Abstract
We determined the genomic sequence of Nocardia farcinica IFM 10152, a clinical isolate, and revealed the molecular basis of its versatility. The genome consists of a single circular chromosome of 6,021,225 bp with an average G+C content of 70.8% and two plasmids of 184,027 (pNF1) and 87,093 (pNF2) bp with average G+C contents of 67.2% and 68.4%, respectively. The chromosome encoded 5,674 putative protein-coding sequences, including many candidate genes for virulence and multidrug resistance as well as secondary metabolism. Analyses of paralogous protein families suggest that gene duplications have resulted in a bacterium that can survive not only in soil environments but also in animal tissues, resulting in disease.
Nocardia are filamentous-growing Gram-positive soil saprophytes that belong to the family Actinomycetales, which also includes clinically and industrially important genera such as Mycobacterium, Streptomyces, Corynebacterium, and Rhodococcus. Many species of Nocardia cause the disease nocardiosis in humans and animals on lung, central nervous system, brain, and cutaneous tissues (1), and a species Nocardia asteroides is suspected to be an etiological agent of Parkinson's disease (2). Nocardiosis is on the rise, with an estimated 109-136 new cases occurring annually in Japan (3) and 500-1,000 in the U.S. (www.cdc.gov/ncidod/dbmd/diseaseinfo/nocardiosis_t.htm). However, there are only a few studies on the mechanisms of nocardial virulence. Nocardia species are resistant to many front-line antibiotics. Because treatment for nocardiosis relies heavily on chemotherapy, their intrinsic multiple drug resistance is a serious problem.
Another feature of the nocardia is that many strains, even clinical isolates, also have the capability to produce bioactive molecules such as antibiotics (4, 5) and enzymes that are industrially important (6). A monobactam antibiotic, nocardicin, was isolated from Nocardia sp. (7), and a terpenoid brasilicardin with immunosuppressive activity was isolated from a clinical isolate (4).
Nocardia farcinica IFM 10152 was isolated from the bronchus of a 68-year-old male Japanese patient. Despite the complicated taxonomy of the nocardia, the species N. farcinica was found to be nearly homogeneous (8). Subsequently, to elucidate the molecular basis of the versatility of the nocardia, we analyzed the genomic sequence of N. farcinica IFM 10152.
Methods
Genome Sequencing and Assembly. The nucleotide sequence of the N. farcinica IFM 10152 genome was determined by a whole-genome shotgun strategy. We constructed small-insert (2 kb) and large-insert (10 kb) genomic libraries and generated 127,077 sequences (giving 9-fold coverage) from both ends of the genomic clones. Sequence assembly was carried out by using the phred/phrap/consed package (9). Remaining gaps were closed by transcriptional sequencing (Nippon Gene, Toyama, Japan) or by primer walking. There were no ambiguous nucleotides in the genomic sequence that we could determine. Consistency of the final assembly was confirmed in terms of restriction fragment patterns. Both AseI and DraI recognition sites estimated from the final assembly were in good agreement with experimental data (not shown). The accuracy of each base was reflected by its phrap score. In the final assembly, 99.85% of the sequence had an error rate of <1 per 10,000 bases (phrap score of ≥40). The number of rrn operons was confirmed by Southern hybridization (data not shown).
Genome Annotation and Analysis. Putative protein-coding sequences were predicted by the glimmer program and then were manually confirmed and corrected by using the blastp (10) and frameplot programs (11), optimized to handle genomic-size sequences. tRNA genes were predicted by the trnascan-se program (12). Sequences were analyzed by using the gcg wisconsin package (Accelrys) and other in-house programs. Clustering of proteomes was done by the blastclust program under the conditions of a minimum of 30% identity and 70% length coverage.
The fully annotated genomic sequence is also available at http://nocardia.nih.go.jp.
Results and Discussion
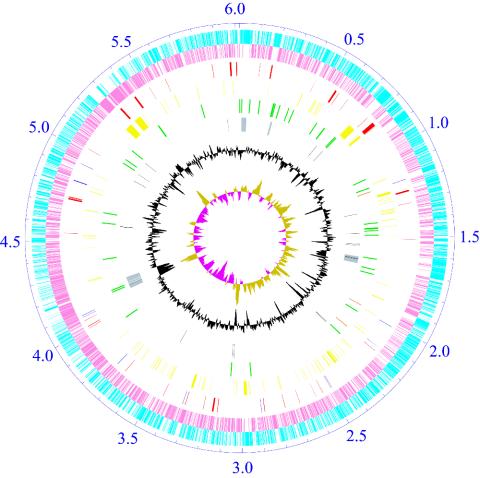
General Features. The chromosome of N. farcinica, unlike that of Streptomyces (13, 14), has a circular topology (Fig. 1) and encodes 53 tRNA genes, three copies of ribosomal RNA operons, and 5,674 predicted protein-coding sequences. Of these, 2,962 (52.2%) can be assigned a putative function, 1,532 (27.0%) matched to hypothetical proteins, and 1,180 (20.8%) have no database match with an E value of <10-10 (Table 1). The replication origin of the chromosome was detectable by the GC skew (15).
Fig. 1.
Schematic representation of the N. farcinica chromosome. The ticks show the scale in megabases, with zero representing the location of the dnaA gene. The outer two circles show the predicted protein-coding sequences on the plus (sky blue) and minus (pink) strands; the third shows the putative virulence (red) and drug-resistance (blue) genes; the fourth shows secondary metabolism genes (yellow); the fifth shows rrn operon (orange) and tRNA genes (green); the sixth shows phage-related (gray) and transposase (black) genes; the seventh shows percentage G+C in relation to mean G+C for the chromosome; and the eighth shows GC-skew (khaki, >0; purple, <0).
Table 1. General features of the genome.
| Chromosome | pNF1 | pNF2 | |
|---|---|---|---|
| Topology | Circular | Circular | Circular |
| Length, bp | 6,021,225 | 184,027 | 87,093 |
| G+C content, % | 70.8 | 67.2 | 68.4 |
| Copy no.* | 1 | 0.7 | 1.8 |
| Protein-coding gene | 5,674 | 160 | 90 |
| Average CDS length, bp | 960 | 963 | 832 |
| Coding density, % | 90.4 | 83.7 | 86.0 |
| Function assigned | 2,962 (52.2%) | 53 (33.1%) | 23 (25.6%) |
| Matched to hypothetical protein | 1,533 (27.0%) | 37 (23.1%) | 26 (28.9%) |
| No database match | 1,179 (20.8%) | 70 (43.8%) | 41 (45.5%) |
| rrn operon | 3 | 0 | 0 |
| tRNA gene | 53 | 0 | 0 |
CDS, protein-coding sequences.
The copy numbers of pNF1 and pNF2 were estimated from the statistical distribution of random reads between the plasmids and the chromosome.
Plasmids pNF1 (184,027 bp) and pNF2 (87,093 bp) were also found in this strain and encoded 160 and 90 predicted protein-coding sequences, respectively. Accurate partitioning of these two low-copy-number plasmids might be facilitated by the parA (pnf110 and pnf210) and parB (pnf120 and pnf220) genes.
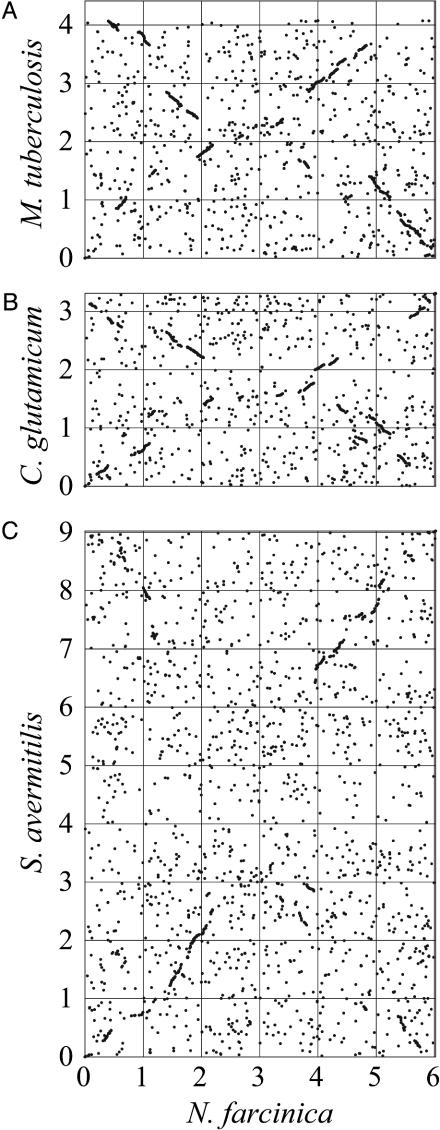
Comparative Study. To characterize the N. farcinica genome, we first compared it with the genomes of closely related bacteria. Ortholog plots indicate that Mycobacterium tuberculosis (16) and Corynebacterium glutamicum are most similar to N. farcinica, whereas Streptomyces avermitilis shows less similarity (Fig. 2). These results are consistent with the taxonomy of this group of bacteria and suggest that all four organisms are derived from a common ancestor. The “Broken-X” patterns indicate that inversions have occurred in the evolutionary process.
Fig. 2.
Ortholog plots of N. farcinica vs. M. tuberculosis (A), C. glutamicum (B), or S. avermitilis (C). Orthologs were identified as reciprocal best hits by using the blastp program. For each genome, the dnaA gene is located at position zero.
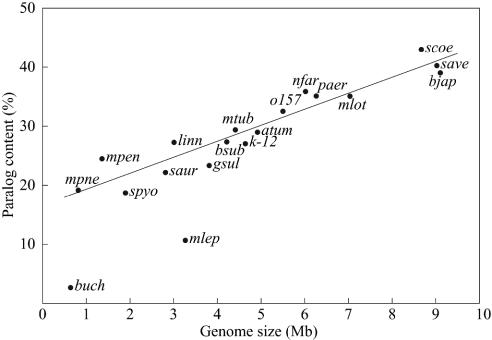
Next, we carried out clustering of all proteins encoded on the N. farcinica chromosome to identify paralogs. The result showed that 36% (2,035) were clustered into 552 paralogous families, with membership ranging from 2 to 86 proteins per family. In general, larger genomes have larger proportions of paralogs (Fig. 3), reflecting that gene duplication is one of the major driving forces for evolution. However, duplications do not occur equally for all genes; therefore, it may be possible to deduce direction in the evolution of genomes from analyzing the composition of paralogous families. As shown in Table 2, the typical soil bacterium S. avermitilis has a relatively large proportion of proteins to adapt to diverse soil environments. These include ABC transporters (families 1 and 16), two-component system proteins (families 4 and 20), and extracytoplasmic function σ factors (family 10). In contrast, the pathogen M. tuberculosis is rich in proteins for pathogenicity and intracellular growth such as PE/PPE/PGRS family proteins (family 5), mammalian cell entry (mce) family proteins (family 58), fibronectin-binding proteins (family 53), short-chain dehydrogenases (family 2), acyl-CoA synthetases (family 6), acyl-CoA dehydrogenases (family 8), enoyl-CoA hydratase/isomerase family proteins (family 11), and lipase/esterase family proteins (family 62). On the other hand, N. farcinica has a large proportion of all of the above families except for the PE/PPE/PGRS family proteins. This characteristic is consistent with the fact that N. farcinica inhabits both soils and animal tissues. These findings suggest that by increasing relevant paralogs, M. tuberculosis and S. avermitilis have evolved to be specialists in their own habitats, whereas N. farcinica has evolved from a common ancestor into a bacterium that can survive in a wider range of environments. The results also suggest that paralog analysis may provide additional clues about the molecular basis of the versatility of N. farcinica.
Fig. 3.
Correlation between paralog content and genome size in various bacteria: atum, Agrobacterium tumefaciens; bjap, Bradyrhizobium japonicum; bsub, Bacillus subtilis; buch, Buchnera sp.; o157, Escherichia coli O-157; k-12, E. coli K-12; gsul, Geobacter sulfurreducens; linn, Listeria innocua; mlot, Mesorhizobium loti; mtub, M. tuberculosis; mlep, Mycobacterium leprae; mpen, Mycoplasma penetrans; mpne, Mycoplasma pneumoniae; nfar, N. farcinica; paer, Pseudomonas aeruginosa; saur, Staphylococcus aureus; spyo, Streptococcus pyogenes; save, S. avermitilis; and scoe, S. coelicolor A3 (2). A linear regression line, which was estimated without using buch and mlep data, is also indicated. Extremely low paralogies in buch and mlep are due to massive decay of genes in these genomes.
Table 2. A selection of paralogous families in related genomes.
| Family no.
|
Occurrence, %
|
|||
|---|---|---|---|---|
| Function | N. farcinica | M. tuberculosis | S. avermitilis | |
| 1 | ABC transporter ATP-binding protein | 57 (1.00) | 21 (0.53) | 100 (1.32) |
| 2 | Short-chain dehydrogenase | 87 (1.53) | 44 (1.12) | 68 (0.90) |
| 3 | TetR transcriptional regulator | 75 (1.32) | 25 (0.64) | 51 (0.67) |
| 4 | Two-component response regulator | 29 (0.51) | 15 (0.38) | 64 (0.84) |
| 5 | PE_PGRS/PPE | 1 (0.02) | 115 (2.93) | 4 (0.05) |
| 6 | Acyl-CoA synthetase | 37 (0.65) | 21 (0.53) | 34 (0.45) |
| 7 | Dehydrogenase | 30 (0.53) | 13 (0.33) | 40 (0.53) |
| 8 | Acyl-CoA dehydrogenase | 40 (0.70) | 24 (0.61) | 25 (0.33) |
| 9 | Transporter | 26 (0.46) | 7 (0.18) | 44 (0.58) |
| 10 | ECF σ factor | 17 (0.30) | 9 (0.23) | 38 (0.50) |
| 11 | Enoyl-CoA hydratase/isomerase | 26 (0.46) | 22 (0.56) | 16 (0.21) |
| 12 | Aldehyde dehydrogenase | 23 (0.41) | 10 (0.25) | 21 (0.28) |
| 13 | Cytochrome P450 | 21 (0.37) | 12 (0.31) | 27 (0.36) |
| 14 | LysR transcriptional regulator | 20 (0.35) | 3 (0.08) | 27 (0.36) |
| 15 | Transporter | 4 (0.07) | 7 (0.18) | 33 (0.44) |
| 16 | ABC transporter ATP-binding protein | 15 (0.26) | 5 (0.13) | 19 (0.25) |
| 17 | Aminotransferase | 14 (0.25) | 7 (0.18) | 19 (0.25) |
| 18 | Serine/threonine protein kinase | 14 (0.25) | 8 (0.20) | 19 (0.25) |
| 19 | Oxidoreductase | 19 (0.33) | 12 (0.31) | 11 (0.15) |
| 20 | Two-component sensor kinase | 7 (0.12) | 7 (0.18) | 21 (0.28) |
| 23 | GntR transcriptional regulator | 11 (0.19) | 2 (0.05) | 23 (0.30) |
| 25 | Glycosyltransferase | 13 (0.23) | 7 (0.18) | 11 (0.15) |
| 46 | YrbE protein | 12 (0.21) | 8 (0.20) | 2 (0.03) |
| 53 | Fibronectin-binding protein | 12 (0.21) | 4 (0.10) | 0 (0.00) |
| 58 | MceB/C protein | 10 (0.18) | 8 (0.20) | 2 (0.03) |
| 62 | Lipase/esterase | 9 (0.16) | 8 (0.20) | 1 (0.01) |
Numbers in parentheses indicate the percentage of the total population in that genome.
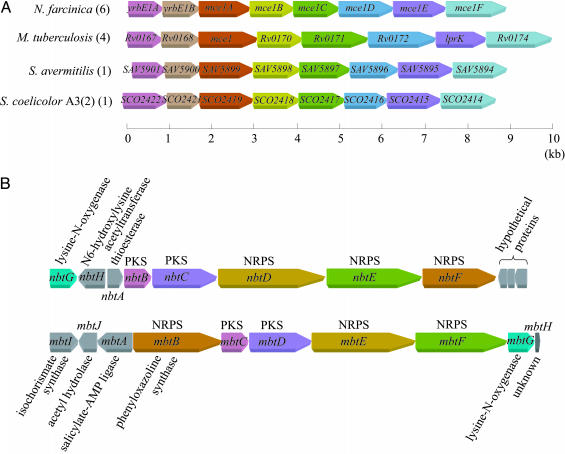
Virulence. Mce proteins are virulence factors of M. tuberculosis (17). There are four copies of the mce operon in M. tuberculosis (16), whereas six copies were identified in N. farcinica (Table 2, families 46 and 58). Mce operons are also found in other actinomycetes (Fig. 4A). S. avermitilis (13) and Streptomyces coelicolor A3 (2, 14), which are nonpathogenic soil bacteria, each have one copy of the mce operon, suggesting that these operons evolved from a common ancestral operon by gene duplication. Similarly, four genes homologous to mycobacterial mce genes (mce1, mce3, and mce4) were identified in an animal pathogen, Rhodococcus equi (18). These data strongly suggest that the multiple copies of mce operons contribute to the virulence of this group of bacteria. Antigen 85 family proteins of M. tuberculosis are known as fibronectin-binding proteins (19). M. tuberculosis has four proteins belonging to this family, whereas N. farcinica was found to have at least 12 proteins. All these proteins belong to the same paralogous family (Table 2, family 53).
Fig. 4.
Conservation of gene organization in actinomycetes. (A) Conservation of mce operons. Orthologous genes are denoted by the same color. Numbers of operons found in each genome are indicated in parentheses. (B) Comparison of gene organization between the putative siderophore (Upper) and the mycobactin (Lower) biosynthesis gene clusters. Orthologous and nonorthologous genes are denoted by the same color or in gray, respectively. Both clusters contain three nonribosomal peptide synthetases (NRPSs) and two polyketide synthases (PKSs).
In addition to the above virulence factors, many candidate genes for virulence were found along the N. farcinica chromosome. Adherence to and invasion into host cells by pathogens are important processes in establishing infection. Nfa34810 is a secreted protein of 49.8 kDa, of which the C-terminal part is highly homologous (72%) to mycobacterial invasins (20). The molecular weight of the mature Nfa34810 is in good agreement with the 43-kDa protein, which plays a dominant role in the adherence and invasion of pulmonary epithelial cells by N. asteroides GUH-2 (21). Nfa52080 is homologous (51%) to the heparin-binding hemagglutinin of M. tuberculosis and may play a role in extrapulmonary dissemination.
Four catalases (KatA, -B, -C, and -G), two superoxide dismutases (SodC and -F), and an alkylhydroperoxidase (AhpD) were also found. These probably act to protect against reactive oxygen species produced by the phagocyte. Although N. farcinica is an obligate aerobe, it contains nitrate reductase genes (narGHIJ and nirBD) for anaerobic respiration. This suggests that nocardia may also survive under low-oxygen conditions, such as in stimulated macrophages (22).
Because iron acquisition is a serious problem in animal tissues, many pathogenic bacteria produce siderophores to survive. The gene cluster from nbtA to -H was found to be very similar to the mycobactin biosynthetic cluster (23) in terms of sequence homology and gene organization, suggesting that the cluster directs biosynthesis of a mycobactin-related siderophore (Fig. 4B). This cluster includes genes coding for two polyketide synthases (NbtB and -C), three nonribosomal peptide synthetases (NbtD, -E, and -F), two lysine modification proteins (NbtG and -H), and a receptor protein (NbtI). Preliminary experiments showed that the cell extracts of N. farcinica have an iron-dependent cytotoxic activity (data not shown).
There were some examples of paralogous families found only in M. tuberculosis. Mycolic acids, very long-chain fatty acids, are unique cell envelope components and are important for the pathogenicity of Mycobacterium, Nocardia, and related genera. In M. tuberculosis, two 3-oxoacyl-acyl carrier protein synthases, KasA and -B, elongate long-chain acyl primers and generate mycolic acids (24). However, only KasA was identified in N. farcinica, suggesting shorter chain lengths (50-60 carbons) of nocardial mycolic acids than those (60-90 carbons) of mycobacterial ones. N. farcinica substantially lacked the PE/PPE/PGRS family proteins (Table 2, family 5), which might confer antigenic variation, and some of which were experimentally shown to be involved in the pathogenicity of M. tuberculosis (25). Their absence might explain why N. farcinica is less infectious than M. tuberculosis.
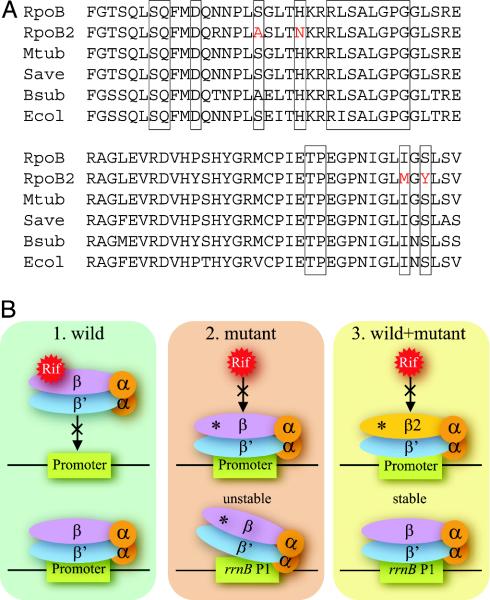
Drug Resistance. Gene duplication may be responsible for the drug resistance of N. farcinica. Surprisingly, the genome possessed two genes for the β subunit of RNA polymerase (RNAP), designated rpoB and rpoB2. To our knowledge, this is the first case of two rpoB genes being present in one bacterial genome. Comparison with other bacterial RpoB proteins suggested that the nocardial RpoB protein is sensitive to rifampin. In contrast, the RpoB2 protein contained substitutions at positions that could convert many bacteria to be rifampin resistant (26) (Fig. 5A). These findings strongly suggest that rpoB2 accounts for the rifampin resistance of this bacterium. It has been shown that mutations in the rpoB gene produce additional effects. RNAPs with rifampin-resistant RpoB show an increase or decrease in the expression of genes controlled by a stringent promoter (27). Therefore, the acquisition of rifampin resistance by the duplication of the rpoB gene may be an elaborate strategy that minimizes the side effects of the rpoB mutation (Fig. 5B). This hypothesis is supported by the fact that the rpoB2 gene is widely distributed among N. farcinica strains and is transcribed in the presence of rifampin (unpublished data).
Fig. 5.
(A) Comparison of the rifampin regions of RNAP β subunits among N. farcinica (RpoB and RpoB2), M. tuberculosis (Mtub), S. avermitilis (Save), B. subtilis (Bsub), and E. coli (Ecol). Boxed regions indicate known positions where mutations lead to rifampin resistance in various bacteria. Substitutions found in the RpoB2 are red-colored. (B) Possible advantage of two types of rpoB genes being present in one genome. 1, Rifampin binds to wild-type RNAP and inhibits its function. 2, Certain mutations (asterisks) on the rpoB gene diminish the binding of rifampin to RNAP, making bacteria resistant in the presence of rifampin. However, rpoB mutants destabilize initiation complexes between RNAP and stringently controlled promoters, for example rrnB P1, even in the absence of rifampin. 3, If both types of rpoB genes are present in one genome, such side effects disappear in the absence of rifampin by the wild-type molecule, and bacteria can survive in the presence of rifampin by the mutant molecule.
We found many other drug-resistance genes on the chromosome (Table 3). IFM 10152 was found to be resistant to at least 18 β-lactam antibiotics. Many of these are a substrate of a β-lactamase (Nfa23080) identical to FAR-1 (28). However, Nfa23080 alone is not sufficient to cause the β-lactam resistance spectra exhibited by IFM 10152, suggesting the existence of other resistance determinants. Actually, two putative β-lactamases (Nfa9770 and Nfa48460) were also found in the genome. IFM 10152 contains three aminoglycoside phosphotransferase (APH) enzymes: nfa38480 encoding an APH(3′), nfa31340 encoding an APH(2″), and nfa38620 encoding an APH(6), leading to resistance to kanamycin, gentamicin, and streptomycin, respectively. From their deduced amino acid sequences, MurA encodes UDP-N-acetylglucosamine 1-carboxyvinyltransferase, and FolP encodes dihydropteroate synthase, which would make IFM 10152 naturally resistant to fosfomycin and sulfamethoxazole, respectively. Ribosomal RNA methyltransferases (Nfa27210 and Nfa47240) conferring resistance to macrolides, and a chloramphenicol efflux ABC transporter (Nfa18410), as well as many other potential drug efflux transporters, were also found.
Table 3. Drug-resistance profile and putative resistance determinants of N. farcinica IFM 10152.
| Drug | MIC, μg/ml | Putative resistance determinant (protein identification) |
|---|---|---|
| Ampicillin | 32 | β-lactamase (Nfa23080, Nfa 19770, and Nfa48460) |
| Aztreonam | >32 | |
| Ceftazidime | >32 | |
| Cefaclor | >32 | |
| Cefditoren | 4 | |
| Cefazolin | >32 | |
| Cefmetazone | 16 | |
| Cefotiam | 8 | |
| Ceftriaxone | 4 | |
| Cefotaxime | 4 | |
| Ceftizoxime | >32 | |
| Flomoxef | 4 | |
| Faropenem | 16 | |
| Latamoxef | >32 | |
| Meropenem | 2 | |
| Penicillin | >8 | |
| Piperacillin | >64 | |
| Imipenem | 0.25 | |
| Arbekacin | <0.25 | |
| Amikacin | <1 | |
| Gentamicin | >16 | APH(2″) (Nfa31340) |
| Kanamycin | 64 | APH(3′) (Nfa38480) |
| Streptomycin | 16 | APH(6) (Nfa38620) |
| Clarithromycin | 4 | rRNA methyltransferase (Nfa27210 and Nfa47240) |
| Erythromycin | >16 | |
| Clindamycin | 16 | |
| Ofloxacin | <0.5 | |
| Tosufloxacin | 0.25 | |
| Chloramphenicol | 64 | Efflux pump (Nfa 18410) |
| Fosfomycin | >32 | MurA (Nfa 10690) |
| Sulfamethoxazole | 19 | FoIP (Nfa4000) |
| Trimethoprim | 1 | |
| Rifampin | >16 | Monooxygenase (Nfa35380) and RpoB2 (Nfa46460) |
| Minocycline | 2 |
MIC, minimum inhibitory concentration.
Secondary Metabolism. Paralogs also likely contribute to the diverse metabolic capabilities of N. farcinica. The genome contains at least 103 oxygenases, 81 of which were found to form only 19 paralogous families. This suggests that N. farcinica may have a higher metabolic potential than Pseudomonas putida KT2440, which is known to have a high metabolic potential and contains 37 oxygenases (29). These oxygenases included homologues to ring-cleavage dioxygenase (Nfa4720 and Nfa40980), 2-nitropropane dioxygenase (Nfa5070, Nfa7150, Nfa30840, Nfa34970, Nfa55800, and Nfa55910), alkane 1-monooxygenase (Nfa33210, Nfa46140, and Nfa46180), steroid terminal dioxygenase (Nfa22480), dihydroxyphenylacetate dioxygenase (Nfa27760), and styrene monooxygenase (Nfa12190, Nfa32440, and Nfa32460). Of 103 oxygenases, 27 are putative cytochrome P450 monooxygenases (P450s). Some may be involved in fatty acid metabolism (e.g., Nfa5180), whereas the rest may play a role in defense against toxic compounds in soil or host organisms. The cyp51 gene probably encodes a 14α-sterol demethylase which is a possible target of azoles (30). Indeed, IFM 10152 is susceptible to clotrimazole, ketoconazole, and miconazole (unpublished data). The sequence data therefore suggest that the sterol pathway may become a target for novel drug development in nocardial infection.
Nocardia species are also known to have the ability to produce bioactive molecules. Most eukaryotes synthesize isoprenoids through the mevalonate pathway, but many bacteria, including pathogens, and plants use the “nonmevalonate pathway.” N. farcinica possesses both pathways. Genes for the mevalonate pathway of N. farcinica are clustered (nfa22070-nfa22120) and probably form an operon with four upstream genes (nfa22030-nfa22060), including isoprenoid-related genes. The operon may direct the biosynthesis of a novel terpenoid with biological activity. The genome possessed 14 nonribosomal peptide synthetase genes, including nfa50330 encoding a huge single polypeptide (1,556 kDa). Three of them (nbtD-nbtF) are located in the siderophore cluster, and four (nfa7170-nfa7200) are clustered at other regions. Although the function of these genes is not clear except for nbtD-nbtF, some of them are possibly involved in the production of novel peptides with biological activity, such as thiazolyl peptide antibiotics previously isolated from Nocardia sp. (31).
Conclusion
The present study suggests that paralogs may contribute not only to the pathogenic and saprophytic features of N. farcinica but also to its versatility. What makes nocardia virulent? The purpose of much of the study discussed here is to help answer this question. Although unfortunately there is no simple answer yet, many potential virulence factors found in this study would greatly facilitate the understanding of the pathogenicity of nocardia. Furthermore, the many similarities to the M. tuberculosis genome should help us understand tuberculosis. N. farcinica is naturally resistant to many antibiotics. The hydrophobic nature of the nocardial cell envelope is believed to act as a permeability barrier and makes nocardia resistant to many drugs, as is mycobacteria. However, the present study reveals that N. farcinica has drug-resistance mechanisms more sophisticated than was previously thought. This finding should help to improve chemotherapy regiments for nocardiosis. None of the drug-resistance genes in N. farcinica were brought in by mobile elements, because no adjacent transposons, phages, or differences in G+C content were found. Although it is still unknown how N. farcinica evolved so many drug-resistance genes without horizontal gene transfer, gene duplication, in part, played a role in the emergence of drug resistance, as shown in the rpoB gene duplication conferring rifampin resistance. Certain rifampin-resistance mutations in rpoB gene have been shown to lead to elevated antibiotic production in Streptomyces (32). This suggests that the rpoB2 gene may also affect the secondary metabolism of N. farcinica. Further analysis of the sequence may provide many other insights into the molecular basis of the versatility of this bacterium.
Acknowledgments
We thank K. Oshima, K. Furuya, C. Yoshino, Y. Yamashita, and A. Nakazawa for technical assistance; and N. Summers, K. Summers, and T. Taylor for help with the text. This study was funded by the Research for the Future Program of the Japan Society for the Promotion of Science (no. 00L01411).
Author contributions: J.I., K.H., M.H., and T.S. designed research; J.I., A.Y., Y.M., Y.H., and H.K. performed research; J.I. analyzed data; J.I. wrote the paper; and Y.M. provided the strains used.
Abbreviations: mce, mammalian cell entry; RNAP, RNA polymerase.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. AP006618-AP006620).
References
- 1.Brown, J. M., McNeil, M. M. & Desmond, E. P. (1999) in Manual of Clinical Microbiology, eds. Murray, P. R., Baron, E. J., Pfaller, M. A., Tenover, F. C. & Yolken, R. H. (Am. Soc. Microbiol., Washington, DC), pp. 370-398.
- 2.Hubble, J. P., Cao, T. Kjelstrom, J. A., Koller, W. C. & Beaman, B. L. (1995) J. Clin. Microbiol. 33, 2768-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kageyama, A., Yazawa, K., Ishikawa, J., Hotta, K., Nishimura, K. & Mikami, Y. (2004) Eur. J. Epidemiol. 19, 383-389. [DOI] [PubMed] [Google Scholar]
- 4.Shigemori, H., Komaki, H., Yazawa, K., Mikami, Y., Nemoto, A., Tanaka, Y., Sasaki, Y., Ishida, T. & Kobayashi, J. (1998) J. Org. Chem. 63, 6900-6904. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka, Y., Komaki, H., Yazawa, K., Mikami, Y., Nemoto, A., Tojyo, T., Kadowaki, K., Shigemori, H. & Kobayashi, J. (1997) J. Antibiot. 50, 1036-1041. [DOI] [PubMed] [Google Scholar]
- 6.Coco, W. M., Levinson, W. E., Crist, M. J., Hektor, H. J., Darzins, A., Pienkos, P. T., Squires, C. H. & Monticello, D. J. (2001) Nat. Biotechnol. 19, 354-359. [DOI] [PubMed] [Google Scholar]
- 7.Hashimoto, M., Komori, T. & Kamiya, T. (1976) J. Am. Chem. Soc. 12, 3023-3025. [DOI] [PubMed] [Google Scholar]
- 8.Laurent, F., Carlotti, A., Boiron, P., Villard, J. & Freney, J. (1996) J. Clin. Microbiol. 34, 1079-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon, D., Desmarais, C. & Green, P. (2001) Genome Res. 11, 614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishikawa, J. & Hotta, K. (1999) FEMS Microbiol. Lett. 174, 251-253. [DOI] [PubMed] [Google Scholar]
- 12.Lowe, T. M. & Eddy, S. R. (1997) Nucleic Acids Res. 25, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda, H., Ishikawa, J., Hanamoto, A., Shinose, M., Kikuchi, H., Shiba, T., Sakaki, Y., Hattori, M. & Omura, S. (2003) Nat. Biotechnol. 21, 526-531. [DOI] [PubMed] [Google Scholar]
- 14.Bentley, S. D., Chater, K. F., Cerdeno-Tarraga, A.-M., Challis, G. L., Thomson, N. R., James, K. D., Harris, D. E., Quail, M. A., Kieser, H., Harper, D., et al. (2002) Nature 417, 141-147. [DOI] [PubMed] [Google Scholar]
- 15.Lobry, J. R. (1996) Mol. Biol. Evol. 13, 660-665. [DOI] [PubMed] [Google Scholar]
- 16.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S., Barry, C. E., III, et al. (1998) Nature 393, 537-544. [DOI] [PubMed] [Google Scholar]
- 17.Arruda, S., Bomfim, G., Knight, R., Huima-Byron, T. & Riley, L. W. (1993) Science 261, 1454-1457. [DOI] [PubMed] [Google Scholar]
- 18.Rahman, M. T., Herron, L. L., Kapur, V., Meijer, W. G., Byrne, B. A., Rena, J., Nicholson, V. M. & Prescott, J. F. (2003) Vet. Microbiol. 94, 143-158. [DOI] [PubMed] [Google Scholar]
- 19.Abou-Zeid, C. Garbe, T., Lathigra, R., Wiker, H. G., Harboe, M., Rook, G. A. & Young, D. B. (1991) Infect. Immun. 59, 2712-2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labo, M., Gusberti, L., De Rossi, E., Speziale, P. & Riccardi, G. (1998) Microbiology 144, 807-814. [DOI] [PubMed] [Google Scholar]
- 21.Beaman, B. L. & Beaman, L. (1998) Infect. Immun. 66, 4676-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wayne, L. G. & Hayes, L. G. (1998) Tuber. Lung Dis. 79, 127-132. [DOI] [PubMed] [Google Scholar]
- 23.Quadri, L. E. N., Sello, J., Keating, T. A., Weinreb, P. H. & Walsh, C. T. (1998) Chem. Biol. 5, 631-645. [DOI] [PubMed] [Google Scholar]
- 24.Schaeffer, M. L., Agnihotri, G., Volker, C., Kallender, H., Brennan, P. J. & Lonsdale, J. T. (2001) J. Biol. Chem. 276, 47029-47037. [DOI] [PubMed] [Google Scholar]
- 25.Banu, S., Honore, N., Saint-Joanis, B., Philpott, D., Prevost, M. C. & Cole, S. T. (2002) Mol. Microbiol. 44, 9-19. [DOI] [PubMed] [Google Scholar]
- 26.Severinov, K., Soushko, M., Goldfarb, A. & Nikiforov, V. (1993) J. Biol. Chem. 268, 14820-14825. [PubMed] [Google Scholar]
- 27.Zhou, Y. N. & Jin, D. J. (1998) Proc. Natl. Acad. Sci. USA 95, 2908-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurent, F., Poirel, L., Naas, T., Chaibi, E. B., Labia, R., Boiron, P. & Nordmann, P. (1999) Antimicrob. Agents Chemother. 43, 1644-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nelson, K. E., Weinel, C., Paulsen, I. T., Dodson, R. J., Hilbert, H., Martins dos Santos, V. A., Fouts, D. E., Gill, S. R., Pop, M., Holmes, M., et al. (2002) Env. Microbiol. 4, 799-808. [DOI] [PubMed] [Google Scholar]
- 30.McLean, K. J., Marshall, K. R., Richmond, A., Hunter, I. S., Fowler, K., Kieser, T., Gurcha, S. S., Besra, G. S. & Munro, A. W. (2002) Microbiology 148, 2937-2949. [DOI] [PubMed] [Google Scholar]
- 31.Li, W., Leet, J. E., Ax, H. A., Gustavson, D. R., Brown, D. M., Turner, L., Brown, K., Clark, J., Yang, H., Fung-Tomc, J. & Lam, K. S. (2003) J. Antibiot. 56, 226-231. [DOI] [PubMed] [Google Scholar]
- 32.Hu, H., Zhang, Q. & Ochi, K. (2002) J. Bacteriol. 184, 3984-3991. [DOI] [PMC free article] [PubMed] [Google Scholar]