) and/or a failure to form the proper inhibitory CC0 conformation at low temperature (Fig. 6C). In any case, both phenotypes are similar to a U1C-depletion extract and indicate that the L13 mutation is a loss-of-function mutation.
A key mechanistic question remaining is how does WT U1C activity promote canonical commitment complex formation? Although we have not measured on rates systematically, our preliminary results suggest that there is no dramatic difference between WT and U1C-depleted extracts (data not shown). Moreover, the mutant complexes have dramatically increased off rates at room temperature relative to WT complexes (Figs. 3 and 4). As there are only modest differences in total complex levels between mutant and WT U1 snRNP, they can be easily explained by differences in complex stability (Figs. 3 and 4). Although this increased off rate makes duplex stabilization a major consequence of U1C activity, it does not distinguish between a direct or an indirect role. Moreover, we have been unable to detect U1C directly by contacting the base-paired region by MB cross-linking (Fig. 5). It is, of course, possible that U1C contacts the duplex region in a MB-undetectable manner (see above). Although not a major focus, we also have not detected striking differences in other protein–5′ss region contacts between stable and unstable complexes (Fig. 4). Taken together, our experiments suggest that U1C contributes only indirectly to stable complex formation. As described above, our interpretation includes sequestering an inhibitory region of U1 snRNA and promoting an intramolecular conformational change.
The temperature requirement recalls experiments on ribosome assembly by Nomura (36) more than 30 years ago. In this system, there is evidence for an energetic barrier to stable complex formation, reflecting a conformational change that requires incubation at elevated temperature. For commitment complex formation, we suggest that elevated temperatures are required to displace U1C from the single-stranded 5′ss, melt the intramolecular RNA–RNA interactions, and promote U1C binding to an autoinhibitory region, perhaps a ps5′. In this view, yeast U1C might act as a dedicated intramolecular chaperone to promote stable base-pairing, as proposed (24).
In HeLa cell nuclear extracts, the U1 snRNA–5′ss interaction is reported to be identical at both high and low temperatures (37), indicating that yeast U1C and human U1C may have somewhat different activities. Although the region of yeast U1C that contacts RNA has not been definitively identified, preliminary chemical mapping experiments suggest that the U1C basic C-terminal tail contacts the 5′ss (unpublished data and e.g., ref. 38). Given the longer tail of yeast U1C and the substantial sequence divergence between yeast and vertebrate U1C in this region (24), the yeast protein may indeed have an additional activity not present in metazoan U1C. This observation also suggests that the conserved N-terminal zinc finger-like structure that contains the L13 mutation may participate only indirectly in RNA binding, or perhaps has another biochemical function.
A somewhat different, perhaps additional function for yeast U1C mirrors the much longer yeast U1 snRNA molecule, 568 vs. 164 nt for humans (33, 34). This size difference may even be related to temperature regulation, which might be much more important for yeast splicing than for many metazoan species. In any case, the putative conformational changes and the effects of temperature should be testable by structure probing yeast U1 snRNA within commitment complexes under different conditions and in different genetic backgrounds (e.g., ref. 35).
Acknowledgments
We thank T. H. Chang for the U1C L13 mutant strain used in this work; K. Dower, K. Abruzzi, and other laboratory members for helpful suggestions on the work and the manuscript; and T. H. Chang, C. Query, and T. Nilsen for comments on the manuscript. This work was supported by National Institutes of Health Grant GM23549.
Author contributions: H.D., D.F.T., and M.R. designed research; H.D. and D.F.T. performed research; H.D., D.F.T., and M.J.M. analyzed data; and H.D. and M.R. wrote the paper.
Abbreviations: snRNP, small nuclear ribonucleoprotein particle; 5′ss, 5′ splice site; ps5′, pseudo-5′ss sequence; snRNA, small nuclear RNA; MB, methylene blue; 4-thio, 4-thiouridine.
References
- 1.Moore, M. J., Query, C. C. & Sharp, P. A. (1993) in The RNA World, eds. Gesteland, R. F. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 303–357.
- 2.Will, C. L. & Luhrmann, R. (1997) Curr. Opin. Cell Biol. 9, 320–328. [DOI] [PubMed] [Google Scholar]
- 3.Burge, C. B., Tuschl, T. & Sharp, P. A. (1999) in The RNA World II, eds. Gesteland, R. R., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 525–560.
- 4.Hartmuth, K., Urlaub, H., Vornlocher, H. P., Will, C. L., Gentzel, M., Wilm, M. & Luhrmann, R. (2002) Proc. Natl. Acad. Sci. USA 99, 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurica, M. S., Licklider, L. J., Gygi, S. R., Grigorieff, N. & Moore, M. J. (2002) RNA 8, 426–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makarov, E. M., Makarova, O. V., Urlaub, H., Gentzel, M., Will, C. L., Wilm, M. & Luhrmann, R. (2002) Science 298, 2205–2208. [DOI] [PubMed] [Google Scholar]
- 7.Rappsilber, J., Ryder, U., Lamond, A. I. & Mann, M. (2002) Genome Res. 12, 1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jurica, M. S. & Moore, M. J. (2003) Mol. Cell 12, 5–14. [DOI] [PubMed] [Google Scholar]
- 9.Zhang, D. & Rosbash, M. (1999) Genes Dev. 13, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puig, O., Gottschalk, A., Fabrizio, P. & Seraphin, B. (1999) Genes Dev. 13, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seraphin, B. & Rosbash, M. (1989) Cell 59, 349–358. [DOI] [PubMed] [Google Scholar]
- 12.Michaud, S. & Reed, R. (1991) Genes Dev. 5, 2534–2546. [DOI] [PubMed] [Google Scholar]
- 13.Neugebauer, K. M. (2002) J. Cell Sci. 115, 3865–3871. [DOI] [PubMed] [Google Scholar]
- 14.Kotovic, K. M., Lockshon, D., Boric, L. & Neugebauer, K. M. (2003) Mol. Cell. Biol. 23, 5768–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevens, S. W., Ryan, D. E., Ge, H. Y., Moore, R. E., Young, M. K., Lee, T. D. & Abelson, J. (2002) Mol. Cell 9, 31–44. [DOI] [PubMed] [Google Scholar]
- 16.Rosbash, M. & Séraphin, B. (1991) Trends Biol. Sci. 16, 187–190. [DOI] [PubMed] [Google Scholar]
- 17.Zhuang, Y. & Weiner, A. M. (1986) Cell 46, 827–835. [DOI] [PubMed] [Google Scholar]
- 18.Seraphin, B., Kretzner, L. & Rosbash, M. (1988) EMBO J. 7, 2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siliciano, P. G. & Guthrie, C. (1988) Genes Dev. 2, 1258–1267. [DOI] [PubMed] [Google Scholar]
- 20.Du, H. & Rosbash, M. (2001) RNA 7, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lund, M. & Kjems, J. (2002) RNA 8, 166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinrichs, V., Bach, M., Winkelmann, G. & Luhrmann, R. (1990) Science 247, 69–72. [DOI] [PubMed] [Google Scholar]
- 23.Will, C. L., Rumpler, S., Klein Gunnewiek, J., van Venrooij, W. J. & Luhrmann, R. (1996) Nucleic Acids Res. 24, 4614–4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang, J., Abovich, N., Fleming, M. L., Seraphin, B. & Rosbash, M. (1997) EMBO J. 16, 4082–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du, H. & Rosbash, M. (2002) Nature 419, 86–90. [DOI] [PubMed] [Google Scholar]
- 26.Chen, J. Y., Stands, L., Staley, J. P., Jackups, R. R., Latus, L. J. & Chang, T. (2001) Mol. Cell 7, 227–232. [DOI] [PubMed] [Google Scholar]
- 27.Staley, J. P. & Guthrie, C. (1999) Mol. Cell 3, 55–64. [DOI] [PubMed] [Google Scholar]
- 28.Umen, J. G. & Guthrie, C. (1995) Genes Dev. 9, 855–868. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Z. R., Sargueil, B. & Smith, C. W. (2000) Methods Enzymol. 318, 22–33. [DOI] [PubMed] [Google Scholar]
- 30.Kohtz, J. D., Jamison, S. F., Will, C. L., Zuo, P., Luhrmann, R., Garcia-Blanco, M. A. & Manley, J. L. (1994) Nature 368, 119–124. [DOI] [PubMed] [Google Scholar]
- 31.Fairman, M. E., Maroney, P. A., Wang, W., Bowers, H. A., Gollnick, P., Nilsen, T. W. & Jankowsky, E. (2004) Science 304, 730–734. [DOI] [PubMed] [Google Scholar]
- 32.Gottschalk, A., Neubauer, G., Banroques, J., Mann, M., Luhrmann, R. & Fabrizio, P. (1999) EMBO J. 18, 4535–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kretzner, L., Rymond, B. C. & Rosbash, M. (1987) Cell 50, 593–602. [DOI] [PubMed] [Google Scholar]
- 34.Siliciano, P. G., Jones, M. H. & Guthrie, C. (1987) Science 237, 1484–1487. [DOI] [PubMed] [Google Scholar]
- 35.Kretzner, L., Krol, A. & Rosbash, M. (1990) Proc. Natl. Acad. Sci. USA 87, 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nomura, M. (1973) Science 179, 864–873. [DOI] [PubMed] [Google Scholar]
- 37.Malca, H., Shomron, N. & Ast, G. (2003) Mol. Cell. Biol. 23, 3442–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, D., Abovich, N. & Rosbash, M. (2001) Mol. Cell 7, 319–329. [DOI] [PubMed] [Google Scholar]