Abstract
The patients with DiGeorge syndrome (DGS), caused by deletion containing dozens of genes in chromosome 22, often carry cardiovascular problem and hearing loss associated with chronic otitis media. Inside the deletion region, a transcription factor TBX1 was highly suspected. Furthermore, similar DGS phenotypes were found in the Tbx1 heterozygous knockout mice. Using ENU-induced mutagenesis and G1 dominant screening strategy, here we identified a nonsynonymous mutation p.W118R in T-box of TBX1, the DNA binding domain for transcription activity. The mutant mice showed deficiency of inner ear functions, including head tossing and circling, plus increased hearing threshold determined by audiometry. Therefore, our result further confirms the pathogenic basis of Tbx1 in DGS, points out the crucial role of DNA binding activity of TBX1 for the ear function, and provides additional animal model for studying the DGS disease mechanisms.
1. Introduction
The two major types of hearing loss, both conductive and sensorineural, are caused by gene mutations that affect the structure and function of the auditory system. Currently, more than 70 nonsyndromic deafness genes have been identified [1]. Usually the nonsyndromic deafness genes participate more specifically in auditory functions such as transduction, ciliogenesis, cell metabolism, and ion homeostasis [2]. For example, GJB2 is a major deafness gene [3] that encodes for a component of gap junctions specifically in the supporting cells of the cochlea and governs potassium recycling [4]. Lhfpl5 was found expressed specifically in hair bundle of cochlear hair cells [5] that modulate the transduction complex location and channel gating [6]. In addition, many more loci have been linked to nonsyndromic deafness though the causative genes have not been characterized yet [1].
However, it is more prevalent that a deafness phenotype always accompanies with other types of disorder(s), which is considered as a syndromic deafness. For example, Pendred syndrome is the most common deaf syndrome that manifests cochlear development abnormalities and sensorineural hearing loss, in parallel with diffuse thyroid enlargement [7]. The Pendred syndrome is caused by a chloride-iodide transport protein malfunction [8]. Patients with Usher syndrome hold both profound hearing loss and retinitis pigmentosa [9]. Hence, the identification of novel deafness gene and the annotation thereafter are appreciated greatly in the research field and clinical community. Strategically the forward genetics and reverse genetics based approaches are widely applied to search novel deafness genes.
For all characterized deafness genes, a large number of them are encoding structural proteins or homeostasis modulators that are mostly linked to nonsyndromic deafness. It may not be difficult to understand, considering that cochlea is such a sophisticated tissue machine endowing ear the hearing function. More importantly, the transcription factors and noncoding RNA molecules are broadly manipulating the development of the ear that often cause syndromic deafness. It has been characterized that a bunch of transcription factors mainly are involved in neural development in addition to inner ear development and morphogenesis. For example, transcription factors including Bmb4, Jag1, Islet1, Lfng, Fgf16, Prox1, and Tbx1 regulate specification of prosensory patches [10]. These transcription factors are expressed in specified temporal and spatial patterns with intermingled interaction to each other.
Mice with Tbx1 mutation showed a reduced expression of Bmp4 and thus resulted in deficiency of sensory epithelia formation [11]. In early inner ear development, Tbx1 is a very important transcription factor [12] that is also one of the candidate genes in pathogenesis of del22q11/DiGeorge syndrome (DGS)/velocardiofacial syndrome (VCFS) (for short DGS below) [13–15]. Usually, DGS patients carry a hemizygous deletion for a 1.5–3 Mb region on human chromosome 22, which includes 24 genes. DGS is complex and manifested by lots of phenotypes including craniofacial anomalies such as external ear defects and hearing impairment in addition to cardiovascular problem [16, 17]. It has been reported that most of the DGS patients carry conductive hearing loss associated with chronic otitis media [18, 19]. However, a minor (15%) of hearing loss is of the sensorineural type with unknown mechanisms [18]. In addition, balance problem was observed in DGS patients [20]. Recent study has given strong evidence that Tbx1 is a critical gene in the pathogenesis of DGS [21–23].
In this study, we have utilized a chemical mutagenesis based N-ethyl-N-nitrosourea (ENU) screening and characterized a mouse line called ENU706 that carried phenotype of deafness and imbalance in a fashion of dominant inheritance. Audiometric analysis demonstrated that the hearing threshold of ENU706 heterozygous mice was elevated around 30 dB SPL by average comparing to control mice. However, the hearing threshold in each heterozygous mouse was randomly elevated for each ear. The genetic analysis pointed out that Tbx1 was the causative gene for the ear problem. A previously unreported nonsynonymous mutation, p.W118R, hits a conserved amino acid in the T-box region of TBX1, a DNA binding domain responsible for the Tbx1 transcription activity. In T-box domain, very close to our mouse mutation, the other 2 human DGS mutations, p.F148Y and p.H194Q, were previously found in familial cases. Therefore, our data further confirm the pathogenic roles of Tbx1 in DGS, pinpoint the mechanistic association of its DNA binding activity and hearing loss, and provide additional animal model for studying the DGS disease mechanisms.
2. Materials and Methods
All procedures were performed in accordance with research guidelines of the institutional animal care and use committee of Tsinghua University. Mice of either sex were used in this study.
2.1. Generation of ENU Mutant Mice
The ENU-mutagenesis protocol and primary phenotypic screen have been described previously [25]. Briefly, C57BL/6J male mice were injected with ENU at the dose of 100 mg/kg according to body weight once every week for three weeks. After recovery of fertility, the mice were mated to female naive C57BL/6J. The offspring G1 mice were applied for neurological phenotypic assays, including learning and memory, movement impairment, hearing loss, and gait analysis. The affected G1 founders were bred to naive C57BL/6J to set up mutant family. The offspring mice carrying the inherited phenotypes were sent for whole exon capture to identify the responsible mutations.
2.2. Whole-Exome Analysis
Exonic single nucleotide polymorphism (SNP) was examined at whole genome level [26]. In brief, the exome captured sequencing library was produced by SeqCap EZ Library SR (Roche). The DNA-seq data was subjected to bioinformatics analysis to recognize the potential variants caused by ENU by comparing mutant (affected) with C57BL/6J database. All the candidates must meet four criteria: (1) the number of supporting reads > 4; (2) the number of supporting reads/the depth of this locus > 0.2; (3) the variant appeared heterozygous in the affected sample but not in the unaffected sample; (4) according to the annotation by ANNOVAR it is a nonsynonymous exonic mutation.
2.3. Audiometry
Auditory brainstem response (ABR) measurement was used to evaluate the hearing threshold of mice in this study as previously described [27]. The measurement was applied on mice with age older than 30 days. To examine developmental effect on hearing progression, the mice were tested with age up to 300 days. Before measurement, the mouse was anesthetized by i.p. injection of pentobarbitone. Then the mouse was transferred into a sound-proof chamber (Shengnuo, Shanghai) for audiometry. The audiometric evaluation was done with a TDT RZ6 system (Tucker-Davis Technologies). The electrodes were placed into the mouse subdermally. The ground electrode was inserted in the back near the hind leg, and the reference electrode was just behind the pinna, and the active electrode was inserted at the vertex. An EC1 close-field speaker was placed onto the external ear canal through a conduct tube. A balanced click stimuli were applied per second, each with a duration of 0.1 ms, starting at 90 dB SPL and decreasing at 10 dB SPL step in intensity. Stimuli and recordings were performed with the BioSigRZ software provided with the TDT workstation. The number of acquisition trials was set at 512 for averaging. Auditory thresholds were analyzed for both ears of mutant mice and single ears of wild-type mice. Wild-type mice were examined for another ear if there was an abnormal hearing. The hearing threshold was defined once a visible ABR emerged in recorded traces with graded click stimuli. Our setup determined the median threshold of wild-type C57BL/6J as 20 dB SPL. This baseline was elevated a bit with aging of mice.
2.4. Scanning Electron Microscopy
Inner ears were dissected out in phosphate buffer (0.1 M Na2HPO4·12H2O, 0.1 M NaH2PO4·2H2O, PH 7.4) and transferred into fixative buffer (2.5% glutaraldehyde, 0.1 M phosphate buffer). A hole was poked at the apex to let the fixative flush through the cochlear labyrinth before the sample was fixed overnight at 4°C. The inner ears were washed by phosphate buffer for 10 minutes with 3 times and fine-dissected to remove the spiral ligament, Reissner's membrane, and tectorial membrane. Samples were dehydrated by 30-minute incubation in 10/20/30/50/70/80/95/100% ethanol, followed by freeze drying (Hitachi ES-2030) and gold coating (Hitachi E-1010). The samples were imaged with FEI Quanta 200.
2.5. Electrophysiology
Cochlear hair cells were observed with an upright microscope (Olympus BX51WI). Borosilicate glass with filament (Sutter) was pulled with a PC-10 pipette puller (Narishige) and polished with MF-830 microforge (Narishige) to resistance of 3–5 MOhm. Hair bundles were deflected with a glass pipette mounted on a P-885 piezoelectric stack actuator (Physik Instrumente). Whole cell currents were sampled at 100 KHz with an EPC 10 USB patch-clamp amplifier operated by Patchmaster software (HEKA). Extracellular solution contains (in mM) 144 NaCl, 0.7 NaH2PO4, 5.8 KCl, 1.3 CaCl2, 0.9 MgCl2, 5.6 glucose, and 10 H-HEPES, pH 7.4. Intracellular solution contains (in mM) 140 KCl, 1 MgCl2, 0.1 EGTA, 2 Mg-ATP, 0.3 Na-GTP, and 10 H-HEPES, pH 7.2. Hair cells were voltage-clamped at −70 mV.
2.6. Data Analysis
Data analysis was performed by software including Excel (Microsoft), Prism (GraphPad), and Igor Pro 6 (WaveMetrics).
3. Results and Discussion
3.1. Generation and Genetic Mapping of ENU706 Mouse Line
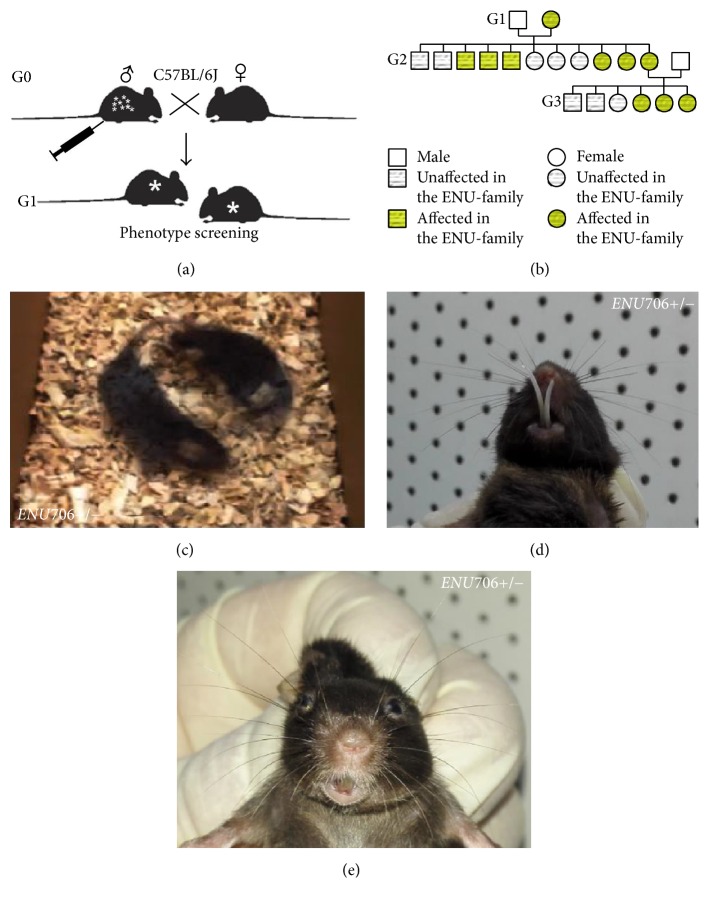
In order to identify the inherited mutations responsible for the mouse neurological phenotypes, we have set up a G1 dominant mutagenesis screening (Figure 1(a)). The ENU706 line appeared obvious circling phenotype in the G1 founders (Figure 1(c)). After crossing to the naive C57BL/6J, we found that the dominant inherited phenotype reoccurred in the family (Figure 1(b)), which met the expected Mendelian ratio (Figure 4(c)).
Figure 1.
Identification of a mutant line ENU706 by ENU-mutagenesis screening. (a) The strategy for our ENU screen on C57BL/6J mice. The male founder mouse was injected with mutagen and cross with female mice. The G1 mice were sent for phenotype screening for dominant phenotypes. (b) The family pedigree for the ENU706 line. (c) Representative image of circling behavior in a heterozygous ENU706 mouse. The syndromic traits were inherited in a dominant manner. Some of the heterozygous mutant mice showed abnormal craniofacial development including (d) elongated teeth and (e) asymmetrical face.
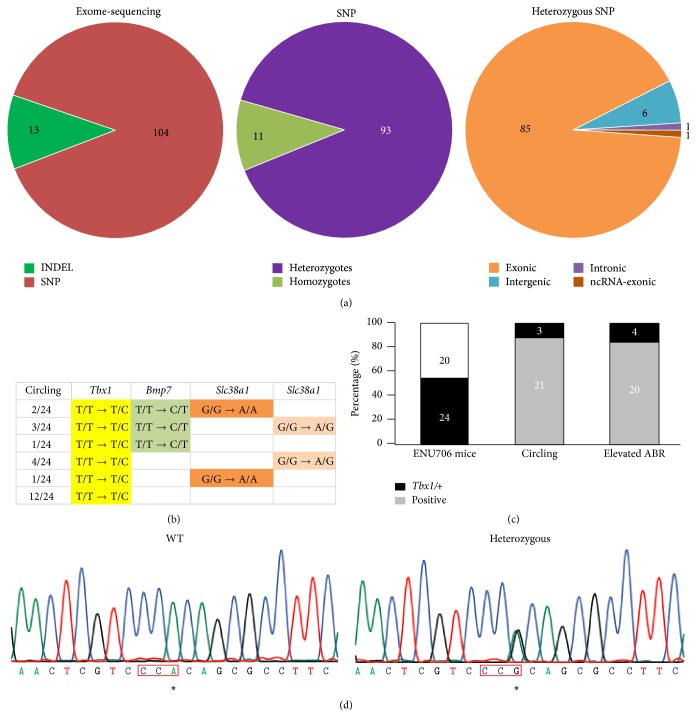
Figure 4.
Genetic analysis of target gene in ENU706 phenotypes. (a) Whole genome sequence identified 104 SNPs in one circling ENU706 mutant mouse, among which 93 SNPs were heterozygous and 85 SNPs were exonic. (b) Top 3 candidate genes were Tbx1, Bmp7, and Slc38a1. The T-to-C mutation in Tbx1 had the high penetration in ENU706 heterozygous mice with the circling phenotype. (c) The genotyping for the T-to-C mutation in Tbx1 showed there were 24 heterozygous mice out of all the 44 mice in this colony. And phenotypic assays showed 21 ENU706 heterozygous mice were circling and 20 ENU706 heterozygous mice had elevated ABR in all the 24 Tbx1+/− mice. (d) A typical DNA chromatogram showing the T-to-C mutation in an ENU706 heterozygous mouse. Note a reverse primer was used for sequencing. ∗ refers to the T-to-C mutation.
3.2. ENU706 Line Possessed Moderate Hearing Loss
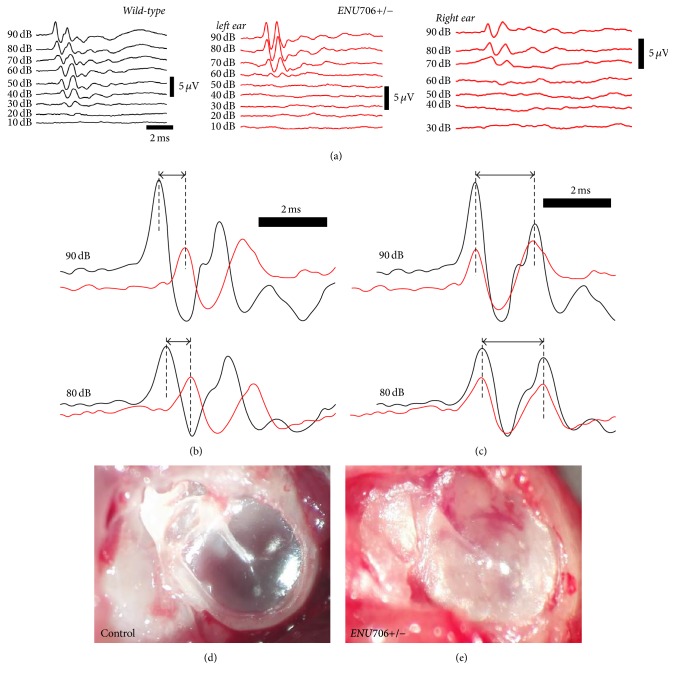
ENU706 heterozygous mice had circling (Figure 1(c)) and head tossing behavior observed as early as 1-month old that was a typical phenotype of vestibular problem. In general, one-half of the mice were circling (Figure 4(c), left bar), which further confirmed the mutation caused imbalance in a dominant inheritance style. Circling is often accompanied with hearing loss in animals suffering from deficit of the inner ear function. We then assessed the hearing threshold for the colony of ENU706 mice. Click ABR test was applied to evaluate the threshold of hearing. A control wild-type mouse started to respond to click sound as low as 20 dB SPL (Figure 2(a), left), which is a typical value for normal hearing. While in an ENU706 heterozygous mouse, the hearing threshold was 50 dB SPL for the left ear and 60 dB SPL for the right ear (Figure 2(a), right). It suggested that this mutant mouse had a moderate hearing loss and might be differentially affected in each ear. Notably, the different hearing threshold for either ear of one mouse was often not identical. We observed in some extreme case that the mouse had one ear normal (20 dB SPL) but with another ear profoundly deaf (70 dB SPL). To determine whether the hearing loss was conductive or sensorineural, the latencies of the ABR responses were analyzed. The onset of ABR was submillisecond slower in the ENU706 heterozygous mouse (Figure 2(b)), but the interpeak seemed not altered (Figure 2(c)). Surgical examinations showed otitis media in ENU706 heterozygous mice if they had hearing loss (Figures 2(d) and 2(e)). In general, this sporadic hearing loss was not related with gender (Figure 3(a)) or age (Figure 3(b)) in ENU706 mice. Indeed, the hearing threshold was significantly elevated about 20 dB by average in ENU706 mice (Figure 3(a)). Consistently, ENU706 mice also shared a parallel trend of progression with wild-type animals but with elevated hearing threshold (Figure 3(b)). To further investigate whether any of sensorineural factors was involved in the hearing loss, we examined mechanotransduction response in cochlear hair cells. Our electrophysiological data showed no obvious change of mechanotransduction currents in ENU706 heterozygous mouse (Figure 3(c)), so did their hair bundle (Figure 3(d)).
Figure 2.
The ENU706 mice possessed moderate hearing loss. (a) A representative case showed the click ABR test in control and ENU706 mice. A control mouse possessed a hearing threshold as low as 20 dB SPL. An ENU706 mouse had 50 dB SPL hearing threshold at the left ear and 60 dB SPL hearing threshold at the right ear. (b) The onset of ABR responses was 0.36 milliseconds later in mutant than that in control mouse. (c) The duration between peaks I and II was not altered obviously in mutant compared to the control. The traces analyzed in (b) and (c) were from the same recordings shown in (a). In (a), (b), and (c), control was shown in black and ENU706 in red. In difference with the control (d), the heterozygous ENU706 mice with elevated ABR responses were found to have otitis media (e).
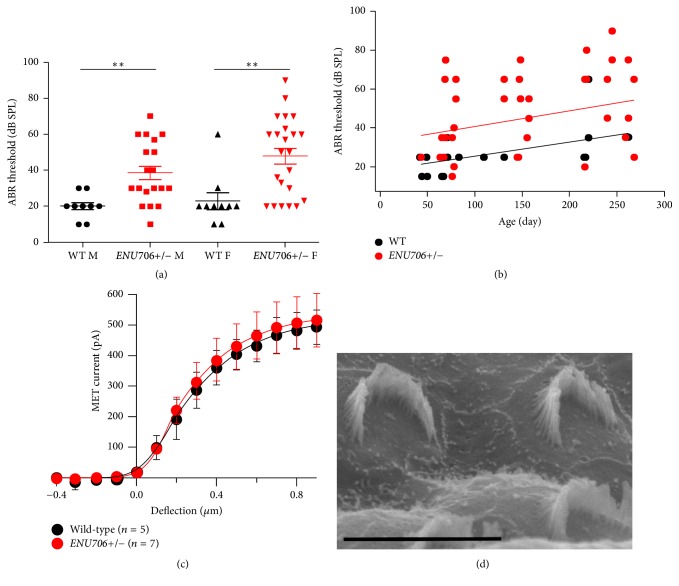
Figure 3.
The ENU706 mice had no obvious defects in hair cell mechanotransduction. (a) The ABR threshold was box-whisker plotted in 4 categories: wild-type male (WT M, 20.0 ± 2.4 dB SPL, 8 males tested, 8 right ears plus 1 left ear), heterozygous male (ENU706+/− M, 38.7 ± 3.9 dB SPL, 11 males tested, 11 right ears, and 8 left ears), wild-type female (WT F, 23.0 ± 4.5 dB SPL, 8 females tested, 9 right ears, and 1 left ear), and heterozygous female (ENU706+/− F, 47.9 ± 4.5 dB SPL, 13 females tested, 13 right ears, and 10 left ears). The “n” numbers were counted twice if both ears were measured. Data shown as mean ± SEM. Statistical significance (∗∗ p < 0.01) was determined by Student's two-tailed unpaired t-test. (b) The ABR threshold was plotted against age. The data pooled from left and right ear were shown (WT, 20 mice tested, 20 right ears, and 3 left ears; ENU706+/−, 24 mice tested, 24 right ears, and 19 left ears). (c) The mechanotransduction currents were measured in outer hair cells of ENU706 heterozygous mice and control littermates (cell number shown in the panel). Data shown as mean ± SD. A set of mechanical deflections from −400 nm to 900 nm at 100 nm step were applied to hair bundle to generate mechanotransduction currents in hair cells. In all panels, control was shown in black and ENU706 in red. (d) Scanning electron microscopy showed the hair bundles of apical-middle outer hair cells were relatively normal in an ENU706 heterozygous mouse. Scale bar: 5 μm.
3.3. Tbx1 Gene Is Linked to the Inner Ear Problem
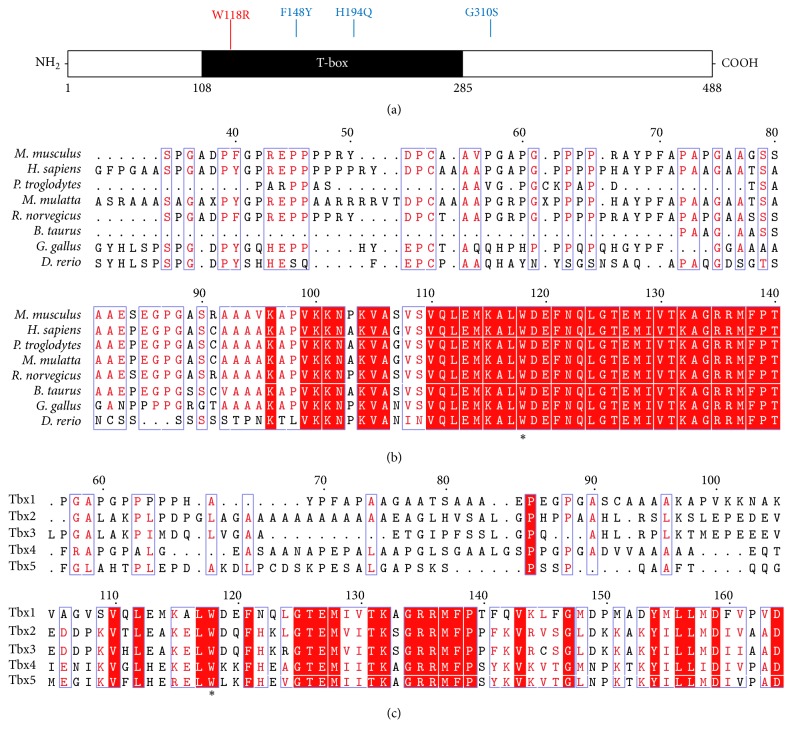
To identify the causative gene linked to the dominant hearing loss and circling phenotype, we performed a whole-exome sequencing. The preliminary analysis indicated that there are 104 SNPs plus 13 insertions and deletions (indels) at the gene coding regions. Considering that the inherited trait is consistent with Mendel's law (Figure 4(c), the 1st bar), the homozygous SNPs were rule out. Then 85 out of 93 heterozygous SNPs were presented in the preliminary analysis to be validated to link to the hearing loss. Based on our previous data-mining experience, 31 out of the 85 SNPs were selected to be studied (Figure 4(a)). We then applied the PCR sequencing to validate each SNP transversion for the 31 candidate genes. It turned out that 3 out of the 31 SNPs were the top candidates that emerged with a high consistency between the genotype and the phenotype. The 3 genes were Tbx1, Bmp7, and Slc38a1, in which Tbx1 and Bmp7 have been previously linked to hearing impairment. Tbx1 has been proposed as a candidate gene for pathogenesis of DGS including hearing impairment [28]. We found a T→C transversion in Tbx1 gene in most of circling ENU706 heterozygous mice. Bmp7 has also been found to relate with development of inner ear and specify the tonotopic cochlea axis [29]. A T→C transversion was found in Bmp7 gene in few ENU706 heterozygous mice. For Slc38a1 gene, there was a G→A mutation and sometimes homozygous mutations were observed at both alleles (Figure 4(b)). With comparison of PCR sequencing result and circling phenotype, we speculated that Tbx1 was the highly possible target gene that carried the mutation T/T→T/C in ENU706 mice (23 out of the 24 circling mice). Among all the 44 mice in breeding, 24 had the T/T→T/C SNP change in Tbx1 gene (Figures 4(c) and 4(d)). In the 24 Tbx1+/− mice, 21 were circling and 20 had elevated ABR (Figure 4(c)). The mutation caused a W to R change at the 118 amino acid in T-box region of TBX1 protein (Figure 5(a)), which is very conserved in different species (Figure 5(b)) and TBX paralogues (Figure 5(c)).
Figure 5.
Conservation analysis of nonsynonymous mutation in TBX1 found in the ENU706 mutant mouse. (a) The ENU706 mutation caused a W118R conversion. F148Y, H194Q, and G310S mutations, previously reported in DGS patients, are also indicated [24]. (b) The W118 is highly conserved across the species. The protein NCBI IDs, NP_035662.1 (M. musculus), NP_542377.1 (H. sapiens), XP_016795078.1 (P. troglodytes), XP_015005141.1 (M. mulatta), NP_001101792.1 (R. norvegicus), XP_002694701.2 (B. taurus), XP_015131167.1 (G. gallus), and NP_899182.1 (D. rerio). (c) The Trp amino acid is also highly conserved in 5 TBX paralogues. The protein NCBI IDs, NP_542377.1 (Tbx1), NP_005985.3 (Tbx2), NP_005987.3 (Tbx3), NP_001308049.1 (Tbx4), and NP_000183.2 (Tbx5). ∗ refers to the W118 in (b) and Trp amino acid in (c).
4. Discussion
All the evidence in this study indicated Tbx1 was linked to the hearing problem of ENU706 mice we generated. ENU706 mice carried a moderate auditory threshold elevation plus vestibular problem, which was induced by a hemizygous SNP transversion in T-box region of Tbx1. Tbx1, as a transcription factor, has been associated to middle and inner ear development and morphogenesis, including conductive and sensorineural hearing loss [18, 19]. It was also the candidate gene concerning the pathogenesis of del22q11/DGS/VCFS [14]. More recent studies indicate that Tbx1 was very likely linked to the hearing defect in DGS patients and engineered mutant mice [21–23]. Moreover, we did notice some syndromic phenotypes in a few ENU706 heterozygous mice (Figures 1(d) and 1(e)) mimicking DGS traits reported in humans and mice. The ENU706 homozygous mice were not found alive in our breeding colony, which also happened in Tbx1 knockout mice [15].
During our gene identification, mutations in BMP7 and Slc38a1 genes had occasionally emerged; BMP7 especially was previously identified critical for cochlear axis specification. These two genes were not likely the causative genes in ENU706 mice. BMP7 is in chromosome 2 and Slc38a1 is in chromosome 15, while Tbx1 is in chromosome 16. The late crossed ENU706 mice, from the 4th generation, did not carry BMP7 and Slc38a1 mutations anymore but still possess the DGS phenotypes. The mechanisms underlying the hearing defect including both conductive and sensorineural hearing loss has been tackled by our audiometry (Figures 2(b) and 2(c)) and electrophysiological recordings (Figure 3(c)). These data indicated that it was a more likely conductive hearing loss in ENU706 mutants. To study the accurate gene function, chromosomally engineered Df1/+ mice and single gene knockout Tbx1/+ mice were used. Heterozygous loss of Tbx1 resulted in major structural abnormalities of the heart similar to those observed in Df1/+ mice and Lgdel/+ mice [16]. Chronic otitis media was also a feature in clinical diagnosis of DGS [28]. The pathogenesis of otitis media is considered resulting from multiple causes, such as deficits of the inflammatory clearance [30, 31] or the mucosa in the middle-ear cavity [32, 33]. Our work was a reminiscent of the study on Df1/+ and Tbx1/+ mice, including dominant inheritance, circling, and partial hearing loss. A recent report further described that a defect in early myogenesis thus resulted in otitis media in mouse models of DGS [23]. Their data showed that the Tbx1 heterozygous mice showed the hearing loss mainly by Eustachian tube problem related with muscle problem. Then it made sense that the hearing thresholds were different between the two ears of a given ENU706 mouse (Figure 2(a)). It also coincided with the observation that the hearing loss in ENU706 mice was moderate and sporadic (Figures 3(a) and 3(b)). Nevertheless, our work pointed out that a single point mutation was responsible for most of the hearing features of DGS. Interestingly, another gene, Comt, in the deleted region of chromosome in DGS patients has also been linked to hearing loss [34].
In early development, Tbx1 as a transcription factor is expressed in the endodermal lining of the first pharyngeal pouch and in the meiosis of the pharyngeal arches [28]. T-box domain has been proposed to be important for T-box protein dimerization and DNA binding activity. It has been reported that F148Y and H194Q in the T-box domain actually induced a gain-of-function effect, plus G310S at the boundary of the T-box [35]. We speculated that W118R in ENU706 mice might also have a gain-of-function as other T-box mutations. More interestingly, recent study indicated that it also binds to chromatin that may function epigenetically [36]. Another study proposed that cortical development was regulated by mesodermal expression of Tbx1 [37]. That raised the complexity of the TBX1 function. This ENU706 mice hence provided a new line of mouse model to finely dissect TBX1 function in development and physiology.
5. Conclusion
To further understand the molecular and physiological relevance of hearing sensation and hearing impairment, we have set out to establish a forward genetics based deafness gene screen in mice. And it was also a major goal in our collaborative effort to identify disease genes linked to neurological disorders. In this study, we characterized a novel mutation in Tbx1 gene, which caused a W118R amino acid change in T-box region of TBX1 protein. This single point missense mutation in the highly conserved region induced a robust phenotype of imbalance in affected mice that also carried moderate hearing loss. More interestingly, the deficiency was in a dominant inherited manner that was consistent with previous study in Tbx1 deficient Df1/+ mice and Tbx1/+ mice. Our study thus provided an alternative model to study Tbx1 function and might help to dissect domain-specific and tissue-specific TBX1 function in the DGS study.
Acknowledgments
This work was funded by the National Natural Science Foundation of China (31571080 and 31522025 to Wei Xiong; 31571097 and 81371361 to Yichang Jia) and the Research Funds of Tsinghua University (to Wei Xiong and Yichang Jia).
Competing Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
Authors' Contributions
Jiaofeng Chen and Xue Zhang contributed equally to this work.
References
- 1.Shearer A. E., Smith R. J. H. Genetics: advances in genetic testing for deafness. Current Opinion in Pediatrics. 2012;24(6):679–686. doi: 10.1097/mop.0b013e3283588f5e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson G. P., De Monvel J. B., Petit C. How the genetics of deafness illuminates auditory physiology. Annual Review of Physiology. 2011;73:311–334. doi: 10.1146/annurev-physiol-012110-142228. [DOI] [PubMed] [Google Scholar]
- 3.Lee S. W., Tomasetto C., Paul D., Keyomarsi K., Sager R. Transcriptional downregulation of gap-junction proteins blocks junctional communication in human mammary tumor cell lines. Journal of Cell Biology. 1992;118(5):1213–1221. doi: 10.1083/jcb.118.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rabionet R., Gasparini P., Estivill X. Molecular genetics of hearing impairment due to mutations in gap junction genes encoding beta connexins. Human Mutation. 2000;16(3):190–202. doi: 10.1002/1098-1004(200009)16:3<190::aid-humu2>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 5.Longo-Guess C. M., Gagnon L. H., Cook S. A., Wu J., Zheng Q. Y., Johnson K. R. A missense mutation in the previously undescribed gene Tmhs underlies deafness in hurry-scurry (hscy) mice. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(22):7894–7899. doi: 10.1073/pnas.0500760102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong W., Grillet N., Elledge H. M., et al. TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell. 2012;151(6):1283–1295. doi: 10.1016/j.cell.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Everett L. A., Glaser B., Beck J. C., et al. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS) Nature Genetics. 1997;17(4):411–422. doi: 10.1038/ng1297-411. [DOI] [PubMed] [Google Scholar]
- 8.Scott D. A., Wang R., Kreman T. M., Sheffield V. C., Karniski L. P. The Pendred syndrome gene encodes a chloride-iodide transport protein. Nature Genetics. 1999;21(4):440–443. doi: 10.1038/7783. [DOI] [PubMed] [Google Scholar]
- 9.Möller C. G., Kimberling W. J., Davenport S. L. H., et al. Usher syndrome: an otoneurologic study. The Laryngoscope. 1989;99(1):73–79. doi: 10.1288/00005537-198901000-00014. [DOI] [PubMed] [Google Scholar]
- 10.Kelley M. W. Regulation of cell fate in the sensory epithelia of the inner ear. Nature Reviews Neuroscience. 2006;7(11):837–849. doi: 10.1038/nrn1987. [DOI] [PubMed] [Google Scholar]
- 11.Raft S., Nowotschin S., Liao J., Morrow B. E. Suppression of neural fate and control of inner ear morphogenesis by Tbx1 . Development. 2004;131(8):1801–1812. doi: 10.1242/dev.01067. [DOI] [PubMed] [Google Scholar]
- 12.Chapman D. L., Garvey N., Hancock S., et al. Expression of the T-box family genes, Tbx1-Tbx5, during early mouse development. Developmental Dynamics. 1996;206(4):379–390. doi: 10.1002/(sici)1097-0177(199608)206:4<379::aid-aja4>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 13.Goldberg R., Motzkin B., Marion R., Scambler P. J., Shprintzen R. J. Velo-cardio-facial syndrome: a review of 120 patients. American Journal of Medical Genetics. 1993;45(3):313–319. doi: 10.1002/ajmg.1320450307. [DOI] [PubMed] [Google Scholar]
- 14.Morrow B., Goldberg R., Carlson C., et al. Molecular definition of the 22q11 deletions in velo-cardio-facial syndrome. The American Journal of Human Genetics. 1995;56(6):1391–1403. [PMC free article] [PubMed] [Google Scholar]
- 15.Jerome L. A., Papaioannou V. E. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nature Genetics. 2001;27(3):286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 16.Merscher S., Funke B., Epstein J. A., et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104(4):619–629. doi: 10.1016/S0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 17.Funke B., Epstein J. A., Kochilas L. K., et al. Mice overexpressing genes from the 22q11 region deleted in velo-cardio-facial syndrome/DiGeorge syndrome have middle and inner ear defects. Human Molecular Genetics. 2001;10(22):2549–2556. doi: 10.1093/hmg/10.22.2549. [DOI] [PubMed] [Google Scholar]
- 18.Digilio M. C., Pacifico C., Tieri L., Marino B., Giannotti A., Dallapiccola B. Audiological findings in patients with microdeletion 22q11 (di George/velocardiofacial syndrome) British Journal of Audiology. 1999;33(5):329–333. doi: 10.3109/03005369909090116. [DOI] [PubMed] [Google Scholar]
- 19.Reyes M. R. T., LeBlanc E. M., Bassila M. K. Hearing loss and otitis media in velo-cardio-facial syndrome. International Journal of Pediatric Otorhinolaryngology. 1999;47(3):227–233. doi: 10.1016/S0165-5876(98)00180-3. [DOI] [PubMed] [Google Scholar]
- 20.Swillen A., Devriendt K., Legius E., et al. The behavioural phenotype in velo-cardio-facial syndrome (VCFS): from infancy to adolescence. Genetic Counseling. 1999;10(1):79–88. [PubMed] [Google Scholar]
- 21.Yagi H., Furutani Y., Hamada H., et al. Role of TBX1 in human del22q11.2 syndrome. The Lancet. 2003;362(9393):1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 22.Arnold J. S., Braunstein E. M., Ohyama T., et al. Tissue-specific roles of Tbx1 in the development of the outer, middle and inner ear, defective in 22q11DS patients. Human Molecular Genetics. 2006;15(10):1629–1639. doi: 10.1093/hmg/ddl084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs J. C., Linden J. F., Baldini A., Tucker A. S. A defect in early myogenesis causes Otitis media in two mouse models of 22q11.2 deletion syndrome. Human Molecular Genetics. 2014;24(7):1869–1882. doi: 10.1093/hmg/ddu604.ddu604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogata T., Niihori T., Tanaka N., et al. TBX1 mutation identified by exome sequencing in a Japanese family with 22q11.2 deletion syndrome-like craniofacial features and hypocalcemia. PLoS ONE. 2014;9(3) doi: 10.1371/journal.pone.0091598.e91598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Probst F. J., Justice M. J. Mouse mutagenesis with the chemical supermutagen ENU. Methods in Enzymology. 2010;477:297–312. doi: 10.1016/s0076-6879(10)77015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fairfield H., Gilbert G. J., Barter M., et al. Mutation discovery in mice by whole exome sequencing. Genome Biology. 2011;12(9, article R86) doi: 10.1186/gb-2011-12-9-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwander M., Sczaniecka A., Grillet N., et al. A forward genetics screen in mice identifies recessive deafness traits and reveals that pejvakin is essential for outer hair cell function. Journal of Neuroscience. 2007;27(9):2163–2175. doi: 10.1523/JNEUROSCI.4975-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liao J., Kochilas L., Nowotschin S., et al. Full spectrum of malformations in velo-cardio-facial syndrome/DiGeorge syndrome mouse models by altering Tbx1 dosage. Human Molecular Genetics. 2004;13(15):1577–1585. doi: 10.1093/hmg/ddh176. [DOI] [PubMed] [Google Scholar]
- 29.Mann Z. F., Thiede B. R., Chang W., et al. A gradient of Bmp7 specifies the tonotopic axis in the developing inner ear. Nature Communications. 2014;5, article 3839 doi: 10.1038/ncomms4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacArthur C. J., Hefeneider S. H., Kempton J. B., Trune D. R. C3H/HeJ mouse model for spontaneous chronic otitis media. Laryngoscope. 2006;116(7):1071–1079. doi: 10.1097/01.mlg.0000224527.41288.c4. [DOI] [PubMed] [Google Scholar]
- 31.Rivkin A. Z., Palacios S. D., Pak K., Bennett T., Ryan A. F. The role of Fas-mediated apoptosis in otitis media: observations in the lpr/lpr mouse. Hearing Research. 2005;207(1-2):110–116. doi: 10.1016/j.heares.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 32.Hilton J. M., Lewis M. A., Grati M., et al. Exome sequencing identifies a missense mutation in Isl1 associated with low penetrance otitis media in dearisch mice. Genome Biology. 2011;12(9, article R90) doi: 10.1186/gb-2011-12-9-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheeseman M. T., Tyrer H. E., Williams D., et al. HIF-VEGF pathways are critical for chronic otitis media in Junbo and Jeff mouse mutants. PLoS Genetics. 2011;7(10) doi: 10.1371/journal.pgen.1002336.e1002336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du X., Schwander M., Moresco E. M., et al. A catechol-O-methyltransferase that is essential for auditory function in mice and humans. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(38):14609–14614. doi: 10.1073/pnas.0807219105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zweier C., Sticht H., Aydin-Yaylagül I., Campbell C. E., Rauch A. Human TBX1 missense mutations cause gain of function resulting in the same phenotype as 22q11.2 deletions. American Journal of Human Genetics. 2007;80(3):510–517. doi: 10.1086/511993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fulcoli F. G., Franzese M., Liu X., Zhang Z., Angelini C., Baldini A. Rebalancing gene haploinsufficiency in vivo by targeting chromatin. Nature Communications. 2016;7 doi: 10.1038/ncomms11688.11688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flore G., Cioffi S., Bilio M., Illingworth E. Cortical development requires mesodermal expression of Tbx1, a gene haploinsufficient in 22q11.2 deletion syndrome. Cerebral Cortex. 2016 doi: 10.1093/cercor/bhw076.bhw076 [DOI] [PubMed] [Google Scholar]