Abstract
KCNQ1 channel is a member of the voltage-gated potassium channel KQT-like subfamily. The KCNQ1 gene has recently been identified as a susceptibility locus for type 2 diabetes mellitus (T2DM). In the present study, we examined the effects of KCNQ1 variants on the therapeutic response to modified-release gliclazide (gliclazide MR) treatment in Chinese patients newly diagnosed with T2DM. A total of 100 newly diagnosed T2DM patients without a history of any anti-diabetic medications were treated with gliclazide MR for 16 weeks, but 91 patients completed the entire study. The anthropometric parameters were determined at baseline and at the final visit, while clinical laboratory tests were performed at baseline and on weeks 2, 4, 6, 12, 16. Two SNPs, rs2237892 and rs2237895, in the region of the KCNQ1 gene were genotyped in all the participants. All calculations and statistical analyses were conducted using SPSS. The rs2237892 TT homozygotes exhibited significantly higher 2-h glucose levels at baseline (P<0.05) and a lower cumulative attainment rate of the target 2-h glucose level (Plog-rank=0.020) than the C allele carriers. Patients with greater numbers of rs2237892 T alleles exhibited larger augmentations (Δ) in the 2-h glucose levels (P=0.027); and patients with the rs2237892 TT genotype exhibited a higher Δ homeostasis model assessment of β-cell function (HOMA-β) than CC and CT genotype carriers (P=0.021 and P=0.043, respectively). Moreover, the rs2237895 C allele was associated with a greater decrement in Δ glycated hemoglobin (HbA1c) (P=0.024); and patients with the CC genotype exhibited greater variance than those with the AA and AC genotypes (P=0.005 and 0.021, respectively). Compared with the C allele, the odds ratio for treatment success among carriers of the rs2237892 T allele was 2.533 (P=0.007); and the rs2237895 C allele was associated with a 2.360-fold decrease in HbA1c compared with the A allele (P=0.009). KCNQ1 polymorphisms are associated with gliclazide MR efficacy in Chinese patients with type 2 diabetes.
Keywords: potassium channel, KCNQ1, type 2 diabetes, sulfonylureas, gliclazide MR, pharmacogenetics
Introduction
Type 2 diabetes mellitus (T2DM) is a genetically heterogeneous group of metabolic disorders that is characterized by chronic hyperglycemia resulting from a deficiency in insulin secretion and insulin resistance1. After metformin, sulfonylureas (SUs) are the most commonly used oral anti-diabetic agents for the treatment of T2DM. In the 2013 Dutch type 2 diabetes guidelines, gliclazide was recommended as the preferred second treatment option over other SUs2. Gliclazide is a second-generation SU that acts exclusively on the pancreatic sulfonylureas receptor 1 (SUR1) and increases plasma insulin concentration via β-cell stimulation3.
Although most type 2 diabetic patients respond well to this agent, there is inter-individual variability in the responses to sulfonylureas. The genetic factors that were involved in drug absorption, distribution, metabolism, and targeting, partly attributed to inter-individual variability of drug response4. Pharmacogenomic studies have proved that variants in ABCC8, KCNJ11, CYP2C9, TCF7L2, PPARG, IRS1, and NOS1AP were associated with the efficacy of sulfonylureas5,6,7,8,9,10,11. Recent studies have linked the voltage-gated potassium channel KQT-like sub-family, member 1 gene (KCNQ1) to T2DM susceptibility in East Asians12,13 and to obesity in China14. Specifically, KCNQ1 channels might play a role in the regulation of insulin secretion and participate in the regulation of cell volume, which is critically important for the regulation of metabolism by insulin13,15,16,17,18.
Moreover, it has been reported recently that KCNQ1 rs2237897 is related to the efficacy of gliclazide monotherapy in Chinese patient who are newly diagnosed with type 2 diabetes19. Our previous pharmacogenetic study of gliclazide demonstrated that KCNJ11 E23K can influence responses to gliclazide due to its effect on insulin secretion in Chinese patients who are newly diagnosed with T2DM20. The potassium inwardly rectifying channel, subfamily J, member 11 (KCNJ11) gene is well known to encode a subunit of the inwardly rectifying potassium channel Kir6.2, which forms the KATP channels in pancreatic β cells20. However, KCNQ1 is the gene that encodes the pore-forming subunit of a voltage-gated K+ channel (KvLQT1) that plays key roles in the repolarization of the cardiac action potential and water and salt transport in epithelial tissues. This channel is expressed in a wide variety of tissues, including the heart, skeletal muscle, liver, and epithelia21. Additionally, KCNQ1 is also expressed in pancreatic islets and cultured insulin-secreting INS-1 cells22. Therefore, both KCNQ1 and KCNJ11 have been associated with β cell function. Furthermore, one study observed a striking increase in insulin sensitivity in a KCNQ1 knockout mouse model, raising the possibility that KCNQ1 may be a novel element that affects insulin sensitivity23. Our previous study demonstrated that single-nucleotide polymorphisms (SNPs) in KCNQ1 are associated with repaglinide efficacy and that the rosiglitazone response is related to improvements in insulin sensitivity rather than β cell function in Chinese patients with type 2 diabetes24. However, it remains unknown whether KCNQ1 SNPs have the same influence on the therapeutic effects of sulfonylureas. Thus, we conducted this study to further explore the association of the KCNQ1 polymorphisms and the therapeutic effect of modified-release (MR) gliclazide in Chinese patients who were newly diagnosed with T2DM.
Materials and methods
Individuals and study design
The study commenced in 2012–2013 with the enrolment of 100 newly diagnosed type 2 diabetic patients without a history of any anti-diabetic medications. All patients were diagnosed according to World Health Organization criteria25 and recruited from the outpatient clinics in Shanghai Jiao Tong University Affiliated Sixth People's Hospital, Shanghai, China. Detailed information in this study has been reported previously20. Diabetes education, including the introduction of T2DM and advice about diet and exercise, were provided to all patients. Then all subjects were treated with gliclazide MR for 16 weeks after a 2-week run-in period, subsequently visited the clinics on weeks 2, 4, 8, 12, and 16, and underwent designed clinical assessments. Nine patients were excluded from the study due to incomplete therapy, resulting in a sample of 91 patients (60 men and 31 women) who completed the 16-week gliclazide MR treatment. The initial minimum dosage of gliclazide was 30 mg per day. The dosage was increased to 60, 90, and 120 mg daily, in successively steps, until achievement of the target fasting plasma glucose (FPG) of ≤7 mmol/L (126 mg/dL) and/or 2-h plasma glucose (2-h PG) of ≤11 mmol/L (200 mg/dL). Patients exhibiting FPG >13 mmol/L (234 mg/dL) or 2-h PG >18 mmol/L (324 mg/dL) at two consecutive visits (at a maximal interval of 6 d) were excluded from the study.
Gliclazide presently is the first-line oral hypoglycemic medicine based on the standards of Care for Type 2 Diabetes in China26. For the purpose of this study, all participants were assigned to receive the gliclazide MR (Servier, Tianjin, China). And the escalating dose of gliclazide MR treatment was considered standard of care treatment for all patients. The study was approved by the Institutional Review Board of the Shanghai Jiao Tong University Affiliated Sixth People's Hospital, Shanghai, China. Written informed consent was obtained from each participant.
Anthropometric and biochemical measurements
The anthropometric measurements and clinical laboratory tests were described in detail previously20. In brief, anthropometric parameters containing height (m), weight (kg), waist and blood pressure were measured at baseline and at the final visit. Standard oral glucose tolerance tests were performed at baseline, and fasting blood samples and 2-h blood samples were collected at each visit for measurement of fasting and 2-h plasma glucose levels. Fasting and 2-h serum insulin levels, as well as serum lipid profiles were determined at baseline and at the final visit. Insulin sensitivity and β cell function were evaluated by homeostasis model assessment (HOMA) for insulin resistance index (HOMA-IR) and β cell function (HOMA-β)27. To evaluate potential β-cell function, the acute insulin response to arginine28 was calculated after arginine stimulation tests at baseline and after 16 weeks of treatment.
Genotyping
Two SNPs in KCNQ1, rs2237892, and rs2237895, which were associated with type 2 diabetes in our previous study, were selected24. One primer set (forward primer: 5′-GCTGCAGCCCGTGTTCCT-3′ reverse primer: 5′-CGCATTCCGGGGGCTTCC-3′) were designed to amply DNA segment containing rs2237892 variant in KCNQ1. Another primer set for rs2237895 variant was 5′-TGGGGCAGGGGTGTCTTTA-3′ (forward primer) and 5′-TCTGCCTCTTGGTCTCATCTTTAC-3′ (reverse primer). Both PCR products were digested with Cfr9I (XmaI) (Thermo Fisher Scientific Inc, Waltham, MA, USA) at 37 °C for 4 h.
Definition of the response to gliclazide MR
In our study, HbA1c were reduced by 22% on average. Other studies involving gliclazide treatment reported a decline of HbA1c by 15%-30%29,30,31,32. Therefore, responders were defined by two criterions. Criterion 1 was a decrease of >20% in the glycated hemoglobin level. Criterion 2 was a decrease of >30% in the glycated hemoglobin level.
Statistical analysis
Data are presented as the mean±SEM or n (%). Paired t-tests were used to compare values at baseline and 16 weeks after gliclazide MR. The allele frequencies were calculated by gene counting, and the results were then subjected to Hardy-Weinberg equilibrium tests. We set a FPG <7.0 mmol/L and a 2-h PG <7.8 mmol/L as attainment. The attainment rates between genotypes were estimated by Kaplan-Meier methodology and compared by the log-rank test and Cox regression model analysis adjusted for age, gender, and BMI at baseline. Data with a skewed distribution were log10-transformed prior to linear regression. The Δ values were calculated as the values at week 16 minus the values at baseline. Kruskal-Wallis test or ANOVA followed by the Student-Newman-Keuls multiple range test as appropriate were performed to analyze the differences among three genotype. After adjusting for age, gender, dosage and pathogenesis at baseline, multiple linear regressions under the additive genetic model were used to evaluate the differences in quantitative traits at baseline, 16 weeks and Δ values. Genotype distribution differences between responders and non-responders were compared by means of Fisher's exact test or χ2-test. The odds ratio (OR) values are presented with 95% confidence intervals (CIs). Statistical significance was considered at P<0.05 (two-tailed). All calculations and statistics were performed using SPSS (version 20; SPSS Statistics, IBM Corporation, Armonk, NY, USA).
Results
Clinic characteristics of the study cohort
Of the 100 enrolled patients, 91 (60 men and 31 women, mean age 55.00±11.00 years old) completed the entire study, and 9 patients lost to follow-up. The baseline and post-therapy clinical characteristics of the study group are summarized in Table 1. Blood pressure, blood glucose, glycated hemoglobin levels and lipid profiles (LDL-C) decreased significantly after 16 weeks of gliclazide MR therapy compared with baseline (all P<0.01). Moreover, there were significant improvements in homeostasis model assessment of insulin resistance (HOMA–IR, P<0.01) and homeostasis model assessment of β-cell function (HOMA–β, P<0.01).
Table 1. Clinical characteristics of patients before and after gliclazide modified release treatment.
| Baseline | 16 weeks | P value | |
|---|---|---|---|
| BMI (kg/m2) | 24.96±0.28 | 25.38±0.26 | 0.160 |
| Waist (cm) | 90.14±0.91 | 90.25±0.79 | 0.641 |
| Waist-hip ratio | 0.93±0.01 | 0.93±0.01 | 0.129 |
| SBP (mmHg) | 136.65±1.91 | 132.00±1.56 | 0.007 |
| DBP (mmHg) | 82.73±1.20 | 79.41±1.13 | 0.004 |
| FPG (mmol/L) | 9.29±0.26 | 6.89±0.15 | <0.001 |
| 2-h PPG (mmol/L) | 16.52±0.46 | 9.37±0.37 | <0.001 |
| HbA1c (%) | 8.45±0.16 | 6.47±0.07 | <0.001 |
| Fasting insulin (pmol/L) | 10.64±0.68 | 11.32±0.84 | 0.184 |
| 2-h insulin (pmol/L) | 46.07±4.25 | 47.05±5.04 | 0.625 |
| HOMA-IR | 4.00±0.30 | 3.37±0.28 | 0.004 |
| HOMA-β | 46.65±3.56 | 80.23±6.07 | <0.001 |
| Acute insulin secretion (μU/mL) | 27.46±2.36 | 27.01±2.39 | 0.263 |
| Total cholesterol (mmol/L) | 5.23±1.71 | 4.91±1.36 | 0.061 |
| HDL-C (mmol/L) | 1.13±1.70 | 1.10±1.34 | 0.107 |
| LDL-C (mmol/L) | 3.06±1.14 | 2.93±1.11 | <0.001 |
| Triglyceride (mmol/L) | 2.16±0.26 | 1.84±0.19 | 0.102 |
Data represent the mean±SEM. BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; FPG, fasting plasma glucose; PPG, post-prandial plasma glucose; HbA1c, glycated hemoglobin; HOMA-IR, homeostasis model assessment of insulin resistance; HOMA-β, homeostasis model assessment of β-cell function; HDL-C, high-density lipoprotein-cholesterol; LDL-C, low-density lipoprotein-cholesterol.
KCNQ1 SNPs and glucose metabolism during gliclazide MR treatment
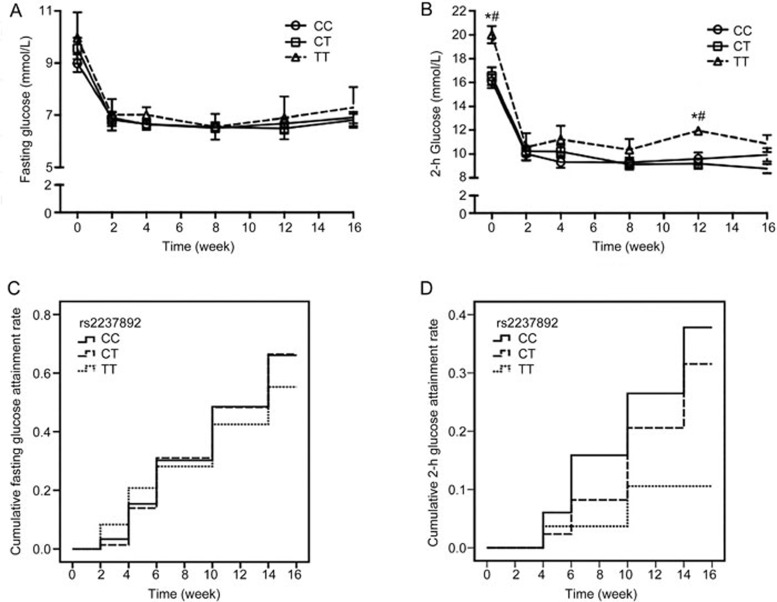
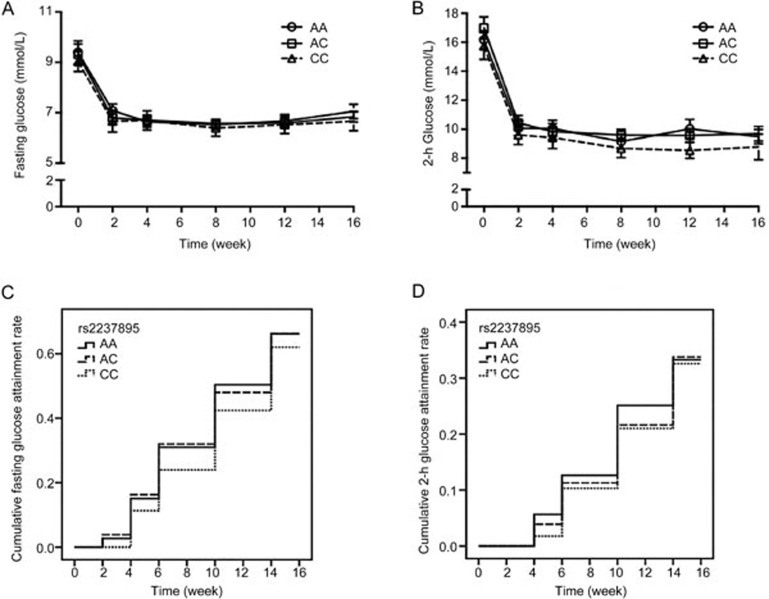
To assess the effects of KCNQ1 genetic polymorphisms (rs2237892 and rs2237895) on blood glucose during treatment, then we analyzed the changes in fasting glucose and 2-h glucose levels during treatment among different genotype groups. The genotype distributions of all SNPs conformed to Hardy-Weinberg equilibrium. The mean values of these parameters over time are displayed in Figure 1. Fasting glucose was not significantly different among patients with the CC, CT, and TT genotypes of rs2237892 (Figure 1A). Patients with the TT genotype of rs2237892 displayed higher 2-h glucose levels than the C allele carriers at baseline and at the fifth visit (all P<0.05, Figure 1B). Regarding rs2237895, there were no apparent variations among carriers of the AA, AC, and CC genotypes in terms of FPG or 2-h PG during the treatment (Figures 2A and 2B). After <7.0 mmol/L and <7.8 mmol/L were set as the fasting glucose and 2-h glucose standards respectively, the survival analyses showed that there was a trend towards a lower attainment rate of 2-h glucose in patients with the rs2237892 TT genotype (Plog-rank=0.020, Figure 1D). The significant difference remained after adjusting for age, gender, and BMI at baseline analyzed by Cox regression (PCox-regression=0.019, Figure 1D). No significant differences in the attainment rate of FPG and 2-h PG among patients with different rs2237895 genotype were found (Plog-rank=0.481 and 0.946, respectively, Figures 2C and 2D).
Figure 1.
Association of rs2237892 with fasting glucose and 2-h glucose levels during gliclazide MR treatment. (A and B) Mean fasting glucose and 2-h glucose levels among carriers of the CC, CT, and TT genotypes. The data are presented as the mean±SEM. *P<0.05 between CC and TT genotype carriers; #P<0.05 between CT and TT genotype carriers. (C and D) Associations of rs2237892 with attainment rates of the target values for fasting glucose and 2-h glucose, respectively, among carriers of the CC, CT, and TT genotypes. (C) Plog-rank=0.752, PCox-regression=0.797; and (D) Plog-rank=0.020, PCox-regression=0.019. The PCox-regression values were adjusted for age, gender, and body mass index at baseline.
Figure 2.
Association of rs2237895 with fasting glucose and 2-h glucose levels during gliclazide MR treatment. (A and B) Mean fasting glucose and 2-h glucose levels among carriers of the CC, CT, and TT genotypes. The data are presented as the mean±SEM. (C and D) The associations between rs2237895 and the attainment rates of target values for fasting glucose and 2-h glucose levels, respectively, among carriers of the CC, CT, and TT genotypes. (C) Plog-rank=0.481, PCox-regression=0.542; and (D) Plog-rank=0.946, PCox-regression=0.891. The PCox-regression values were adjusted for age, gender, and body mass index at baseline.
Next, we further explored the relevance of KCNQ1 SNPs to quantitative traits. After adjusting for age, gender, dosage and pathogenesis at baseline, there were significant linear relationships between the number of rs2237892 T alleles and the augmentations in Δ2-h glucose levels according to multiple stepwise regressions (P=0.027, Table 2). Additionally, trends towards such associations were observed in ΔHbA1c (P=0.058), ΔHOMA-β (P=0.094), and (Δ) acute insulin response (P=0.058), as illustrated in Table 2. Significant differences in ΔHOMA-β were detected; ie, the patients with the TT genotype exhibited higher variance than those with the CC and CT genotypes (P=0.021 and P=0.043, respectively, Table 2). Similarly, as illustrated in Table 3, there were obvious linear relationships between the number of rs2237895 C alleles and the decrease in ΔHbA1c (P=0.024). Significant differences in the changes in ΔHbA1c were detected among the three groups; patients with the CC genotype exhibited a higher variance than those with the AA and AC genotypes (P=0.005 and 0.021, respectively; Table 3). Patients with the rs2237895 AC genotype exhibited a lower 2-h insulin level after treatment than AA homozygotes (P=0.014, Table 3).
Table 2. Associations of the single-nucleotide polymorphism rs2237892 in KCNQ1 with the clinical features of the gliclazide cohort.
| Parameter | CC | CT | TT | P value | |
|---|---|---|---|---|---|
| Age (year) | 55.80±1.77 | 53.88±1.76 | 55.57±5.41 | 0.743 | |
| Gender (male/female) | 31/13 | 23/17 | 6/1 | 0.266 | |
| Dosage (mg/d) | 45.39±3.43 | 41.03±2.95 | 54.20±13.59 | 0.984 | |
| BMI (kg/m2) | Baseline | 25.26±0.30 | 24.74±0.48 | 24.78±1.15 | 0.607 |
| 16 Weeks | 25.40±0.30 | 25.20±0.46 | 25.72±1.20 | 0.884 | |
| Δ value | 0.03±0.15 | 0.33±0.23 | 0.33±0.38 | 0.463 | |
| Waist (cm) | Baseline | 91.46±0.98 | 89.57±1.74 | 92.00±1.06 | 0.463 |
| 16 Weeks | 90.76±1.06 | 89.39±1.31 | 91.25±3.33 | 0.770 | |
| Δ value | −0.70±0.70 | −0.18±0.96 | 0.75±0.85 | 0.764 | |
| Waist-hip ratio | Baseline | 0.94±0.01 | 0.93±0.01 | 0.96±0.03 | 0.429 |
| 16 Weeks | 0.93±0.01 | 0.92±0.01 | 0.97±0.02 | 0.154 | |
| Δ value | −0.01±0.01 | −0.02±0.01 | 0.01±0.02 | 0.605 | |
| Fasting glucose (mmol/L) | Baseline | 8.99±0.34 | 9.56±0.41 | 9.96±1.37 | 0.162 |
| 16 Weeks | 6.90±0.21 | 6.81±0.22 | 7.30±0.77 | 0.531 | |
| Δ value | −2.06±0.27 | −2.78±0.39 | −2.90±1.11 | 0.153 | |
| 2-h glucose (mmol/L) | Baseline | 16.11±0.57 | 16.54±0.75 | 20.02±0.73*,# | 0.184 |
| 16 Weeks | 9.94±0.57 | 8.76±0.38 | 10.88±0.71 | 0.879 | |
| Δ value | −6.10±0.66 | −7.92±0.78 | −10.45±1.72 | 0.041 | |
| Glycated hemoglobin (%) | Baseline | 8.05±0.16 | 8.81±0.30 | 8.60±0.57 | 0.195 |
| 16 Weeks | 6.46±0.10 | 6.49±0.10 | 6.40±0.46 | 0.649 | |
| Δ value | −1.59±0.19 | −2.33±0.31* | −2.40±0.45 | 0.058 | |
| Fasting insulin (pmol/L) | Baseline | 10.22±0.77 | 10.83±0.87 | 9.29±2.90 | 0.909 |
| 16 Weeks | 10.96±0.84 | 11.74±1.30 | 11.18±3.25 | 0.268 | |
| Δ value | 0.73±0.58 | 0.90±0.96 | 1.89±2.67 | 0.247 | |
| Log10 (2-h insulin) (pmol/L) | Baseline | 1.59±0.05 | 1.53±0.05 | 1.70±0.08 | 0.758 |
| 16 Weeks | 1.57±0.06 | 1.61±0.05 | 1.69±0.05 | 0.341 | |
| Δ value | −0.03±0.05 | 0.08±0.04 | −0.03±0.04 | 0.257 | |
| Log10 HOMA-IR | Baseline | 0.52±0.03 | 0.55±0.03 | 0.45±0.15 | 0.616 |
| 16 Weeks | 0.47±0.03 | 0.45±0.04 | 0.50±0.14 | 0.736 | |
| Δ value | −0.05±0.02 | −0.11±0.03 | 0.03±0.07 | 0.887 | |
| Log10 HOMA-β | Baseline | 1.60±0.04 | 1.56±0.05 | 1.50±0.11 | 0.395 |
| 16 Weeks | 1.84±0.04 | 1.82±0.05 | 1.92±0.08 | 0.789 | |
| Δ value | 0.23±0.03 | 0.26±0.04 | 0.46±0.06*,# | 0.094 | |
| Log10 (acute insulin response) (pmol/L) | Baseline | 1.38±0.04 | 1.37±0.04 | 1.34±0.10 | 0.609 |
| 16 Weeks | 1.33±0.04 | 1.36±0.05 | 1.38±0.09 | 0.451 | |
| Δ value | −0.06±0.03 | −0.01±0.03 | 0.04±0.06 | 0.058 |
Data are presented as the mean±SEM. The Δ values were calculated as the values at weeks 16 minus the values at baseline.
*P<0.05 compared with the CC genotype (one-way ANOVA followed by the Student-Newman-Keuls multiple range test).
#P<0.05, compared with the CT genotype (one-way ANOVA followed by the Student-Newman-Keuls multiple range test). P values were adjusted for age, gender, dosage, and pathogenesis at baseline when linear regression models were adopted. BMI, body mass index; HOMA-β, homeostasis model assessment of β-cell function; HOMA-IR, homeostasis model assessment of insulin resistance.
Table 3. Associations of the single-nucleotide polymorphism rs2237895 in KCNQ1 with the clinical features of the gliclazide cohort.
| Parameter | AA | AC | CC | P value | |
|---|---|---|---|---|---|
| Age (year) | 55.65±2.21 | 55.05±1.69 | 52.85±2.87 | 0.755 | |
| Gender (male/female) | 19/14 | 30/15 | 10/3 | 0.435 | |
| Dosage (mg/d) | 46.01±4.71 | 43.58±2.84 | 40.63±4.94 | 0.902 | |
| BMI (kg/m2) | Baseline | 24.95±0.47 | 25.33±0.40 | 24.91±0.67 | 0.973 |
| 16 Weeks | 25.09±0.48 | 25.50±0.36 | 25.23±0.53 | 0.771 | |
| Δ value | 0.14±0.22 | 0.16±0.19 | 0.32±0.35 | 0.894 | |
| Waist (cm) | Baseline | 89.86±1.59 | 91.46±1.44 | 89.33±1.74 | 0.856 |
| 16 Weeks | 89.46±1.50 | 90.91±1.15 | 89.67±1.39 | 0.594 | |
| Δ value | −0.40±0.78 | −0.55±0.89 | 0.33±1.22 | 0.859 | |
| Waist-hip ratio | Baseline | 0.93±0.01 | 0.94±0.01 | 0.92±0.02 | 0.551 |
| 16 Weeks | 0.92±0.01 | 0.93±0.01 | 0.91±0.02 | 0.861 | |
| Δ value | 0.00±0.01 | −0.02±0.01 | −0.01±0.01 | 0.599 | |
| Fasting glucose (mmol/L) | Baseline | 9.39±0.46 | 9.32±0.40 | 9.03±0.39 | 0.947 |
| 16 Weeks | 7.05±0.28 | 6.84±0.21 | 6.66±0.37 | 0.335 | |
| Δ value | −2.31±0.35 | −2.50±0.38 | −2.51±0.35 | 0.338 | |
| 2-h glucose (mmol/L) | Baseline | 16.16±0.62 | 16.99±0.75 | 15.73±0.92 | 0.730 |
| 16 Weeks | 9.48±0.49 | 9.68±0.51 | 8.77±0.89 | 0.569 | |
| Δ value | −6.97±0.70 | −7.15±0.83 | −7.41±1.21 | 0.742 | |
| Glycated hemoglobin (%) | Baseline | 8.15±0.21 | 8.40±0.24 | 9.29±0.54* | 0.071 |
| 16 Weeks | 6.55±0.11 | 6.48±0.11 | 6.15±0.18 | 0.500 | |
| Δ value | −1.59±0.17 | −1.94±0.27 | −3.13±0.49*,# | 0.024 | |
| Fasting insulin (pmol/L) | Baseline | 11.75±1.02 | 9.40±0.78 | 10.53±1.16 | 0.241 |
| 16 Weeks | 12.69±1.14 | 10.56±1.18 | 10.37±0.85 | 0.207 | |
| Δ value | 0.93±0.82 | 1.15±0.84 | −0.16±1.23 | 0.733 | |
| Log10 (2-h insulin) (pmol/L) | Baseline | 1.62±0.06 | 1.56±0.05 | 1.50±0.06 | 0.131 |
| 16 Weeks | 1.72±0.07 | 1.51±0.05* | 1.60±0.03 | 0.088 | |
| Δ value | 0.04±0.06 | −0.01±0.04 | 0.07±0.06 | 0.948 | |
| Log10 HOMA-IR | Baseline | 0.57±0.04 | 0.48±0.03 | 0.58±0.04 | 0.825 |
| 16 Weeks | 0.52±0.04 | 0.43±0.04 | 0.43±0.05 | 0.313 | |
| Δ value | −0.05±0.03 | −0.05±0.03 | −0.15±0.05 | 0.383 | |
| Log10 HOMA-β | Baseline | 1.59±0.05 | 1.54±0.04 | 1.63±0.08 | 0.958 |
| 16 Weeks | 1.85±0.05 | 1.81±0.03 | 1.89±0.05 | 0.513 | |
| Δ value | 0.26±0.04 | 0.26±0.03 | 0.26±0.07 | 0.583 | |
| Log10 (acute insulin response) (pmol/L) | Baseline | 1.43±0.04 | 1.34±0.04 | 1.34±0.07 | 0.146 |
| 16 Weeks | 1.38±0.05 | 1.31±0.04 | 1.36±0.08 | 0.642 | |
| Δ value | −0.04±0.04 | −0.03±0.03 | 0.02±0.05 | 0.286 |
Data are presented as the mean±SEM. The Δ values were calculated as the values at weeks 16 minus the values at baseline.
*P<0.05 compared with the AA genotype (one-way ANOVA followed by the Student Newman Keuls multiple range test).
#P<0.05 compared with the AC genotype (one-way ANOVA followed by the Student-Newman-Keuls multiple range test). P values were adjusted for age, gender, dosage, and pathogenesis at baseline when linear regression models were adopted. BMI, body mass index; HOMA-β, homeostasis model assessment of β-cell function; HOMA-IR, homeostasis model assessment of insulin resistance.
KCNQ1 SNPs and response to gliclazide MR treatment
To identify the association of the KCNQ1 genetic polymorphisms with the response rate to gliclazide MR treatment, the genotype frequencies according to the therapeutic responses are illustrated in Table 4. Based on the first criterion, no significant effects of the variances in rs2237892 or rs2237895 on gliclazide MR treatment were observed; however, the rs2237892 TT homozygotes and rs2237895 CC homozygotes exhibited good responses to gliclazide MR therapy. According to the second criterion, both rs2237892 and rs2237895 were associated with the response to gliclazide MR treatment. Regarding rs2237892, there were more responders among the rare TT allele homozygotes TT; 57.1% of the TT homozygotes responded, compared with only 15.9% of the CC homozygotes. The heterozygote CT group exhibited an intermediate response rate. The odds ratio for the T allele with respect to treatment success was 2.533 (95% CI: 1.283–4.999, P=0.007) compared with the rs2237892 C allele. Similarly, the rs2237895 C allele was associated with a 2.360-fold decrease in glycated hemoglobin compared with the A allele (95% CI: 1.225–4.550, P=0.009).
Table 4. Genotype and allele distributions of responders and non-responders carrying the KCNQ1 rs2237892 and rs2237895 variants.
| Criterion 1 | Criterion 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| rs2237892 | CC | CT | TT | P value | CC | CT | TT | P value |
| Responder (%) | 21 (47.7) | 21 (52.5) | 5 (71.4) | 0.563 | 7 (15.9) | 15 (37.5) | 4 (57.1) | 0.014 |
| Non-responder (%) | 23 (52.3) | 19 (47.5) | 2 (28.6) | 37 (84.1) | 25 (62.5) | 3 (42.9) | ||
| rs2237895 | AA | AC | CC | P value | AA | AC | CC | P value |
| Responder (%) | 16 (48.5) | 22 (48.9) | 9 (69.2) | 0.391 | 6 (18.2) | 12 (26.7) | 8 (61.5) | 0.014 |
| Non-responder (%) | 17 (51.5) | 23 (51.1) | 4 (30.8) | 27 (81.8) | 33 (73.3) | 5 (38.5) | ||
Criterion 1: Responders were defined by a greater than 20% decrease in HbA1c. Criterion 2: Responders were defined by a greater than 30% decrease in HbA1c.
Discussion
Gliclazide MR is a new formulation of a second-generation SU drug that was designed for once-daily administration33. Gliclazide MR initiates insulin secretion by closing potassium channels. Our previous research indicated that the KCNJ11 E23K variant is associated with the therapeutic effect of gliclazide in Chinese patients with T2DM20. However, the relationship between KCNQ1 and gliclazide has rarely been reported. Based on the original genome-wide association scans and replication studies, common variants in KCNQ1 (rs2237892, rs2237895, and rs2237897) demonstrate the strongest associations with T2DM12,13,18,34. A recent study focusing on Chinese patients with T2DM confirmed that rs2237897 but not rs2237892 is related to the efficacy of gliclazide monotherapy according to FPG alone19. Thus far, no studies have reported data for the relationship between rs2237895 and SUs. Rs2237892 and rs2237895, in moderate linkage disequilibrium (LD), are located in intron 15 of the KCNQ1 gene on chromosome 11p15, and encode the pore-forming α subunit of the KvLQT1 channel. It has previously been shown that various KCNQ1 polymorphisms (rs2237892 and rs2237895) are associated with an increased risk of T2DM due to not only decreasing levels of various measures of insulin secretion13,15,16,17,18 but also effects on insulin sensitivity23,24. Therefore, we aimed to identify the relationships between KCNQ1 polymorphisms (rs2237892 and rs2237895) and the therapeutic effect of gliclazide in Chinese populations.
Our present study revealed that the KCNQ1 SNPs were associated with the response to gliclazide MR therapy in newly diagnosed T2DM patients. Regarding rs2237892, the TT homozygotes exhibited a lower treatment failure rate, and there was a significant linear relationship between the number of rs2237892 T alleles and the augmentation of Δ2-h glucose levels. Therefore, even if the 2-h glucose levels and cumulative attainment rate of the target 2-h glucose level among TT patients were higher than those of CC and CT patients, the patients with the T alleles exhibited a greater reduction in 2-h glucose levels following gliclazide treatment. Furthermore, we also identified some genotype-related differences with respect to improvements in blood glucose (HbA1c) and β-cell function (HOMA-β and acute insulin response); the patients with the T alleles displayed lower HbA1c and improved β-cell function. These findings suggest that the rs2237892 T allele improved the efficiency of gliclazide. The KV channels encoded by KCNQ1 play important roles in mediation of the repolarization of the membrane that terminates Ca2+ influx and insulin secretion in pancreatic β-cells35, and the rs2237892 T allele has been found to be associated with higher second-phase glucose-stimulated insulin secretion via the hyperglycemic clamp36. Therefore, we hypothesized that the KCNQ1 rs2237892 T allele may increase insulin release after gliclazide treatment. Nevertheless, how the KCNQ1 rs2237892 polymorphism affects the electrical change of the β-cell membrane following gliclazide MR treatment remains unknown and should be further investigated.
Recently, some researchers indicated that the Kcnq1 molecule affected insulin sensitivity via glucose metabolism23. In our previous study investigating KCNQ1 SNPs and repaglinide efficacy, the rs2237892 TT genotype was associated with better 2-h glucose normalization accompanied by greater improvements in insulin sensitivity24. However, in the present study, we failed to detect an influence of KCNQ1 rs2237892 on insulin sensitivity following gliclazide MR treatment, which was most likely due to the ability of repaglinide to improve insulin sensitivity37 or to the small sample size assessed in the present study. Based on all these findings, we propose the tentative hypothesis that the better 2-h glucose normalization observed in individuals with the rs2237892 TT genotype might be due to greater improvements in islet β-cell function in these individuals. Specifically, the rs2237892 T allele could improve the gliclazide response with respect to stimulation of insulin secretion and lowering of blood glucose levels; however, the underlying mechanism remains to be elucidated.
Regarding rs2237895, Jonsson et al conducted a prospective population-based study and found that the rs2237895 C allele indeed increased the risk of future T2DM and that this effect was due to failure of β-cell function15. In the present study, the carriers of greater numbers of C alleles at rs2237895 consistently exhibited higher HbA1c at baseline, which is in agreement with our previous research on the efficacy of repaglinide24. Furthermore, the rs2237895 CC patients exhibited greater augmentation in ΔHbA1c following treatment, potentially accounting for the lower failure rate of gliclazide therapy among the CC homozygotes. However, we also failed to detect a change in insulin sensitivity or β-cell function in the rs2237895 carriers following gliclazide treatment. Thus far, to the best of our knowledge, two studies have assessed the KCNQ1 gene and the therapeutic response to SU treatment. The first study evaluated KCNQ1 (rs163184) and the therapeutic response (FPG, HbA1c) to sulfonylurea treatment38. However, the patients who were not newly diagnosed with T2DM in this study were not treated with SU monotherapy, and other parameters related to insulin sensitivity and β-cell function were not assessed. In the other study published latest, Duan et al found that KCNQ1 rs2237897 was associated with the efficacy of gliclazide accessed by FPG after 8-week monotherapy in Chinese type 2 diabetic patients. Specifically, the FPG reduction and treatment success rate were significantly higher in carriers of rs2237897 CT and TT genotypes, whereas no significant difference was found in the FPG reduction and treatment success rate among different genotype of rs223789219. Similar with our study, the enrolled patients were newly diagnosed T2DM and treated with gliclazide monotherapy for perspective research. And the FPG reduction had no significant difference for rs2237892, which also confirmed by our present study. However, Duan et al measured only FPG but no other glucose metabolic parameters, such as 2-h PG, HbA1c, insulin sensitivity and islet β-cell function. Besides, the 8-week follow-up was not enough to observe a long-term effect of KCNQ1 polymorphisms on the efficacy of gliclazide treatment. And rs22237895 was not analyzed in Duan's study. So our study was not a replication study and added more findings about KCNQ1 rs2237892 and rs2237895 to this field.
To sum up, we found that the newly diagnosed T2DM patients carrying the KCNQ1 rs2237892 T allele and the rs2237895 C allele were inclined to achieve better therapeutic results with gliclazide following prospective monotherapy for 16 weeks. Interestingly, consistent with our findings, Dai et al found that T2DM patients with the rs2237892 T allele and the rs2237895 C allele following repaglinide treatment were more likely to have a positive effect on postprandial glucose levels than patients with the rs2237892 CC and rs2237895 AA genotype39. In addition, our previous study demonstrated that patients with rs2237892 TT homozygotes had lower glucose levels following repaglinide treatment, while the rs2237895 C risk allele in those patients was associated with greater increments in fasting insulin24. Because the rs2237892 and rs2237895 markers are located near the outside of a KCNQ1 exon, they do not change the amino acid sequence. These SNPs acted as biological markers may play a more direct role in gliclazide metabolism by affecting the gene's function. However, only biological markers can not explain gene function, which need more intelligent guesswork. Therefore, how the rs2237892 and rs2237895 KCNQ1 polymorphisms affect the outcomes of gliclazide treatment remain unknown, and why the non-risk T allele of rs2237892 is associated with a better therapeutic outcome but the risk C allele of 2237895 is related to a greater beneficial effect require further investigation.
Several limitations to this study should be considered. First, the sample size was relatively small, which implies that the statistical power was insufficient and may account for the failure to detect an association between the KCNQ1 SNPs and insulin sensitivity. But the effect of an inadequate follow-up period also cannot be ruled out. Second, the possibility of a false-positive finding cannot be excluded due to the lack of adjustment for multiple comparisons. However, because the analyzed SNPs were present in modest linkage disequilibrium and the quantitative traits were highly related, the influences of multiple comparisons may be limited. Third, no placebo group were included in the present study, so the effects of lifestyle modifications cannot be completely eliminated. Nevertheless, all patients were provided the same diabetes education according to guidelines on diet and exercise. And in the view of ethics, drug intervention as soon as possible to target blood glucose will benefit glycemic control as well as delay the occurrence of diabetic complications, lacking of placebo group in pharmacogenomics research focusing on well established anti-diabetic drug is reasonable and in accordance with study design performed by international counterparts3,8,40,41,42,43,44,45,46,47. Fourth, the glucose metabolic parameters like HbA1c, insulin sensitivity and islet β-cell function during the 16-week treatment were not measured, which might further strengthen our findings. However, in our previous pharmacogenetics study24, blood glucose level decreased obviously within 12 weeks after treatment and kept at a steady state then. So the follow-up time were shortened to 16 weeks in this study design. And in our opinion, as for the newly diagnosed T2D patients without any anti-diabetic drug, the changes of blood glucose are sensitive and insulin secretions are unstable at the beginning of monotherapy. In the current study, there were two major visit time-point (baseline and 16 weeks) and four intermediate visit time-point (2, 4, 8, and 12 weeks). We determine the basic clinic traits at four intermediate visits, and comprehensive assessment at two major visits, this design can greatly save research cost as well as avoid too much blood drawing from the patients. Fifth, the mechanism underling is still unclear due to lacking of functional study.
In conclusion, the KCNQ1 polymorphism is associated with the therapeutic response to sulfonylurea treatment in Chinese patients who are newly diagnosed with type 2 diabetes. Further investigations with a lager sample size and a placebo control group are necessary to confirm our findings.
Author contribution
Cheng HU and Wei-ping JIA conceived and designed research; Qing LI, Ting-ting TANG, Feng JIANG, Rong ZHANG, and Miao CHEN performed the experiments; Jun YIN, Yu-qian BAO, and Xiang CHENG contributed new reagents or analytic tools; Qing LI, Ting-ting TANG, and Cheng HU analyzed the data; Qing LI, Ting-ting TANG, Cheng HU, and Wei-ping JIA wrote the paper.
Acknowledgments
We thank the individuals who participated in the present study. We gratefully acknowledge the skillful technical support of all the nursing and medical staff at the Shanghai Clinical Center for Diabetes. This work was supported by grants from the “Personalized Medicines — Molecular Signature-based Drug Discovery and Development”, Strategic Priority Research Program of the Chinese Academy of Sciences (XDA12000000), the National Natural Science Foundation of China (81322010 and 81570713), the national 863 program (2015AA020110), Gaofeng Clinical Medicine Grant Support of Shanghai Municipal Education Commission (20152527), and the National Program for Support of Top-notch Young Professional.
References
- Doria A, Patti ME, Kahn CR. The emerging genetic architecture of type 2 diabetes. Cell Metab 2008; 8: 186–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landman GW, de Bock GH, van Hateren KJ, van Dijk PR, Groenier KH, Gans RO, et al. Safety and efficacy of gliclazide as treatment for type 2 diabetes: a systematic review and meta-analysis of randomized trials. PLoS One 2014; 9: e82880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Mao G, Ren X, Xing H, Tang G, Li Q, et al. Ser1369Ala variant in sulfonylurea receptor gene ABCC8 is associated with antidiabetic efficacy of gliclazide in Chinese Type 2 diabetic patients. Diabetes Care 2008; 31: 1939–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and individualized drug therapy. Annu Rev Med 2006; 57: 119–37. [DOI] [PubMed] [Google Scholar]
- Xu H, Murray M, Mclachlan AJ. Influence of genetic polymorphisms on the pharmacokinetics and pharmaco-dynamics of sulfonylurea drugs. Curr Drug Metab 2009; 10: 643–58. [DOI] [PubMed] [Google Scholar]
- Holstein A, Hahn M, Stumvoll M, Kovacs P. The E23K variant of KCNJ11 and the risk for severe sulfonylurea-induced hypoglycemia in patients with Type 2 diabetes. Horm Metab Res 2009; 41: 387–90.19214942 [Google Scholar]
- Kirchheiner J, Roots I, Goldammer M, Rosenkranz B, Brockmöller J. Effect of genetic polymorphisms in cytochrome p450 (CYP) 2C9 and CYP2C8 on the pharmacokinetics of oral antidiabetic drugs: clinical relevance. Clin Pharmacokinet 2005; 44: 1209–25. [DOI] [PubMed] [Google Scholar]
- Pearson ER, Donnelly LA, Kimber C, Whitley A, Doney AS, McCarthy MI, et al. Variation in TCF7L2 influences therapeutic response to sulfonylureas: a GoDARTs study. Diabetes 2007; 56: 2178–82. [DOI] [PubMed] [Google Scholar]
- Swen JJ, Guchelaar HJ, Baak-Pablo RF, Assendelft WJ, Wessels JA. Genetic risk factors for type 2 diabetes mellitus and response to sulfonylurea treatment. Pharmacogenet Genomics 2011; 21: 461–8. [DOI] [PubMed] [Google Scholar]
- Sesti G, Marini MA, Cardellini M, Sciacqua A, Frontoni S, Andreozzi F, et al. The Arg972 variant in insulin receptor substrate-1 is associated with an increased risk of secondary failure to sulfonylurea in patients with Type 2 diabetes. Diabetes Care 2004; 27: 1394–8. [DOI] [PubMed] [Google Scholar]
- Becker ML, Aarnoudse AJ, Newton-Cheh C, Hofman A, Witteman JC, Uitterlinden AG, et al. Common variation in the NOS1AP gene is associated with reduced glucose-lowering effect and with increased mortality in users of sulfonylurea. Pharmacogenet Genomics 2008; 18: 591–7. [DOI] [PubMed] [Google Scholar]
- Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet 2008; 40: 1098–102. [DOI] [PubMed] [Google Scholar]
- Hu C, Wang C, Zhang R, Ma X, Wang J, Lu J, et al. Variations in KCNQ1 are associated with type 2 diabetes and beta cell function in a Chinese population. Diabetologia 2009; 52: 1322–5. [DOI] [PubMed] [Google Scholar]
- Yu W, Ma RC, Hu C, So WY, Zhang R, Wang C, et al. Association between KCNQ1 genetic variants and obesity in Chinese patients with type 2 diabetes. Diabetologia 2012; 55: 2655–9. [DOI] [PubMed] [Google Scholar]
- Jonsson A, Isomaa B, Tuomi T, Taneera J, Salehi A, Nilsson P, et al. A variant in the KCNQ1 gene predicts future type 2 diabetes and mediates impaired insulin secretion. Diabetes 2009; 58: 2409–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JT, Nurbaya S, Gardner D, Ye S, Tai ES, Ng DP. Genetic variation in KCNQ1 associates with fasting glucose and beta-cell function: a study of 3,734 subjects comprising three ethnicities living in Singapore. Diabetes 2009; 58: 1445–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmkvist J, Banasik K, Andersen G, Unoki H, Jensen TS, Pisinger C, et al. The type 2 diabetes associated minor allele of rs2237895 KCNQ1 associates with reduced insulin release following an oral glucose load. PLoS One 2009; 4: e5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig K, Staiger H, Machicao F, Kirchhoff K, Guthoff M, Schäfer SA, et al. Association of type 2 diabetes candidate polymorphisms in KCNQ1 with incretin and insulin secretion. Diabetes 2009; 58: 1715–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan F, Guo F, Zhang L, Chen P, Wang X, Liu Z, et al. Association of KCNQ1 polymorphisms with gliclazide efficacy in Chinese type 2 diabetic patients. Pharmacogenet Genomics 2016. DOI: 10.1097/FPC.0000000000000204. [DOI] [PubMed]
- Li Q, Chen M, Zhang R, Jiang F, Wang J, Zhou J, et al. KCNJ11 E23K variant is associated with therapeutic effect of sulfonylureas in Chinese type 2 diabetic patients. Clin Exp Pharmacol Physiol 2014; 41: 748–54. [DOI] [PubMed] [Google Scholar]
- Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. KVLQT1 and IsK (minK) proteins associate to form the IKS cardiac potassium current. Nature 1996; 384: 78–80. [DOI] [PubMed] [Google Scholar]
- Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet 2008; 40: 1092–7. [DOI] [PubMed] [Google Scholar]
- Boini KM, Graf D, Hennige AM, Koka S, Kempe DS, Wang K, et al. Enhanced insulin sensitivity of gene-targeted mice lacking functional KCNQ1. Am J Physiol Regul Integr Comp Physiol 2009; 296: R1695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Hu C, Zhang R, Wang C, Qin W, Lu J, et al. Effects of KCNQ1 polymorphisms on the therapeutic efficacy of oral antidiabetic drugs in Chinese patients with type 2 diabetes. Clin Pharmacol Ther 2011; 89: 437–42. [DOI] [PubMed] [Google Scholar]
- Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med 1998; 15: 539–53. [DOI] [PubMed] [Google Scholar]
- Chinese Diabetes Society. China's prevention and treatment guideline for type 2 diabetes Mellitus (2013 edition). Chin J Diab Mellitus 2014; 6: 447–98. [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985; 28: 412–9. [DOI] [PubMed] [Google Scholar]
- Larsson H, Ahrén B. Glucose-dependent arginine stimulation test for characterization of islet function: studies on reproducibility and priming effect of arginine. Diabetologia 1998; 41: 772–7. [DOI] [PubMed] [Google Scholar]
- Schernthaner G, Grimaldi A, Di Mario U, Drzewoski J, Kempler P, Kvapil M, et al. GUIDE study: double-blind comparison of once-daily gliclazide MR and glimepiride in type 2 diabetic patients. Eur J Clin Invest 2004; 34: 535–42. [DOI] [PubMed] [Google Scholar]
- Ceriello A, Johns D, Widel M, Eckland DJ, Gilmore KJ, Tan MH. Comparison of effect of pioglitazone with metformin or sulfonylurea (monotherapy and combination therapy) on postload glycemia and composite insulin sensitivity index during an oral glucose tolerance test in patients with type 2 diabetes. Diabetes Care 2005; 28: 266–72. [DOI] [PubMed] [Google Scholar]
- Wang H, Ni Y, Yang S, Li H, Li X, Feng B. The effects of gliclazide, metformin, and acarbose on body composition in patients with newly diagnosed type 2 diabetes mellitus. Curr Ther Res Clin Exp 2013; 75: 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hissa MR, Cavalcante LL, Guimarães SB, Hissa MN. A 16-week study to compare the effect of vildagliptin versus gliclazide on postprandial lipoprotein concentrations and oxidative stress in patients with type 2 diabetes inadequately controlled with metformin monotherapy. Diabetol Metab Syndr 2015; 7: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouin P, Standl E. Diamicron MR Study Group. Gliclazide modified release: results of a 2-year study in patients with type 2 diabetes. Diabetes Obes Metab 2004; 6: 414–21. [DOI] [PubMed] [Google Scholar]
- Liu Y, Zhou DZ, Zhang D, Chen Z, Zhao T, Zhang Z, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes in the population of mainland China. Diabetologia 2009; 52: 1315–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald PE, Ha XF, Wang J, Smukler SR, Sun AM, Gaisano HY, et al. Members of the Kv1 and Kv2 voltage-dependent K+ channel families regulate insulin secretion. Mol Endocrinol 2001; 15: 1423–35. [DOI] [PubMed] [Google Scholar]
- van Vliet-Ostaptchouk JV, van Haeften TW, Landman GW, Reiling E, Kleefstra N, Bilo HJ, et al. Common variants in the type 2 diabetes KCNQ1 gene are associated with impairments in insulin secretion during hyperglycaemic glucose clamp. PLoS One 2012; 7: e32148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tian H, Li Q, Wang N, Wu T, Liu Y, et al. Improvement of insulin sensitivity and beta-cell function by nateglinide and repaglinide in type 2 diabetic patients — a randomized controlled double-blind and double-dummy multicentre clinical trial. Diabetes Obes Metab 2007; 9: 558–65. [DOI] [PubMed] [Google Scholar]
- Schroner Z, Dobrikova M, Klimcakova L, Javorsky M, Zidzik J, Kozarova M, et al. Variation in KCNQ1 is associated with therapeutic response to sulphonylureas. Med Sci Monit 2011; 17: CR392–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai XP, Huang Q, Yin JY, Guo Y, Gong ZC, Lei MX, et al. KCNQ1 gene polymorphisms are associated with the therapeutic efficacy of repaglinide in Chinese Type 2 diabetic patients. Clin Exp Pharmacol Physiol 2012; 39: 462–8. [DOI] [PubMed] [Google Scholar]
- Tkáč I, Klimčáková L, Javorský M, Fabianová M, Schroner Z, Hermanová H, et al. Pharmacogenomic association between a variant in SLC47A1 gene and therapeutic response to metformin in type 2 diabetes. Diabetes Obes Metab 2013; 15: 189–91. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Yanagawa T, Shibasaki T, Kaniwa N, Hasegawa R, Tohkin M, et al. Effect of CYP2C9 genentic polymorphisms on the efficacy and pharmacokinetics of glimepiride in subjects with type 2 diabetes. Diabetes Res Clin Pract 2006; 72: 148–54. [DOI] [PubMed] [Google Scholar]
- Zhou K, Donnelly L, Burch L, Tavendale R, Doney AS, Leese G, et al. Loss-of-function CYP2C9 variants improve therapeutic response to sulfonylureas in type 2 diabetes: a Go-DARTS study. Clin Pharmacol Ther 2010; 87: 52–6. [DOI] [PubMed] [Google Scholar]
- Becker ML, Visser LE, Trienekens PH, Hofman A, van Schaik RH, Stricker BH. Cytochrome P450 2C9*2 and *3 polymorphisms and the dose and effect of sulfonylurea in type II diabetes mellitus. Clin Pharmacol Ther 2008; 83: 288–92. [DOI] [PubMed] [Google Scholar]
- Schroner Z, Javorsky M, Tkacova R, Klimcakova L, Dobrikova M, Habalova V, et al. Effect of sulphonylurea treatment on glycaemic control is related to TCF7L2 genotype in patients with type 2 diabetes. Diabetes Obes Metab 2011; 13: 89–91. [DOI] [PubMed] [Google Scholar]
- Javorsky M, Klimcakova L, Schroner Z, Zidzik J, Babjakova E, Fabianova M, et al. KCNJ11 gene E23K variant and therapeutic response to sulfonylureas. Eur J Intern Med 2012; 23: 245–9. [DOI] [PubMed] [Google Scholar]
- Zhang H, Liu X, Kuang H, Yi R, Xing H. Association of sulfonylurea receptor 1 genotype with therapeutic response to gliclazide in type 2 diabetes. Diabetes Res Clin Pract 2007; 77: 58–61. [DOI] [PubMed] [Google Scholar]
- Javorský M, Babjaková E, Klimčáková L, Schroner Z, Zidzik J, Stolfová M, et al. Association between TCF7L2 genotype and glycemic control in diabetec patients treated with glicalzide. Int J Endocrinol 2013; 2013: 374858. [DOI] [PMC free article] [PubMed] [Google Scholar]