Abstract
Many proteins select special DNA sequences to form functional complexes. In one possible mechanism, protein molecules would scan DNA sequences by tracking a groove without complete dissociation. Upon dragging single molecules of DNA over a surface carrying fixed Escherichia coli RNA polymerase holoenzyme, we detected rotation of individual DNA molecules, providing direct evidence that a DNA-binding protein can track a DNA groove. These results confirm our previous observations of longitudinal movement of RNA polymerase along fixed, extended DNA and, moreover, imply that groove tracking facilitates scanning of DNA sequences.
Some DNA-binding proteins can move along DNA, in a behavior generically called 1D diffusion, by several mechanisms (1–3). These movements have been given various names but are classified here into sliding and intersegment transfer mechanisms. The former implies lateral movement along the surface of DNA, whereas the latter involves direct jumps between DNA segments that are momentarily adjacent in space. The sliding mechanism can be further subdivided into helical or nonhelical movement of protein molecules along the DNA, called “sliding” and “hopping,” respectively, in an early report (1). The mechanism of helical movement could involve tracking of a DNA groove or another helical component, such as a row of phosphate residues. If a protein molecule, to exert its function, must bind to a specific site on DNA, tracking a groove could facilitate its search for such a site. Because a groove is helical, longitudinal movement of a protein molecule should be accompanied by rotational motion relative to the DNA. Thus, if DNA molecules are dragged over a surface carrying fixed protein molecules, any groove tracking should rotate the DNA. By observing the movements of a bead fixed to one DNA end during such an experiment, we tested for the existence of groove tracking during sliding. Among proteins that can diffuse one-dimensionally along DNA, we chose to examine one that has a single DNA-binding site, Escherichia coli RNA polymerase holoenzyme (4, 5). This enzyme has to find specific promoter sites on DNA before initiating transcription. We avoided proteins that have two DNA-binding sites, several of which have been shown to carry out intersegment transfer (2, 3).
Materials and Methods
Preparation of DNA. In preparation of the 13-kb DNA, a 10.1-kb fragment (corresponding to base pairs 3836–13983 of T7 DNA) was amplified by PCR with LA Taq (Takara Bio, Otsu, Japan) by using a primer that created a BamHI site at base pair 13983. This fragment contains no strong promoters. A 2.1-kbp DNA fragment constructed by PCR and harboring a high-affinity site for RNA polymerase (6) consisted of base pairs 4746–5096, followed by base pairs 1–1680 of pRLG3749, with a BamHI target introduced alongside position 1680. The 2.1-kbp DNA fragment was digested with BamHI and ligated to the similarly digested 10.1-kbp T7 DNA fragment. A 0.8-kbp DNA fragment of pACYC177 (base pairs 2420–3940) was amplified by PCR with Ex Taq (Takara Bio) in the presence of primers, one of which had a biotinylated 5′-OH end, and the product was further biotinylated by using Carbobiotin (Nisshinbo, Tokyo). This multibiotinylated DNA fragment was ligated to the end of the T7 DNA distal to the BamHI site, exploiting the HindIII site of pACYC177 and a corresponding site introduced next to position 3836 of the T7 DNA. The DNA was treated with T4 DNA ligase (Takara Bio) before use to reduce nicks.
Preparation of Beads. As markers we used 93-nm-diameter orange fluorescent carboxylated spheres [Molecular Probes, Lot 6942 (some other lots improperly aggregated during reactions)]. We suspended 0.2 mg of these spheres in 50 μl of 10 mM Na-phosphate buffer (pH 7.3) containing 26 mM 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) (Pierce) and 3 M 1,2-diaminoethane and incubated them for 1 h in the dark. The mixture was then passed through a water-equilibrated SPIN+TE-10 Column (BD Biosciences Clontech) to remove small molecules. Separately, 910-nm-diameter carboxylated beads (Polysciences) in 50 μl of a 2.5% suspension were washed with 0.1 M 2-morpholinoethanesulfonic acid buffer (Dojin, Kumamoto, Japan; pH 7.0) and resuspended in 50 μl of the same buffer. An equal volume of 0.1 M EDC in this buffer was then added dropwise, with each addition followed by vortex mixing. The mixture was incubated for an additional hour with rotation in a sample tube. The activated beads were then washed three times with 0.2 M Na-borate buffer (pH 8.5) and resuspended in 50 μl of the buffer. The suspension of fluorescent spheres (1–5 μl) was added, and the mixture was rotated in the dark. After 15 min, 20 μg of streptavidin (Pierce) in 200 μl of the buffer was added and incubated for another 2 h. The reaction was quenched by adding glycine (to 35 mM) and incubating for 2 h. Finally, the beads were washed with and stored in 50 mM Tris·HCl buffer (pH 7.9) containing 50 mM KCl and 10 mg/ml BSA. The latter prevents aggregation of the beads. Beads prepared as above were loaded with DNA by incubation overnight with an equal number of molecules of the 13-kbp DNA fragment in buffer A (50 mM Tris·HCl, pH 7.5/50 mM KCl/0.5 mM MgCl2/0.1 mM DTT). Unbound DNA was then removed by washing with buffer A containing 1 mg/ml BSA.
Immobilization of Holoenzyme Molecules. E. coli RNA polymerase holoenzyme was prepared as described in ref. 7. The 5-μl observation chamber was composed of two coverslips (22 × 40 mm and 18 × 18 mm) with two strips of Parafilm as spacers. The coverslips had been dipped in a 0.1% collodion solution in isoamyl alcohol and then air-dried. RNA polymerase at 320 nM in buffer A was injected into the chamber and incubated for 20 min. The chamber was then washed with buffer A containing 1 mg/ml partially hydrolyzed casein.
Measurements. All measurements and reactions were performed at room temperature. The DNA-linked beads were introduced into an observation chamber and incubated for 20 min, after which unfixed beads were washed away with buffer A containing 1 mg/ml casein. The beads were tethered to the surface by the 12-kbp linker part of the 13-kbp DNA. More than 95% of the beads could be removed if treated with 50 units of DNase I (Sigma) for 1 min, confirming the tethering.
The homogeneity of the tethered beads was checked before each assay of rotation. To avoid beads tethered by more than one DNA molecule, we first tried to move a bead linearly in both directions by using laser tweezers. Half of the beads did not move irrespective of movements of the trapping center, presumably because they had been nonspecifically adsorbed on the surface. Most of the rest showed a movement limited to 6–7 μm when the trapping center was moved farther than that distance. Because this is the limiting distance expected for beads tethered as shown in Fig. 1B, we selected these beads for rotational analysis. Less than 10% of beads showed abnormally small limiting distances, probably because of tethering by two or more molecules of DNA. There were also small numbers of beads with nicked DNA linkers, which showed a maximum moving distance much greater than 7 μm, because the single-stranded part could be stretched even by the low tension that was applied in this experiment (<5 pN).
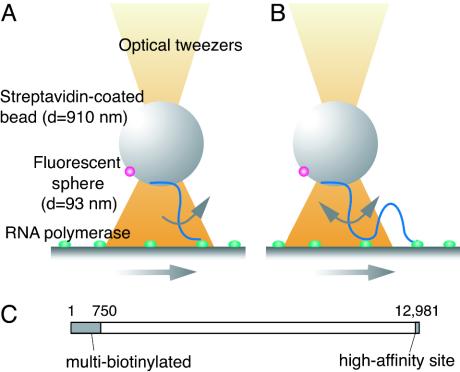
Fig. 1.
Detection of groove tracking. (A) Basic system: A streptavidin-coated bead was held above the glass surface on which holoenzyme molecules were fixed. When the surface was moved, the RNA polymerase molecules could slide along the DNA fixed to the bead, generating a torque to rotate the bead. (B) The improved system: A high-affinity RNA polymerase-binding site (6) was introduced into a 150-bp segment at the end of the DNA distal from the biotin residues so that this end was effectively fixed to the glass surface. (C) The 13-kbp linker DNA.
The microscope (Axiovert 135 TV, Zeiss), illuminated by a Hg lamp, was equipped with a Plan Neofluar ×100 oil-immersion objective (numerical aperture 1.4) and a silicon-intensified-target video camera (C2400, Hamamatsu Photonics, Hamamatsu City, Japan). The fluorescence images were obtained through an ARGUS-20 processor (Hamamatsu Photonics). A laser beam of 1,064 nm from a Nd: yttrium/aluminum garnet laser (DPY 421II, Coherent Radiation, Santa Clara, CA) with a power of 66 mW at the back aperture of the objective was used for the optical tweezers, with a trap stiffness of 200 pN/μm. The microscope stage was moved by a piezo electric driver (P-720:E-662, Physik Instrumente, Karlsruhe, Germany) at a constant rate. At every video frame, the x–y coordinate of the center of the fluorescent sphere was determined as the average over the bright pixels, with their fluorescent intensities as weights.
Rotational Coherence. A sequence of 1,800 video frames (recorded over 1 min) was divided into clusters in which the polar coordinate θ increased or decreased monotonously. The sizes of clusters and the distances between the positions of the fluorescent sphere in two adjacent frames were then determined. The rotational coherence was calculated as a weighted sum of the distances over clusters. We calculated this index by using different definitions of the weights and the clusters (see Supporting Text, which is published as supporting information on the PNAS web site). We also attempted replacement of the cluster by a time window of 1 s, which was reported to be the lifetime of a sliding complex (8). All nine calculations of rotational coherence gave essentially the same result explained below.
Results
Detection of Groove Tracking. If a holoenzyme molecule tracks a DNA groove, it will rotate relative to the DNA axis. This groove tracking will produce no net rotation when averaged over a long time period, because sliding consists microscopically of bidirectional thermal motions. However, if we introduce an external perturbation that distorts the distribution of the microscopic thermal motions, a net macroscopic rotation should be observed. Specifically, a gentle unidirectional macroscopic movement of the DNA relative to the holoenzyme molecule should produce either a significant net macroscopic rotation or none, depending on whether the protein tracks a groove. We have built the system shown in Fig. 1 A to detect such an asymmetric rotation by the motion of a 910-nm bead that is bound to one end of a DNA molecule through several biotin residues incorporated into that end (Fig. 1C) so that DNA can transmit a torque to the bead. A 93-nm fluorescent sphere is covalently attached to the bead so that its rotation can be visualized. The center of the bead is held by laser tweezers, but the bead is free to rotate. It is kept in position 1–1.5 μm above a surface on which holoenzyme molecules are immobilized to allow the 4-μm DNA to reach the holoenzyme. The external perturbation is introduced by moving the enzyme-laden surface at 8 nm/s. This perturbation is 10–8-fold smaller than the average thermal velocity of the protein molecules in solution, the velocity calculated from equal thermal energy per freedom, and thus could not disturb DNA–protein interactions.
There are three factors that could prevent detection of the putative groove tracking. First, rotational Brownian motion of the bead will work as a microscopic noise, obscuring the expected macroscopic rotation. Second, any torque generated may not be efficiently transmitted to the bead, because DNA is moderately flexible toward twist. Finally, the lifetime of a sliding complex of E. coli RNA polymerase holoenzyme is estimated to be ≈1 s (8). Torque can therefore be accumulated for only 1 s, generating a macroscopic rotation much smaller than that predicted from the pitch of the DNA helix and the velocity of the surface. If the formation of a sliding complex is infrequent and the DNA molecules are mostly free, there may be too little generated torque to detect. In fact, the device illustrated in Fig. 1 A could not distinguish any rotational movements of the beads from Brownian motion. Among the three factors listed, the first two are inevitable, but the third can be mitigated. We introduced at the free end of DNA a tight binding site for holoenzyme (6) that effectively fixes that end to an enzyme molecule on the surface (Fig. 1B). This new arrangement increases the frequency of formation of sliding complexes by other enzyme molecules, enlarging their contribution to the movements of the bead. A drawback is that the consequence of the putative groove tracking is no longer a unidirectional macroscopic rotation, although the tracking rotates the DNA macroscopically in the same direction in either case. In the new arrangement, after the sliding complex breaks down, the bead will rotate macroscopically in the opposite direction, driven by the twisting and supercoiling that have been accumulated in the DNA segment between the end fixed on the surface and the final position of the sliding complex. Therefore, the consequence of the putative groove tracking is alternating macroscopic rotational motions, rather than a unidirectional one. We here call the index of such macroscopic motions “rotational coherence.”
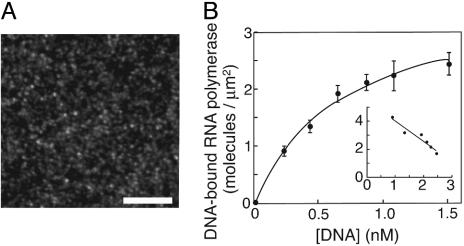
Homogeneity of Samples for Statistical Analysis. Because the putative macroscopic motions are superimposed on random Brownian motions, the index must be reliably evaluated by averaging signals based on statistics, using homogeneously active polymerase molecules. To check the molecules' homogeneity, we examined their affinities for the high-affinity site on a 4-kbp DNA (6). The individual bound DNA molecules were visualized by staining and then counted (Fig. 2A). The highest concentration of DNA used in this experiment was 1.5 nM, because individual DNA molecules are no longer observable as discrete spots at higher concentrations. The numbers of DNA-bound holoenzyme molecules per unit area fitted a curve consistent with a simple hyperbolic dependence on DNA concentrations (Fig. 2B), as predicted for a simple binding equilibrium between two homogeneous components. This is the mass-action law in a single-molecule version, although the measured dissociation constant may be different from that in a perfect solution. [Such a difference from solution has been observed in measurements with surface plasmon resonance (9).] The mean distance between nearest-neighbor molecules, assuming the closest packing, was calculated from the saturation level to be 0.6 μm, large enough to avoid steric hindrance in DNA binding.
Fig. 2.
Homogeneous DNA-binding activity of the immobilized holoenzyme. (A) Anepifluorescence image of DNA harboring the high-affinity site bound to the fixed holoenzyme. The linearized plasmid pRLG3749 was injected into the chamber on which the holoenzyme had been fixed, incubated for 20 min, and washed with the casein-containing buffer A. The bound DNA molecules were stained with YOYO-1 (Molecular Probes). (Scale bar, 10 μm.) (B) The densities of DNA-bound holoenzyme molecules. The curve is the best-fit hyperbola, C[DNA]/(Kd + [DNA]), where C is the density of active holoenzyme and Kd is the dissociation constant. An excellent fit was confirmed by the linearity of the Scatchard-type plot shown in Inset (DNA-bound RNA polymerase)/[DNA] against (DNA-bound RNA polymerase) in the units of the main graph, which would tend to exaggerate any deviation from a hyperbola. The calculated saturation level, C, was 3.5 ± 0.2 molecules per μm2, and the dissociation constant, Kd, was 0.6 ± 0.1 nM.
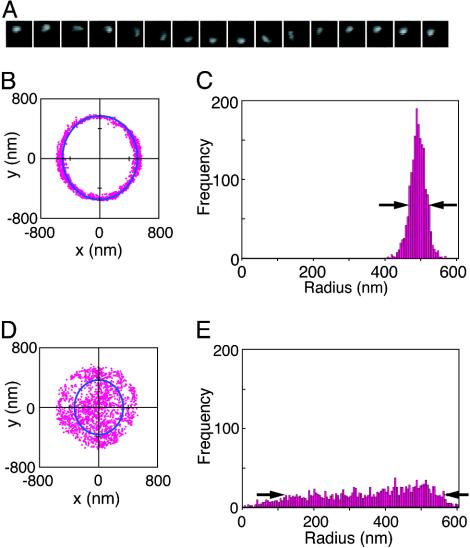
Indices of Rotational Motion. Fig. 3 shows the analysis of the movements in a standard system and a control system lacking DNA, and their rotational movements are shown in Movies 1 and 2, respectively, which are published as supporting information on the PNAS web site. The x–y coordinate of the center of the fluorescent sphere was determined and plotted for 1 min in the x–y plane in Fig. 3 A and C for the standard and control systems, respectively. Rotation of the beads, if any, was generally observed as an oval trace, because the rotational axis was not necessarily parallel to the light axis. The x–y coordinates were best fitted to an oval and converted into polar coordinates, r–θ, on the plane where the oval becomes a circle. The narrower the distribution of r values, the more well defined is the axis (Fig. 3 B and D). Therefore, the half-width of the histogram was used as the first index to measure how well the movements of the bead approximated to rotation around a fixed axis. The rotational coherence is large when θ repeats continuous increase and decrease but small when θ rapidly fluctuates. Because our definition of rotational coherence was somewhat arbitrary, we tested nine different definitions (see Materials and Methods) but obtained essentially the same results.
Fig. 3.
Indices of the movements of the beads in two cases. (A) A series of microscopic images of a fluorescent sphere; an example in the standard system covering 1.1 s. (B and D) The 1,800 coordinates (x, y) of the fluorescent sphere during 1 min in the standard system (B) and a control system (D). The movements are shown in Movies 1 and 2. The solid lines are the best-fit ovals. (C and E) Distributions of the polar coordinate r for B and D, respectively. The half-widths are shown by the arrows.
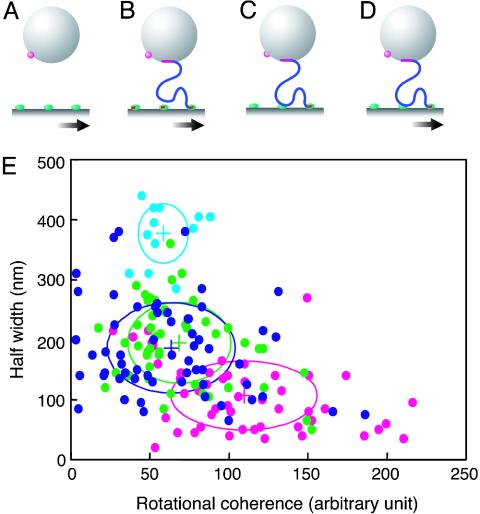
Observed Rotational Movements of Beads. We prepared three control systems lacking essential substances or perturbation. The first control system (Fig. 4A) had no DNA but was otherwise the same as the standard system (Fig. 4D), including the movement of the surface, which displayed only Brownian motion. In the second control (Fig. 4B), we inhibited the formation of sliding complexes by adding either 0.1 mg/ml heparin or 100 nM 150-bp DNA harboring the high-affinity site to the otherwise standard system. The observed effects of the two inhibitors were indistinguishable, so the results were pooled into a single category. The third control system was as the standard, except that the surface was not moved (Fig. 4C).
Fig. 4.
Values of two indices obtained for the three controls and the standard system. (A) The control with no DNA. (B) The control with inhibition of formation of the sliding complex. (C) The control with no movement of the surface. (D) The standard system. (E) The values of the half-width and of the rotational coherence are plotted for all of the measurements. The crosses denote the average values for the controls (A, sky blue; B, green; C, blue) or the standard system (D, magenta). The ovals denote the standard deviations of the indices. Note that the average values for the standard system are outside all three control ovals, indicating that the movement in that system is statistically distinct from that in the controls.
The two indices were determined for each experiment by using a control or the standard system and are plotted in Fig. 4E. In the first control, lacking any DNA tether, rotational Brownian motion of the beads gave a large half-width, as expected. In the second control, lacking any sliding complex but including tethering with DNA, the half-width decreased 2-fold, but the rotational coherence was as small as in the first control. Therefore, the tethering provided a more definite axis of rotation by reducing its spherical symmetry but did not induce any macroscopic rotation. The half-width was further decreased in the third control system, lacking movement of the surface. This reduction of the half-width suggests that the formation of a sliding complex shortened the length of the linker, further increasing axial symmetry. It should be noted that the mere formation of a sliding complex did not increase the rotational coherence, because the rotation was still driven only by incoherent Brownian motion. This result shows that the rotational coherence does not reflect the putative torque generated by microscopic bidirectional sliding. The standard system included the movement of the surface and showed a rotational coherence distinctly larger than that of any of the controls, as expected if the enzyme is tracking a DNA groove. The standard system showed the smallest half-width, probably because the axial symmetry of the bead was increased by a bulk flow induced by the surface movement. In conclusion, these results provide evidence for the existence of groove tracking by RNA polymerase during its sliding along DNA.
Discussion
In the single-molecule experiments, where each result reflects a different molecule, the results obtained become relevant only after statistical analysis. Statistical analysis requires a homogeneous population of observed molecules, although examination of homogeneity is often neglected. In this study, the promoter-binding activity of the holoenzyme was shown to be homogenous, and homogeneity of tethering was ensured by checking in situ before measurements. Therefore, statistical analysis using all data, without subjective exclusion, would make the obtained conclusion significant.
The use of groove tracking for scanning DNA sequences has been a controversial issue: Its employment by restriction enzymes was proposed from the kinetics of cleavage at sites with a juxtaposed site or star site (10, 11), as well as from a calculation by molecular dynamics (12), but negative kinetic evidence was also reported (13). Here we provide direct evidence for its existence. Such tracking could be a universal mechanism of searching for specific sequences. From this viewpoint, it is interesting that the rotary motor used by phage φ29 for packaging DNA into its capsid, where sequence recognition is not required, has an aperture whose size suggests interaction with the helical row of phosphate residues rather than with a groove (14).
Strictly speaking, this work provides evidence for the helical movement that is consistent with tracking a groove. Holoenzyme molecules read sequences mostly during their 1D diffusion along DNA, because their binding to promoters is always preceded by such diffusion, not by direct access from bulk (4). The inclusion of helical movement in 1D diffusion thus can naturally be interpreted as tracking a groove that allows the reading. In our full system, the macroscopic movement of holoenzyme relative to a bead was kept slow (8 nm/s), because the resultant tension along the DNA should not stretch it enough to change its internal and external structures. When the holoenzyme keeps tracking for 1 s [estimated to be the mean lifetime of a sliding complex (8)], DNA should be rotated by 2.3 turns. However, the beads do not rotate to this extent, because DNA is not a solid rod. In our experimental condition, DNA behaves like a thin rubber cord: The torque generated at a site is mostly consumed in making loops of the cord, and only a small portion is transmitted to the bead (15). DNA fragments too short (150 bp or less) to form a loop could transmit torque to the bead with less attenuation. However, an experimental system in which such short DNA is used is not practicable, because sliding complexes would rarely form on such a short DNA and the bead would be in danger of contracting the holoenzyme and surface directly.
By using DNA molecules fixed in parallel, we previously showed that fluorescently labeled polymerase molecules driven by flow frequently moved in parallel to the DNA rather than to the bulk flow (4). Recently, some have tried to dismiss this evidence of sliding as a hydrodynamic artifact (16). However, any such artifact was already explicitly excluded in the work (4): The movement assigned as sliding was absent when the polymerase molecules were prebound to a short DNA fragment or to heparin or when they were replaced by small fluorescent particles. What we can now deny, but could not previously, is the possibility that what we had interpreted as sliding events could all be consecutive intersegment transfer. There is evidence of intersegment transfer, sometimes called direct (17) or 3D (18) transfer, during 1D diffusion along DNA by several proteins possessing two DNA-binding sites (3). However, the existence of intersegment transfer has been illogically interpreted as evidence against sliding (16). In this context, the present results that indicate helical sliding do not exclude its coexistence with nonhelical sliding or intersegment transfer (1, 2).
As a DNA-binding protein, holoenzyme, but not a core enzyme lacking the σ70 subunit, recognizes promoter sequences. Region 4.2 of σ70 has been suggested to interact with the –35 sequence on double-stranded DNA (19), and in the x-ray structural model (20, 21), the helix–turn–helix motif of region 4.2 is in the major groove at the site. In a sliding complex, this motif could be a part of the notch generating the observed helical motion between holoenzyme and nonspecific DNA segments. If so, this mechanism of sliding would enable RNA polymerase to continuously scan sequences of DNA and, presumably, first recognize the –35 promoter element, a prediction that can be tested in future work.
The helical movements by protein molecules relative to DNA clarify the existence of groove tracking during their sliding along DNA. This finding suggests not only how they search for their specific sequences but also the importance of their structural interaction with nonspecific sites. A sliding complex must maintain a structure where a part of a protein is always kept in the groove in an arrangement that enables the protein to check base sequences. Such a fine configuration most likely is realized by structural, rather than electrostatic, interaction. In conclusion, the finding of DNA groove tracking not only consolidates evidence for the existence of sliding but may also have wider biological significance.
Supplementary Material
Acknowledgments
We thank Dr. R. S. Hayward (Edinburgh University, Edinburgh) and Dr. J. Tomizawa (National Institute of Genetics) for useful comments and critical reading of the manuscript. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to N.S. and K.S.-S.) and the Hayashi Memorial Foundation for Female Natural Scientists (to K.S.-S.).
Author contributions: K.S.-S. and N.S. designed research; K.S.-S. performed research; N.S. and K.S.-S. contributed new reagents/analytic tools; N.S. and K.S.-S. analyzed data; and N.S. and K.S.-S. wrote the paper.
References
- 1.Berg, O. G., Winter, R. B. & von Hippel, P. H. (1981) Biochemistry 20, 6929–6948. [DOI] [PubMed] [Google Scholar]
- 2.von Hippel, P. H. & Berg, O. G. (1989) J. Biol. Chem. 264, 675–678. [PubMed] [Google Scholar]
- 3.Shimamoto, N. (1999) J. Biol. Chem. 274, 15293–15296. [DOI] [PubMed] [Google Scholar]
- 4.Kabata, H., Kurosawa, O., Arai, I., Washizu, M., Margarson, S. A., Glass, R. E. & Shimamoto, N. (1993) Science 262, 1561–1563. [DOI] [PubMed] [Google Scholar]
- 5.Guthold, M., Zhu, X., Rivetti, C., Yang, G., Thomson, N. H., Kasas, S., Hansma, H. G., Smith, B., Hansma, P. K. & Bustamante, C. (1999) Biophys. J. 77, 2284–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaal, T., Ross, W., Estrem, S. T., Nguyen, L. H., Burgess, R. R. & Gourse, R. L. (2001) Mol. Microbiol. 42, 939–954. [DOI] [PubMed] [Google Scholar]
- 7.Kubori, T. & Shimamoto, N. (1996) J. Mol. Biol. 256, 449–457. [DOI] [PubMed] [Google Scholar]
- 8.Singer, P. T. & Wu, C.-W. (1988) J. Biol. Chem. 263, 4208–4214. [PubMed] [Google Scholar]
- 9.Schuck, P. (1997) Annu. Rev. Biophys. Biomol. Struct. 26, 541–566. [DOI] [PubMed] [Google Scholar]
- 10.Jeltsh, A., Alves, J., Wolfes, H., Maass, G. & Pingoud, A. (1994) Biochemistry 33, 10215–10219. [DOI] [PubMed] [Google Scholar]
- 11.Berkhout, B. & van Wamel, J. (1996) J. Biol. Chem. 271, 1837–1840. [DOI] [PubMed] [Google Scholar]
- 12.Sun, J., Viadiu, H., Aggarwal, A. K. & Weinstein, H. (2003) Biophys. J. 84, 3317–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kampmann, M. (2004) J. Biol. Chem. 279, 38715–38720. [DOI] [PubMed] [Google Scholar]
- 14.Guasch, A., Pous, J., Ibarra, B., Gomis-Rüth, F. X., Valpuesta, J. M., Sousa, N., Carrascosa, J. L. & Coll, M. (2002) J. Mol. Biol. 315, 663–676. [DOI] [PubMed] [Google Scholar]
- 15.Strick, T., Allemand, J.-F., Croquette, V. & Bensimon, D. (2000) Prog. Biophys. Mol. Biol. 74, 115–140. [DOI] [PubMed] [Google Scholar]
- 16.Halford, S. E. & Szczlkum, M. D. (2002) Eur. Biophys. J. 31, 257–267. [DOI] [PubMed] [Google Scholar]
- 17.Hsieh, M. & Brenowitz, M. (1997) J. Biol. Chem. 272, 22092–22096. [DOI] [PubMed] [Google Scholar]
- 18.Gowers, D. M. & Halford, S. E. (2003) EMBO J. 22, 1410–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.deHaseth, P. L., Zupancic, M. L. & Record, M. T., Jr. (1998) J. Bacteriol. 180, 3019–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Campbell, E. A., Muzzin, O., Chlenov, M., Sun, J. L., Olson, C. A., Weinman, O., Trester-Zedlitz, M. L. & Darst, S. A. (2002) Mol. Cell 9, 527–539. [DOI] [PubMed] [Google Scholar]
- 21.Murakami, K. S., Masuda, S., Campbell, E. A., Muzzin, O. & Darst, S. A. (2002) Science 296, 1285–1290. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.