Abstract
In Drosophila, the product of the fs (2)cup gene (Cup) is known to be crucial for diverse aspects of female germ-line development. Its functions at the molecular level, however, have remained mainly unexplored. Cup was found to directly associate with eukaryotic translation initiation factor 4E (eIF4E). In this report, we show that Cup is a nucleocytoplasmic shuttling protein and that the interaction with eIF4E promotes retention of the Cup protein in the cytoplasm. Cup is required for the correct accumulation and localization of eIF4E within the posterior cytoplasm of developing oocytes. We furthermore show that cup and eIF4E interact genetically, because a reduction in the level of eIF4E activity deteriorates the development and growth of ovaries bearing homozygous cup mutant alleles. Our results reveal a crucial role for the Cup–eIF4E complex in ovary-specific developmental programs.
Keywords: ovary development, translational regulation, oogenesis
The mechanisms directing germ-line formation and maintenance, just like those controlling gonad development, are essential for metazoan reproduction. In Drosophila melanogaster and many other organisms, primordial germ cell determination takes place early in embryogenesis through the segregation of maternally inherited germ-line factors to a specialized region of cytoplasm, named germ plasm (1). Primordial germ cells subsequently invaginate into the embryo and migrate to the gut to reach the mesoderm where the somatic gonadal precursor cells are located.
In female flies, an ovary consists of 14–16 ovarioles each made of a germarium and a series of developing oocytes (2). The asymmetric division of each germ-line stem cell contained within the germarium gives rise to a new stem cell and a daughter cell that becomes first a cystoblast, then a 16-cell cyst. Fifteen of these cells develop into nurse cells, and the most posterior one develops into an oocyte.
Genetic screens in Drosophila have identified essential genes involved in oogenesis. A key player during Drosophila female germ-line development is the product of the fs (2)cup gene (Cup). Cup is required in oogenesis to assure cyst formation, germ-line chromosome structure, oocyte development, and growth (3). cup RNA and protein are found in the cytoplasm of all germ-line cells but are not detectable inside nurse cell or oocyte nuclei or in the somatic follicle cells (3, 4). cup mutant ovaries display aberrant nurse cell nuclear morphology and immature egg chambers, whose development arrests between stages 5 and 14 (3).
cup genetically interacts with ovarian tumor (otu) to attain normal egg chamber formation and growth (3). We have previously shown that Cup also directly interacts with Nanos (Nos) to promote normal development and survival of the female germ-line stem cells (4). However, the functions of Cup at the molecular level essentially have remained unexplored.
We performed a yeast two-hybrid screen for factors that could either mediate or modulate Cup activity during ovarian development and isolated the product of the eukaryotic translation initiation factor 4E (eIF4E) gene as a specific interactor. In this report, we show that Cup is able to shuttle between the nucleus and the cytoplasm and that this process is blocked by its interaction with eIF4E. Cup colocalizes with eIF4E to the posterior cytoplasm of developing oocytes and is required for the correct accumulation and localization of eIF4E within the oocyte. cup and eIF4E furthermore interact genetically to control ovary development and growth. Our results demonstrate that the interaction between the general translation initiation factor eIF4E and an organ-specific interactor is essential to achieve the correct level of translational activity required for the normal development of an organ.
Materials and Methods
Yeast Interaction Assays. A GAL4 transcriptional activation domain (AD) fusion library (5) was screened by using the full-length Cup protein (amino acids 2–1,132) as bait (pGBT9-Cup). eIF4E cDNA fragments were tested against the GAL4 DNA-binding domain (DBD) Cup fusion used to screen the cDNA library. The fragment encoding the AD–Cup100–MEBS mutant protein was generated by PCR. Cup cDNA fragments, cloned into pACT2 (4), were tested against a DBD-eIF4E fusion encoding amino acids 89–259. Protein–protein interactions were assessed by streaking transformants on selective media lacking Leu, Trp, and His with the addition of 3–10 mM 3-aminotriazole.
In Vitro Binding Assays and Phosphatase Treatment. Protein extracts from ovary pairs were obtained as described in ref. 6. Pull-down binding reactions were performed as described in ref. 4 by incubating ovarian extracts with 5 μg of either GST or GST-eIF4E. To obtain GST-eIF4E, a NotI-filled fragment containing amino acids 2–259 of eIF4E was cloned into pGEX-2T digested with SmaI. Cup was detected in a Western blot with an α-Cup antibody raised against amino acids 211–501 (4).
For coimmunoprecipitations, proteins were obtained by in vitro transcription/translation and incubated with α-hemagglutinin (-HA) or α-Cup antisera as described in ref. 4. Protein G Sepharose (Amersham Pharmacia) was added and incubated at 4°C for 1 h. Beads were washed, and eluted proteins were subjected to SDS/PAGE and visualized by autoradiography. The fragment encoding amino acids 199–399 of eIF4G (eIF4G-200) was generated by PCR.
Ovaries from 3- to 5-day-old flies were lysed in 50 mM Tris, pH 8.0/150 mM NaCl/5% glycerol/1% Nonidet P-40 supplemented with protease inhibitors (Roche, Gipf-Oberfrick, Switzerland) in the presence or absence of phosphatase inhibitors (5 mM NaF/0.1 mM Na3VO4/50 mM β-glycerophosphate). Protein extracts were incubated with 20 units of alkaline phosphatase (New England Biolabs) for 1 h at 37°C. The eIF4E protein was detected in a Western blot with an α-eIF4E antibody (7).
Drosophila Strains. cup21 and cup8 are described in refs. 3 and 4; eIF4E07238 is described in ref. 7 and was provided by the Bloomington Stock Center at Indiana University.
Expression Constructs, Transfections, and Immunocytochemistry. For expression in Drosophila Schneider (S2) cells, the cDNAs encoding full-length eIF4E (as a blunted EcoRI-AseI fragment) and Cup (as a blunted ScaI-XhoI fragment) were cloned into the EcoRV site of the pAC5c expression vector. Drosophila S2 cells were grown in Schneider's Drosophila medium (GIBCO) supplemented with 10% FCS and antibiotics. Cells were transiently transfected with 5 μg of the various expression constructs by CaPO4 precipitation. Leptomycin B (LMB) treatment of S2 cells was performed as described in ref. 8.
α-Cup, α-eIF4E, and α-Orb antibodies were described in refs. 4, 7, and 9, respectively. Mouse α-HA tag (Santa Cruz Biotechnology) and α-rabbit, α-mouse, or α-rat FITC- or tetramethylrhodamine isothiocyanate-conjugated secondary antibodies (Sigma) were used according to the manufacturer's instructions. For S2 immunocytochemistry, cells were treated as described in ref. 8. For ovary immunocytochemistry, ovaries were treated by standard methods. Nuclei were visualized by staining with Hoechst 33258 (Sigma) or TOTO-3 (Molecular Probes).
Results
Cup Interacts with eIF4E. To identify factors interacting with Cup during ovarian development, we performed a yeast two-hybrid screen. The full-length Cup protein fused to the GAL4 DBD was used as bait to screen a Drosophila GAL4 AD fusion cDNA library. We isolated a total of 24 potential interactors, 11 of which were revealed to be different portions of the eIF4E protein (Fig. 1A). The alignment of the different eIF4E clones shows that the distal carboxyl-terminal part of eIF4E (residues 89–259) is the region responsible for efficient binding to full-length Cup.
Fig. 1.
Cup interacts with eIF4E in yeast and in vitro. (A) eIF4E clones isolated from the yeast two-hybrid screen. Interaction of the various isolated eIF4E fragments (amino acids are indicated on the left) with full-length Cup was determined by streaking transformants on selective media containing 10 mM 3-aminotriazole. (B) eIF4E binds to ovary-derived Cup. GST-eIF4E and GST proteins were challenged with wild-type fly ovary extracts; retained proteins were eluted and separated by SDS/PAGE. An arrow indicates the Cup protein (≈145 kDa).
To validate the interaction between Cup and eIF4E observed in yeast, we used a GST pull-down approach on wild-type ovarian extracts. A GST fusion protein containing amino acids 2–259 of eIF4E (GST-eIF4E) retained ≈5% of endogenous Cup (Fig. 1B). Cup was not retained by GST alone. The binding specificity also was confirmed by the lack of interaction between eIF4E and full-length Nos in a yeast two-hybrid assay (data not shown).
Cup Is a Nucleocytoplasmic Shuttling Protein and Colocalizes with eIF4E in the Cytoplasm of S2 Cells. Because eIF4E is partially localized to the nuclei of mammalian and yeast cells (10, 11), we verified whether Drosophila eIF4E and Cup shuttle between the nucleus and the cytoplasm. We transfected S2 cells with expression constructs for the two proteins. Immunostainings using an α-Cup antibody showed that Cup is exclusively cytoplasmic in S2 cells (Fig. 2). Similarly, eIF4E was found mainly in the cytoplasm. Immunostaining and analysis by confocal microscopy of S2 cells coexpressing Cup and eIF4E showed that the two proteins colocalize in the cytoplasm (Fig. 2).
Fig. 2.
Cup is a nucleocytoplasmic shuttling protein. The upper two rows show that Cup and eIF4E colocalize to the cytoplasm of expressing Drosophila S2 cells. The lower two rows show that nuclear relocalization of Cup is blocked by eIF4E. Transfected S2 cells, with or without LMB treatment, were stained by immunofluorescence with α-Cup and α-HA (detecting HA-tagged eIF4E) antibodies and analyzed by confocal microscopy. Colocalization of Cup and eIF4E appears in yellow. Nuclei were stained with Hoechst.
Transfected S2 cells then were treated with LMB, which selectively blocks nuclear export mediated by the exportin1/CRM1 receptor (12). As shown in Fig. 2, LMB induced nuclear relocalization of most of the Cup protein, whereas it induced only a partial relocalization of eIF4E. LMB-treated S2 cells coexpressing eIF4E and Cup, conversely, displayed reduced Cup nuclear relocalization when compared with cells expressing Cup alone (Fig. 2). Thus, a fraction of Cup appears to be still colocalized with eIF4E in the cytoplasm and perinuclear region upon LMB treatment, suggesting that Cup is retained by eIF4E in the cytoplasm.
Taken together, these data indicate that Cup, known to be transiently in contact with the periphery of the nurse cell nuclei (3), and eIF4E are associated in the cytoplasm and that this association blocks the nucleocytoplasmic shuttling of Cup.
Cup and eIF4E Proteins Colocalize to the Posterior Cytoplasm of Developing Oocytes. To test the colocalization of Cup and eIF4E during oogenesis, we performed double immunofluorescent antibody stainings. In wild-type (3) and cup21/+;eIF4E07238/+ (this work) ovaries, Cup was found throughout the cytoplasm of all germ-line cells starting in the germarium and appeared to be excluded from the nuclei of either nurse cell or oocyte and from the follicular cells (Fig. 3 A, B, E, G, and I). During oogenesis, the distribution of Cup inside 16-cell cysts changes remarkably: It accumulates preferentially in the cytoplasm at the posterior end of the forming oocyte (as early as region 2a of the germarium and up to stage 10). On the contrary, in nurse cells, Cup becomes progressively cytoplasmic. Interestingly, eIF4E accumulates and colocalizes with Cup to the posterior cytoplasm of the developing oocytes. The colocalization of the two proteins is evident also in the nurse cells, where eIF4E migrates to the internal borders of the cells (Fig. 3 A and B). In contrast to Cup, eIF4E appears to concentrate also in the follicular cells (Fig. 3 A, B, E, G, and I).
Fig. 3.
Cup and eIF4E colocalize to the posterior cytoplasm in growing oocytes. (A, B, and E–J) Confocal microscopy of cup21/+;eIF4E07238/+ (A, B, E, G, and I) and cup21/cup21 (F, H, and J) egg chambers to visualize eIF4E (green), Cup (red), and colocalization (yellow). Both cup21/cup21 and cup21/cup21;eIF4E07238/+ ovaries show comparable expression of the two proteins. (A and B) Germarium and early egg chambers, respectively. Protein enrichment can be seen mainly in the developing oocyte. (E and F) Stage 3 egg chambers. (G and H) Stage 5 egg chambers. (I and J) Stage 7 egg chambers. Arrowheads indicate the oocytes. Similar images were obtained with single antibodies. (C and D) Stage 8 cup21/+;eIF4E07238/+ and cup21/cup21 egg chambers, respectively, immunostained with α-eIF4E and α-Orb to mark the developing oocyte.
We then analyzed the localization of the Cup and eIF4E proteins in either cup21/cup21 or cup21/cup21; eIF4E07238/+ egg chambers. Cup levels were reduced drastically in all ovarian compartments (Fig. 3 F, H, and J). eIF4E, conversely, showed a specific clearing from the forming oocytes, whereas its localization inside nurse cells and the other cellular compartments remained substantially unaltered (Fig. 3 F, H, and J). This result indicates that Cup is required for the posterior localization of eIF4E within the developing oocyte. Confocal microscopy of double immunostainings with α-eIF4E and α-Orb antibodies (9, 13) confirmed that eIF4E localizes within the posterior cytoplasm of cup21/+;eIF4E07238/+ forming oocytes, whereas it clears from cup21/cup21 forming oocytes (Fig. 3 C and D). The analysis of multiple focal planes furthermore confirmed that the eIF4E signal is not present in a different focal plane within the oocytes in a cup21 homozygous mutant background.
Cup Binds eIF4E and Nos by Means of Distinct Regions and Contains a Functional eIF4E-Binding Motif. To define the minimal region of Cup required for interaction with eIF4E, we tested 10 different terminal deletions in yeast. Amino acids 342–453 (hereafter referred to as Cup100) represent the shortest fragment of Cup examined capable of binding eIF4E, thus identifying a functional domain lying upstream of the Nos-binding region (amino acids 593–962) (Fig. 4A). Expression of all Cup deletions was verified by Western blots on yeast extracts (4).
Fig. 4.
Cup interacts with eIF4E (4E) through an eIF4E-binding motif. (A) Cup deletions tested in a yeast two-hybrid assay. Growth on selective media containing 4 mM 3-aminotriazole (+) is indicated on the right. Amino acids 342–453 (Cup100) represent the minimal region of Cup interacting with eIF4E (black box). The striped box indicates the Nos-binding domain (amino acids 593–962) (4). (B) Cup100 interacts with eIF4E in a GST pull-down assay. Retained proteins were eluted and subjected to SDS/PAGE. (C) Coimmunoprecipitation assay using an α-HA antibody of 35S-Met-labeled Cup100 and HA-tagged eIF4E produced in reticulocyte lysates. Cup100 coimmunoprecipitates with HA-eIF4E (Cup100 + 4E). (Right) Control immunoprecipitations with α-HA antibody show specific immunoprecipitation of HA-eIF4E but not of Cup100. (D) Yeast two-hybrid assay using Cup100 (wild-type) and Cup100-MEBS (mutant). Yeast transformants were streaked on selective media containing 4 mM 3-aminotriazole. AD, GAL4 AD; DBD, GAL4 DBD.
We next tested 35S-Met-labeled Cup100 for interaction with eIF4E via GST pull-down. A GST-eIF4E fusion retains ≈50% of the Cup-100 protein (Fig. 4B). We also performed coimmunoprecipitations by using 35S-Met-labeled HA-tagged full-length eIF4E and Cup100, produced in reticulocyte lysates. By using an α-HA antibody, eIF4E is able to specifically coimmunoprecipitate Cup100 (Fig. 4C).
Because the eIF4E-interacting region within Cup was found to be distinct from the Nos-binding domain, we tested in yeast whether a trimeric Nos–Cup–eIF4E complex could be assembled. Cup (amino acids 29–962) was coexpressed with either full-length Nos fused to the GAL4 AD (AD–Nos) or eIF4E fused to the GAL4 DBD (DBD–eIF4E), or vice versa. No interaction was detected between AD-Nos and DBD–eIF4E (or DBD–Nos and AD–eIF4E), and no interaction was observed between the two couples of fusion proteins in the presence of Cup (data not shown), suggesting that Cup cannot form a trimeric complex with Nos and eIF4E in yeast. Expression of all fusion constructs used was verified by Western blots of yeast extracts (data not shown).
Cup100 contains a potential eIF4E-binding motif (YTRSRLM, residues 342–348) (Fig. 4D), matching the consensus YXXXXLΦ (where X is any amino acid and Φ is a hydrophobic one) (14). To test the functionality of the binding motif, three amino acids that are conserved among all eIF4E-binding motifs were changed into Ala residues (Cup100–MEBS). Cup100–MEBS failed to interact with eIF4E in a yeast two-hybrid assay (Fig. 4D). Immunoblots on yeast extracts showed that Cup100 and Cup100–MEBS were expressed at similar levels (data not shown).
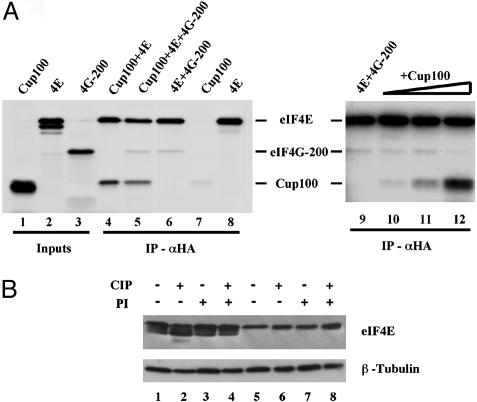
Cup Competes with Eukaryotic Translation Initiation Factor 4G (eIF4G) for Binding to eIF4E. eIF4G is an eIF4E-binding protein that positively regulates cap-dependent translation initiation. To test whether Cup competes with eIF4G for eIF4E binding, we coimmunoprecipitated HA-tagged full-length eIF4E and a portion of eIF4G (eIF4G-200), including its eIF4E-binding motif, in the presence of increasing amounts of Cup100. Cup100 was able to antagonize the binding of eIF4G-200 to eIF4E. A 13× molar excess of Cup 100 added to eIF4E and eIF4G-200 reduced the amount of coimmunoprecipitated eIF4G-200 by 45% (Fig. 5A, lane 12). Repetitions of this experiment gave virtually identical results to and confirmed those obtained by Nelson et al. (15). We tested in addition the ability of an excess of eIF4G-200 to antagonize Cup100 binding to eIF4E. eIF4G-200 antagonized the binding of Cup100 to eIF4E. A 2.5× molar excess of eIF4G-200 added to Cup100 and eIF4E reduced the amount of coimmunoprecipitated Cup100 by 25% (Fig. 5A, lane 5).
Fig. 5.
Cup modulates eIF4E functions. (A) Cup competes with eIF4G (4G-200) for binding to eIF4E (4E). Coimmunoprecipitation assays using an α-HA antibody on HA-tagged eIF4E, Cup100, and a portion of eIF4G produced in reticulocyte extracts. A 2.5× molar excess of eIF4G added to Cup100 and eIF4E reduced the amount of bound Cup100 (lane 5) by 25%; reciprocally, a 1×,3×, and 13× molar excess of Cup100 added to eIF4E and eIF4G-200 reduced the amount of bound eIF4G-200 (lanes 10–12) by a maximum of 45%. Lanes 6–9, control immunoprecipitations. Densitometry was performed by using image-quant software. (B) Effect of phosphatase treatment on the mobility of eIF4E isoforms. Wild-type (lanes 1–4) and cup21/cup21 (lanes 5–8) ovary extracts were treated with calf intestinal phosphatase (CIP) in the absence (lanes 2 and 6) or presence (lanes 4 and 8) of phosphatase inhibitors (PI). Samples were analyzed by Western blot with α-eIF4E antibody. α-β-Tubulin antibody was used as loading control.
The Expression of eIF4E Isoforms Is Changed Within cup Mutant Ovaries. We compared the phosphorylation status of eIF4E in wild-type and cup21 homozygous flies by Western blot on protein ovary extracts. In wild-type ovaries, eIF4E displays three bands (Fig. 5B, lane 1), which were previously shown to correspond to different isoforms of the protein (7). In homozygous cup21 ovaries, conversely, we could detect mainly the slower-migrating band, corresponding to the eIF4EI isoform (7), which was present at lower levels than in wild-type ovaries (Fig. 5B, lane 5). Phosphatase treatment of wild-type extracts led to an increase in the abundance of the faster migrating band (Fig. 5B, compare lanes 1 and 2) and the appearance of an additional band of a lower molecular weight. Phosphatase treatment of cup21/cup21 ovary extracts caused only minor changes in the mobility of eIF4E (Fig. 5B, compare lanes 5 and 6), suggesting a reduction in the fraction of phosphorylated eIF4E. The addition of phosphatase inhibitors (Fig. 5B, lanes 3, 4, 7, and 8) prevented any alteration in the relative abundance of the bands.
cup and eIF4E Genetically Interact to Modulate Ovary Development and Growth. We next determined whether lowering the level of eIF4E altered any of the ovarian phenotypes associated with perturbed Cup function. The cup alleles behave like a continuous allelic series and fall into three major classes representing different grades of severity of the ovary phenotype (3). eIF4E07238 is a P element insertion within the large first intron of the gene and displays a homozygous lethal phenotype (7), whereas heterozygous eIF4E mutant alleles behave as wild-type flies. Two different cup alleles, cup21 (a weak class III allele) and cup8 (a strong class I allele), were tested in combination with eIF4E07238. Ovaries obtained from cup21/cup21;eIF4E07238/+ (Fig. 6 Upper) and cup8/cup8;eIF4E07238/+ (data not shown) flies are reduced in size when compared with ovaries derived from control flies (homozygous cup21 and cup8, respectively) of the same age and grown under identical conditions.
Fig. 6.
cup and eIF4E interact genetically to modulate ovary growth and development. (Upper) Nomarski photomicrographs (all at the same magnification) of ovaries dissected 3–5 days after eclosion. (Lower) Fluorescence photomicrographs (all at the same magnification) of Hoechst-stained ovarioles to visualize DNA. Note that flies carrying heterozygous cup21 or eIF4E07238 mutant alleles, as well as a transheterozygous combination of these two alleles, display normal ovaries.
To analyze the effect on ovary development of a reduction of eIF4E levels in homozygous cup21 mutant flies, we performed Hoechst stainings of egg chambers. The oogenesis defects displayed by the class III cup21 mutant allele are rendered more severe by lowering the level of eIF4E (Fig. 6 Lower). Indeed, in cup21/cup21;eIF4E07238/+ ovaries, the chromosome morphology remains almost unaltered, but egg chamber development is blocked at an earlier stage (around stage 6) than in homozygous cup21, thus phenocopying a class II cup mutant allele. In addition, the growth rate of the egg chambers is affected. Egg chambers have smaller sizes that are noticeable soon after budding from the germarium when compared with cup homozygous mutant alleles, suggesting that the genetic interaction is also specific for ovary growth. No other defects were observed in cup21/cup21;eIF4E07238/+ flies. Indeed, germaria derived from homozygous cup mutant flies with a heterozygous eIF4E gene dosage appeared normal, suggesting that the germ-line stem cells were viable and developed properly.
To confirm the specificity of the cup-eIF4E genetic interaction, we obtained cup21/cup21;nosRC/+ flies and performed Hoechst stainings on their ovaries. Nos is an established translational regulator (16), and the nosRC mutant allele was shown to specifically affect oogenesis (4, 17, 18). No difference was observed between cup21/cup21;nosRC/+ and cup21/cup21 ovaries (data not shown), thus demonstrating that it is not any gene function affecting ovarian development that alters the cup phenotype.
Discussion
In this report, we show that Cup functionally interacts with eIF4E, thus pointing to a crucial role for Cup in the control of translation initiation during ovary development.
Cup Interacts with eIF4E by Means of a Canonical eIF4E-Binding Motif. In agreement with recently published results, we found that Cup interacts in yeast and in vitro with eIF4E (15, 19, 20). eIF4E is the rate-limiting component for cap-dependent translation initiation and therefore represents the major target for translational control (21, 22).
Various positive and negative translational regulators, such as eIF4G (23), 4E-binding proteins (24, 25), maskin (26, 27), and Bicoid (28), control cap-dependent translation initiation by directly interacting with eIF4E through an eIF4E-binding motif having the consensus YXXXXLΦ (29). We identified Cup as an additional eIF4E-binding protein, confirming recently published results (15, 19, 20). Interestingly, the eIF4E-binding motif we identified within Cup and the region mediating the interaction with Nos are distinct and are 245 amino acids apart. However, Cup is apparently unable in yeast to interact simultaneously with Nos and eIF4E to form a ternary complex. Further work will be required to elucidate the potential significance of this finding.
Cup Is a Nucleocytoplasmic Shuttling Protein. In agreement with the identified protein–protein interactions between Cup and eIF4E, we found that Cup colocalizes with eIF4E to the cytoplasm in transiently expressing S2 cells. Treatment with LMB revealed that at least part of the Cup protein pool can enter the cell nucleus and is then efficiently exported to the cytoplasm through the CRM1/exportin 1 pathway.
eIF4E has been described to be also partially nuclear, but its nuclear functions are unknown (10, 11, 30). Accordingly, we detected eIF4E mainly, but not exclusively, within the cytoplasm in transfected S2 cells. Upon LMB treatment, eIF4E is only marginally relocalized to the nucleus, showing, as reported in ref. 11, that it does not enter efficiently into the nucleus. Strikingly, we found that coexpression of eIF4E with Cup significantly reduces the fraction of Cup that is relocalized to the nucleus upon LMB treatment, indicating that the formation of a Cup::eIF4E complex actually blocks Cup relocalization to the nucleus.
Within mammalian cells, the eIF4E-transporter protein (4E-T) was reported to mediate the nuclear import of eIF4E (29). Dostie et al. (29) have shown 4E-T to be a nucleocytoplasmic shuttling protein. Interestingly, whereas Cup shares a limited but high degree of similarity with a short segment of the 4E-T protein (amino acids 565–630, which span a portion of the Nos-binding domain), the nuclear localization signal and nuclear export signals found within 4E-T do not have an obvious counterpart within Cup. Moreover, in contrast with the Cup::eIF4E interaction, the 4ET::eIF4E association causes nuclear accumulation of eIF4E in the presence of LMB. Cup is therefore unlikely to act solely as a carrier for eIF4E nuclear import.
Cup Mediates the Proper Localization of eIF4E Within the Growing Oocyte. Cup and eIF4E colocalize mainly inside the cytoplasm of nurse cells and the posterior cytoplasm of forming oocytes. In ovaries derived from either cup21/cup21 or cup21/cup21;eIF4E07238/+ female flies, we observed a clearing of eIF4E from the posterior cytoplasm of the forming oocyte, confirming the results reported by Wilhelm et al. (19) on cup1/cup4506 egg chambers. These results show that Cup is required for the localization of eIF4E at the posterior end of the oocyte cytoplasm. We found that Cup affects both the total amount of eIF4E and its isoform expression profile within the ovary. Cup does not appear to be generally required for the localization of proteins within the developing oocyte because previous reports have shown that the distributions of Gurken (19, 20) and of a kinesin-β-galactosidase (20) fusion protein are normal within cup mutant egg chambers.
Genetic Interaction Between cup and eIF4E Is Required for the Correct Development of the Ovary. Data from different laboratories recently have identified Cup as an eIF4E-binding protein acting in localization-dependent translation: Wilhelm et al. (19) showed that Cup represses translation of oskar (osk) mRNA in the oocyte (reviewed in ref. 31); Nakamura et al. (20) showed that Cup acts together with Bruno (Bru) to repress osk mRNA translation in oogenesis; and Nelson et al. (15) showed that Smaug represses translation in the embryo by means of a Cup-dependent activity.
In this work, we show that by lowering the level of functional eIF4E, the ovary defects displayed by the cup21 mutant allele are aggravated. Cup is specifically required for the proper accumulation of eIF4E in the developing oocyte and plays a pivotal role(s) in modulating ovary-specific translation by directly interacting with eIF4E in competition with eIF4G.
Phosphorylation is one of the mechanisms by which eIF4E activity is controlled, because eIF4E hyperphosphorylation correlates with high translational activity (32, 33). The function of phosphorylated eIF4E was shown to be necessary for the proper growth and development of Drosophila (7). Our results suggest that Cup also may play a role in the control of the phosphorylation status of eIF4E within the developing ovary, because in cup21/cup21 ovaries the fraction of phosphorylated eIF4E appears to be reduced. Further work will be required to unravel the link between Cup function, eIF4E phosphorylation, and translation efficiency within the ovary.
In view of the recently reported data on the role of Cup in the translational control of specific mRNAs such as osk (reviewed in ref. 34) and of our results on the genetic interaction between Cup and eIF4E, a model can be proposed for the function of Cup within the ovary (Fig. 7, which is published as supporting information on the PNAS web site). Cup, through its interaction with eIF4E, was shown to modulate the localization and translational repression during transport of selected mRNAs (e. g., osk) at the posterior end of the oocyte (19). Transport of the mRNAs, and of eIF4E, would be ensured by means of a localization factor, such as Barentz (19). During transport, Cup would inhibit translation by antagonizing the interaction between eIF4G and eIF4E. Upon localization of the transcript, the interaction between eIF4E and Cup would be weakened, e.g., because of the action of a local trigger mechanism(s), thus allowing eIF4G to interact with eIF4E and translation to start. In this context, impairment of Cup function would lead to a dramatic decrease in the localization of the eIF4E protein and of the bound mRNAs at the posterior end of the oocyte. Indeed, in cup21/cup21 ovaries, we observed a clearing of eIF4E from the posterior oocyte, which also was noticed by Wilhelm et al. (19) in cup1/cup4506 ovaries, and a reduction in the overall eIF4E protein levels in the ovary. In accordance with this model, mutations in cup would cause premature translation and little posterior translation of the transported mRNAs, which was indeed observed by Wilhelm et al. (19) and Nakamura et al. (20) with osk mRNA.
In agreement with this scenario, an impairment in eIF4E function (e. g., the eIF4E07238 mutation) in the context of a cup mutant, by further reducing the amount of functional eIF4E and, hence, of the translation of the residual localized mRNAs, would lead to a worsening of the cup21/cup21 phenotype, as we indeed observed in cup21/cup21;eIF4E07238/+ ovaries. Because of the reduction in the overall levels of eIF4E within the ovary, it cannot be excluded that the translation of other mRNAs might be affected also, thus contributing to the growth defect phenotype. Moreover, our finding that Cup also possesses nucleocytoplasmic shuttling properties indicates the existence of an uncharacterized function(s) of Cup in the cell nucleus. These activities could be exerted in association with nuclear eIF4E (10, 11, 30). Consequently, part of the cup21/cup21;eIF4E07238/+ phenotype also could reflect an impairment of the nuclear functions of Cup and/or of Cup-eIF4E during ovary development.
Supplementary Material
Acknowledgments
We thank F. Salvatore, F. Blasi, and P. Buono for support, encouragement, and help; F. Graziani and S. Gigliotti [both at the Institute of Genetics and Biophysics, Consiglio Nazionale delle Ricerche (IGB-CNR), Naples] for providing α-Orb and α-β-tubulin antibodies, reagents, and suggestions and for critical reading of the manuscript; P. Lasko (McGill University, Montreal) for providing the α-eIF4E antibody and fly stocks; R. P. Wharton (Howard Hughes Medical Institute, Duke University, Durham, NC) for providing the cDNA library; K. Delaval (Department of Biological and Technical Science, San Raffaele Scientific Institute, Milan) for generating pGBT9-Cup; M. Chiariello and C. Iavarone for advice with the phosphatase experiment; and L. Muzio and S. Arbucci for assistance in microscopy. Confocal imaging was performed at the Biological and Technological Research Department of San Raffaele Scientific Institute and IGB-CNR. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro and Italian Telethon (to V.Z.).
Abbreviations: cup, fs (2)cup; eIF4E, eukaryotic translation initiation factor 4E; eIF4G, eukaryotic translation initiation factor 4G; S2, Schneider; AD, activation domain; DBD, DNA-binding domain; LMB, leptomycin B; HA, hemagglutinin.
References
- 1.Saffman, E. E. & Lasko, P. (1999) Cell. Mol. Life Sci. 55, 1141–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spradling, A. C. (1993) in The Development of Drosophila melanogaster, eds. Bate, M. & Arias, A. (Cold Spring Harbor Lab. Press, Plainview, NY), Vol. I, pp. 1–70. [Google Scholar]
- 3.Keyes, L. N. & Spradling, A. C. (1997) Development (Cambridge, U.K.) 124, 1419–1431. [DOI] [PubMed] [Google Scholar]
- 4.Verrotti, A. C. & Wharton, R. P. (2000) Development (Cambridge, U.K.) 127, 5225–5232. [DOI] [PubMed] [Google Scholar]
- 5.Dahanukar, A., Walker, J. A. & Wharton, R. P. (1999) Mol. Cell 4, 209–218. [DOI] [PubMed] [Google Scholar]
- 6.Kim-Ha, J., Kerr, K. & Macdonald, P. M. (1995) Cell 81, 403–412. [DOI] [PubMed] [Google Scholar]
- 7.Lachance, P. E., Miron, M., Raught, B., Sonenberg, N. & Lasko, P. (2002) Mol. Cell. Biol. 22, 1656–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kilstrup-Nielsen, C., Alessio, M. & Zappavigna, V. (2003) EMBO J. 22, 89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lantz, V., Chang, J. S., Horabin, J. I., Bopp, D. & Schedl, P. (1994) Genes Dev. 8, 598–613. [DOI] [PubMed] [Google Scholar]
- 10.Lejbkowicz, F., Goyer, C., Darveau, A., Neron, S., Lemieux, R. & Sonenberg, N. (1992) Proc. Natl. Acad. Sci. USA 89, 9612–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dostie, J., Lejbkowicz, F. & Sonenberg, N. (2000) J. Cell Biol. 148, 239–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolff, B., Sanglier, J. J. & Wang, Y. (1997) Chem. Biol. 4, 139–147. [DOI] [PubMed] [Google Scholar]
- 13.Christerson, L. B. & McKearin, D. M. (1994) Genes Dev. 8, 614–628. [DOI] [PubMed] [Google Scholar]
- 14.Strudwick, S. & Borden, K. L. (2002) Differentiation 70, 10–22. [DOI] [PubMed] [Google Scholar]
- 15.Nelson, M., Leidal, A. & Smibert, C. (2004) EMBO J. 23, 150–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sonoda, J. & Wharton, R. P. (1999) Genes Dev. 13, 2704–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhat, K. M. (1999) Genetics 151, 1479–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forbes, A. & Lehmann, R. (1998) Development (Cambridge, U.K.) 125, 679–690. [DOI] [PubMed] [Google Scholar]
- 19.Wilhelm, J., Hilton, M., Amos, Q. & Henzel, W. (2003) J. Cell Biol. 163, 1197–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura, A., Sato, K. & Hanyu-Nakamura, K. (2004) Dev. Cell. 6, 69–78. [DOI] [PubMed] [Google Scholar]
- 21.Mader, S. & Sonenberg, N. (1995) Biochimie 77, 40–44. [DOI] [PubMed] [Google Scholar]
- 22.McKendrick, L., Pain, V. M. & Morley, S. J. (1999) Int. J. Biochem. Cell Biol. 31, 31–35. [DOI] [PubMed] [Google Scholar]
- 23.Keiper, B. D., Gan, W. & Rhoads, R. E. (1999) Int. J. Biochem. Cell Biol. 31, 37–41. [DOI] [PubMed] [Google Scholar]
- 24.Raught, B., Gingras, A. C., Gygi, S. P., Imataka, H., Morino, S., Gradi, A., Aebersold, R. & Sonenberg, N. (2000) EMBO J. 19, 434–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachs, A. B. & Varani, G. (2000) Nat. Struct. Biol. 7, 356–361. [DOI] [PubMed] [Google Scholar]
- 26.Stebbins-Boaz, B., Cao, Q., de Moor, C. H., Mendez, R. & Richter, J. D. (1999) Mol. Cell 4, 1017–1027. [DOI] [PubMed] [Google Scholar]
- 27.Richter, J. D. & Theurkauf, W. E. (2001) Science 293, 60–62. [DOI] [PubMed] [Google Scholar]
- 28.Niessing, D., Blanke, S. & Jackle, H. (2002) Genes Dev. 16, 2576–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dostie, J., Ferraiuolo, M., Pause, A., Adam, S. A. & Sonenberg, N. (2000) EMBO J. 19, 3142–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lang, V., Zanchin, N. I., Lunsdorf, H., Tuite, M. & McCarthy, J. E. (1994) J. Biol. Chem. 269, 6117–6123. [PubMed] [Google Scholar]
- 31.Lasko, P. (2004) J. Cell Biol. 163, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhoads, R. E. (1993) J. Biol. Chem. 268, 3017–3020. [PubMed] [Google Scholar]
- 33.Gingras, A. C., Gygi, S. P., Raught, B., Polakiewicz, R. D., Abraham, R. T., Hoekstra, M. F., Aebersold, R. & Sonenberg, N. (1999) Genes Dev. 13, 1422–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Macdonald, P. M. (2004) Curr. Biol. 14, R282–R283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.