Abstract
Previous studies have shown that microRNA-1304 (miR-1304) is dysregulated in certain types of cancers, including non-small cell lung cancer (NSCLC), and might be involved in tumor survival and/or growth. In this study we investigated the direct target of miR-1304 and its function in NSCLC in vitro. Human lung adenocarcinoma cell lines (A549 and NCI-H1975) were studied. The cell proliferation and survival were investigated via cell counting, MTT and colony-formation assays. Cell apoptosis and cell cycle were examined using annexin V-PE/7-AAD and PI staining assays, respectively. The dual-luciferase reporter assay was used to verify post-transcriptional regulation of heme oxygenase-1 (HO-1) by miR-1304. CRISPR/Cas9 was used to deplete endogenous miR-1304. Overexpression of MiR-1304 significantly decreased the number and viability of NSCLC cells and colony formation, and induced cell apoptosis and G0/G1 phase cell cycle arrest. HO-1 was demonstrated to be a direct target of miR-1304 in NSCLC cells. Restoration of HO-1 expression by hemin (20 μmol/L) abolished the inhibition of miR-1304 on cell growth and rescued miR-1304-induced apoptosis in A549 cells. Suppression of endogenous miR-1304 with anti-1304 significantly increased HO-1 expression and promoted cell growth and survival in A549 cells. In 17 human NSCLC tissue samples, miR-1304 expression was significantly decreased, while HO-1 expression was significantly increased as compared to normal lung tissues. MicroRNA-1304 is a tumor suppressor and HO-1 is its direct target in NSCLC. The results suggest the potential for miR-1304 as a therapeutic target for NSCLC.
Keywords: miR-1304, NSCLC, heme oxygenase-1, hemin
Introduction
Lung cancer is one of the leading causes of cancer-related death. Non-small cell lung cancer (NSCLC) accounts for >80% of lung cancer cases and currently has an overall five-year survival rate of only 15%1. Although treatment strategies such as surgical resection, chemotherapy and radiotherapy have made great progress in recent decades, the survival rates for NSCLC have not been effectively improved2. Therefore, the discovery of early, safe and noninvasive detection methods is important for screening and diagnosis, and new therapies and treatment targets are in demand.
MicroRNAs (miRNAs) are endogenous, small non-coding RNAs that are approximately 22 nucleotides in length. MiRNAs regulate gene expression at the post-transcriptional level by binding to the 3′ untranslated regions (3′UTR) of target mRNAs3,4,5. In mammals, miRNAs are predicted to modulate the expression of more than 60% of all protein-coding genes and thus play an important regulatory role in almost every cellular process, including cell differentiation, proliferation, apoptosis, invasion and migration6,7,8. Numerous studies have reported that expression of certain miRNA(s) is dysregulated in different tumor types, including lung cancer, and miRNAs have emerged as important players in tumorigenesis and cancer development9.
Previous studies have reported that miR-1304 expression is dysregulated in paclitaxel-treated hypopharynx cancer and bcl-xl silenced NSCLC10,11. Those studies suggested that miR-1304 might be involved in tumor survival and/or growth. However, the role of miR-1304 in NSCLC remains unclear. Although certain target genes of miR-1304 have been predicted and detected in other types of cells10,12, the direct target of miR-1304 and its function in NSCLC remain to be clarified. In this study, we investigated the function and target of miR-1304 in NSCLC.
Materials and methods
Cell lines and culture conditions
Human lung adenocarcinoma cell lines (A549 and NCI-H1975) were acquired from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) medium supplemented with 10% fetal bovine serum (FBS) and kept in a humidified incubator at 37 °C with 5% CO2.
Transfection of microRNA mimics and HO-1 siRNA
The miR-1304 mimics (5′-UUUGAGGCUACAGUGAGAUGUG-3′), its mutant (5′-UAACUCCCUACAGUGAGAUGUG-3′), and HO-1 siRNA duplexes were purchased from GenePharma (Shanghai, China). The synthetic siRNA target sequence is 5′-CAGUUGCUGUAGGGCUUUATT-3′. Transient transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions.
Cell counting and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay
Cells were counted using a Vi-cell XR cell viability analyzer (Beckman Coulter, Brea, CA, USA). Cell viability was determined using the MTT assay. In brief, cells were harvested following 24 h of transfection and plated at 2×103 cells per well in 96-well plates. Following incubation, 20 μL of MTT reagent (5 mg/mL) was added into each well, and the cells were incubated in the dark at 37 °C for 4 h. Then, 100 μL of dissolution buffer was added into each well and incubated overnight. Absorbance was measured at 570 nm using a microtiter plate reader (Bio-Tek Instruments, Winooski, VT, USA).
Colony-formation assay
Twenty-four hours after transfection with miR-1304 mimics or negative control oligonucleotides, the NSCLC cells were seeded in 6-well plates and grown for ten days for the colony-formation assay. The cells were washed with PBS, fixed with methyl alcohol, stained by Geisma and photographed using a Typhoon FLA 9500 instrument (GE Healthcare, Little Chalfont, UK). Colonies were counted using ImageQuant TL (GE Healthcare, Little Chalfont, UK).
Annexin V-PE/7-AAD apoptosis and cell cycle assay
Apoptotic cells were examined using the PE Annexin V Apoptosis Detection Kit I (BD Biosciences, San Jose, CA, USA). The cells were harvested and stained with 5 μL of annexin V-PE and 5 μL of 7-AAD for 15 min at room temperature in the dark. Cell cycle distribution was analyzed by PI staining assay. Fixed cells were treated with RNase A and stained with PI. The cells were measured using a BD FACS flow cytometer (BD Biosciences, San Jose, CA, USA) according to the manufacturer's instructions.
Western blot
Cells were harvested, and protein lysates were separated using 12% SDS-PAGE, transferred to PVDF membranes (Millipore, Billerica, MA, USA) and incubated with a primary antibody against HO-1 (Enzo Life Sciences, ADI-SPA-896-D, USA) or β-actin (sc-47778; Santa Cruz Biotechnology, Dallas, TX, USA) as an internal control. The density of the bands was quantified using ImageQuant 5.2 software (GE Healthcare, Little Chalfont, UK).
RNA isolation and quantitative RT-PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. For HO-1 expression, reverse transcription was performed with PrimeScript RT Master Mix (TaKaRa Biotechnology, Dalian, China) following the manufacturer's instructions. Reverse transcription (RT) reactions contained 0.5 μg of total RNA samples and 4 μL of PrimeScript RT Master and were topped off to 20 μL with DEPC treated water. The thermal profile for RT consisted of incubation at 37 °C for 15 min and termination of the reaction at 85 °C at 5 s. One microliter of undiluted cDNA was used in the RT-qPCR protocol. Quantitative real-time PCR (RT-qPCR) was performed with the QuantiNova SYBR Green PCR kit (Qiagen, Valencia, CA, USA). RT-qPCR reactions contained 10 μL of SYBR Green PCR Master Mix, 0.4 μL of forward primer (10 μmol/L), and 0.4 μL of reverse primer (10 μmol/L) and were topped off to 20 μL with DEPC-treated water. RT-qPCR was analyzed on Rotor-Gene Q 2plex HRM System (Qiagen, Valencia, CA, USA). PCR reactions were performed in triplicate for each sample. The PCR thermal profile consisted of one activation step (90 °C for 1 min), followed by 35 cycles of denaturation at 95 °C for 10 s, annealing and extension at 60 °C for 40 s and subsequent melting curve analysis. The relative HO-1 mRNA levels were analyzed by normalizing the threshold cycle (Ct) value to that of the internal loading control, β-actin. The following primers were used: HO-1 forward, 5′-GCCAGCAACAAAGTGCAAG-3′ and reverse, 5′-GAGTGTAAGGACCCATCGGA-3′ β-actin forward, 5′-GGCTACAGCTTCACCACCAC-3′ and reverse, 5′-GAGTACTTGCGCTCAGGAGG-3′.
To quantify miR-1304, the expression level of miR-1304 was measured using the NCode™ VILO miRNA cDNA Synthesis Kit and EXPRESS SYBR GreenER miRNA qRT-PCR Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. The PCR thermal profile consisted of two activation steps (50 °C for 2 min, 90 °C for 2 min), followed by 40 cycles of denaturation at 95 °C for 15 s, annealing and extension at 60 °C for 40 s and subsequent melting curve analysis. U6 snRNA were used as an internal loading control.
Dual luciferase reporter assay
A full length of human HO-1 3′UTR was amplified and cloned into the Renilla psi-CHECK2 vector (Promega). Mutations in the 3′UTR were generated using a KOD-Plus-Mutagenesis kit (Toyobo).
Cells seeded in 6-well plates were co-transfected with miR-1304 mimics (25 nmol/L) or negative control and reporter constructs (0.5 μg) using Lipofectamine 2000. Cell extracts were prepared 48 h after transfection, and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega).
CRISPR/Cas9-mediated miR-1304 knockdown
CRISPR/Cas9-mediated nucleotide deletion was performed as previously described13. Two sgRNAs (5′-caccgtcactgtagcctcaaaccgc-3′ 5′-caccgaggctacagtgagatgtggc-3′) were cloned into px330-mCherry and px330-GFP vectors, respectively. The plasmids were co-transfected into A549 cells, and positively transfected cells were isolated using flow cytometry and used in the following experiments.
Statistical analysis
All statistical analysis were performed using GraphPad Prism software (version 5.01; GraphPad Software, Inc, CA, USA). The data are shown as the mean values with standard error of the mean (SEM), and P<0.05 was considered statistically significant. All experiments were performed independently at least three times. The significance of differences between two groups was measured using Student's t-test. One-way analysis of variance (ANOVA) was used to measure the significance of comparisons for more than two groups.
Results
Ectopic miR-1304 expression suppresses cell growth and survival of NSCLC cells by inducing cell apoptosis and cell cycle arrest
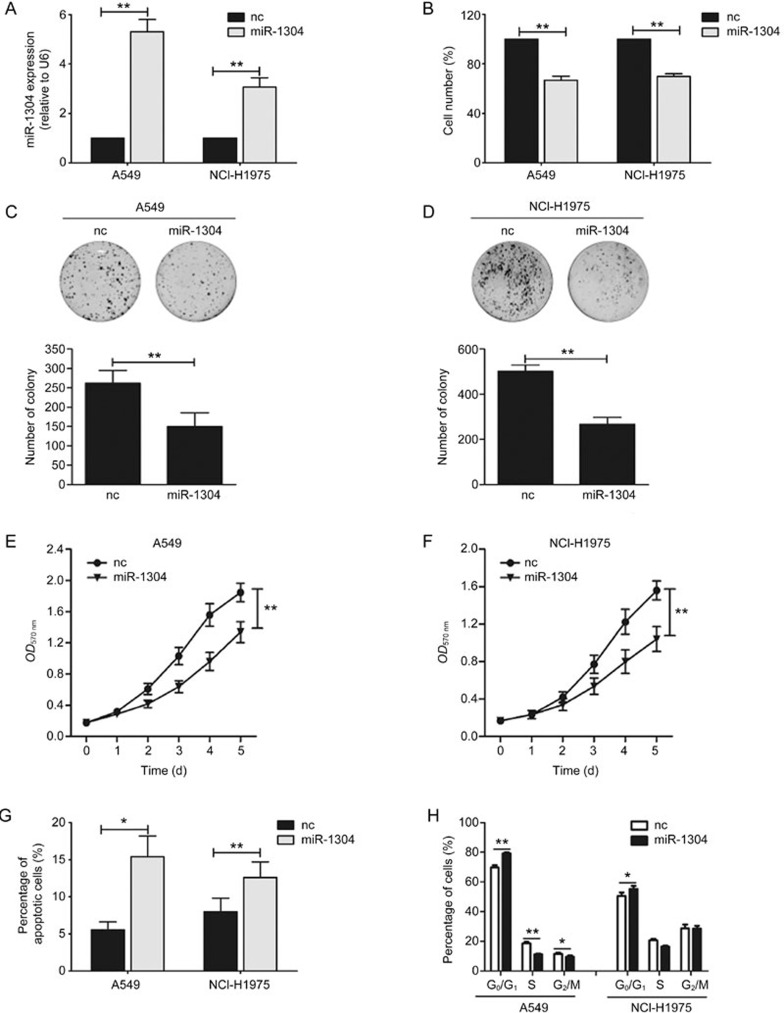
To characterize the function of miR-1304 in NSCLC, A549 and NCI-H1975 cells were transiently transfected with miR-1304 mimics. RT-qPCR analysis confirmed that mature miR-1304 was successfully overexpressed in cells transfected with miR-1304 mimics (Figure 1A). Ectopic miR-1304 expression markedly inhibited growth of A549 and NCI-H1975 cells (Figure 1B). In the colony-formation assay, the colony-forming activities of both A549 and NCI-H1975 cells transfected with miR-1304 mimics were inhibited compared with negative control oligonucleotides (Figure 1C, 1D). To further investigate the function of miR-1304 in the survival of NSCLC cells, we performed an MTT assay to examine the viability of NSCLC cells. The results showed that the viability of A549 and NCI-H1975 cells transfected with miR-1304 mimics was clearly decreased (Figure 1E, 1F). These data demonstrated that miR-1304 was able to suppress the growth of NSCLC cells.
Figure 1.
Ectopic miR-1304 expression suppresses cell growth and survival of NSCLC cells by inducing cell apoptosis and cell cycle arrest. (A) miR-1304 expression in A549 and NCI-H1975 cells transfected with miR-1304 mimics or negative control oligonucleotides (nc). (B) The number of A549 and NCI-H1975 cells transfected with miR-1304 mimics or nc. (C, D) Colony formation in A549 and NCI-H1975 cells transfected with miR-1304 mimics compared with nc. Top: Representative image of the colony formation. Bottom: Statistical results. (E, F) MTT analysis of A549 and NCI-H1975 cells transfected with miR-1304 mimics or nc. Data are presented as the mean±SEM of three independent experiments. (G) Annexin V-PE/7-AAD apoptosis assay in A549 and NCI-H1975 cells measuring the percentage of apoptotic cells at 48 h post-transfection with miR-1304 mimics or nc. (H) PI staining assay and flow cytometry detecting the cell cycle in the indicated cells transfected with miR-1304 mimics or nc. Data are presented as the mean±SEM of three independent experiments. *P<0.05, **P<0.01.
Flow cytometry assays were performed to determine the effects of miR-1304 on apoptosis and the cell cycle of NSCLC cells. As shown in Figure 1G, miR-1304 overexpression led to a significantly increased ratio of apoptotic cells. Cell cycle analysis by flow cytometry based on propidium iodide (PI) staining revealed that transfection with miRNA-1304 mimics in A549 and NCI-H1975 cells increased the percentage of cells in the G0/G1 phase (Figure 1H). Taken together, these data suggested that miR-1304 suppressed cell growth and survival by inducing cell apoptosis and cell cycle arrest in the G0/G1 phase.
HO-1 is a direct target gene of miR-1304
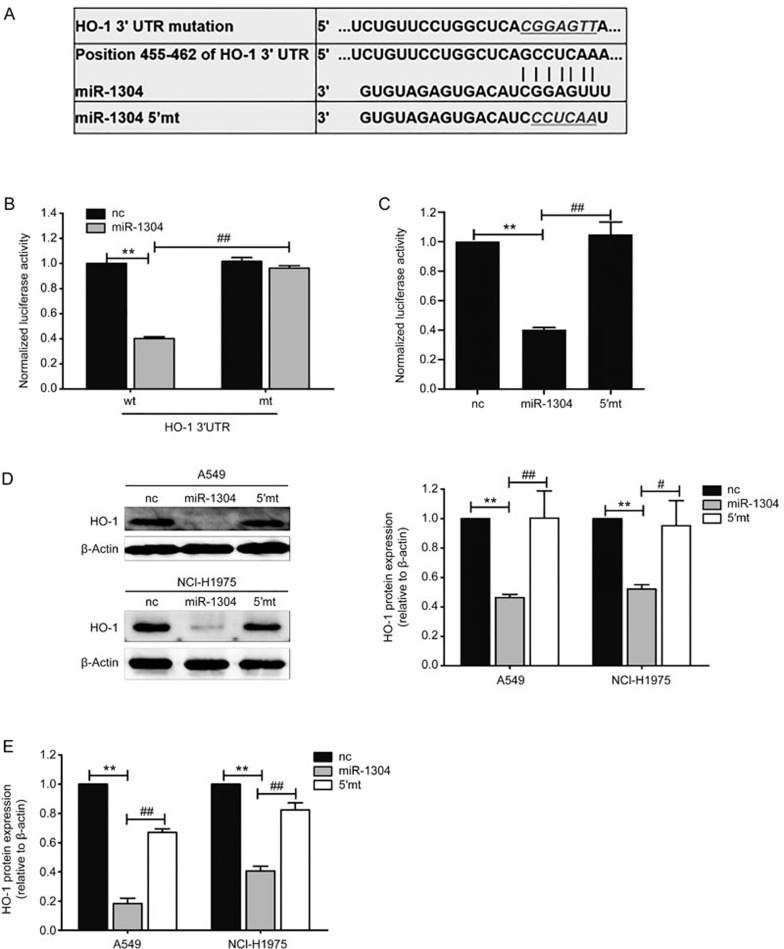
Next, we sought the target of miR-1304, which mediates its inhibitory effects on cell growth and apoptosis. In silico analysis using the database TargetScan (http://www.targetscan.org/) identified HO-1 as a direct target gene of miR-1304. Heme oxygenase-1 (HO-1) catalyzes the rate-limiting step in heme degradation by converting heme to carbon monoxide (CO), free iron and biliverdin. Numerous reports have shown that the expression of HO-1 was higher in cancer cells than in the surrounding healthy tissue. Additionally, HO-1 is believed to exert a cancer cell growth-promoting effect. As shown in Figure 2A, the 3′UTR of HO-1 mRNA contains a putative site that is complementary to the seed region of miR-1304. To confirm that HO-1 is the direct target of miR-1304, a dual luciferase reporter assay was performed. Our results showed that miR-1304 significantly inhibited the luciferase activity of the wild-type (wt) 3′UTR of HO-1 but had no inhibitory effect on the mutant form (mt) (Figure 2B). Similarly, the inhibition effect on HO-1 3′UTR activity was abolished when miR-1304 was mutated in the seed region (5′mt) (Figure 2C). Western blot and RT-qPCR analyses were performed to detect the effects of miR-1304 on HO-1 expression at the protein and mRNA levels, respectively. The results showed that miR-1304 mimics but not its mutant form significantly inhibited HO-1 protein and mRNA expression (Figure 2D, 2E) in A549 and NCI-H1975 cells. These results indicated that HO-1 is a direct target of miR-1304 and is negatively regulated by miR-1304 in NSCLC cells.
Figure 2.
HO-1 is a direct target gene of miR-1304. (A) Sequence complementarity between the 3′UTR of HO-1 mRNA and the seed region of miR-1304. Mutant sequences of HO-1 3′UTR (mt) and miR-1304 (5′ PubMed mt) were used in Figure 3B–3E. (B) Luciferase activity in cells transfected with miR-1304 and reporter plasmids containing wt or mt HO-1 3′-UTR normalized to activity in cells transfected with nc. (C) Luciferase activity in cells transfected with miR-1304 or its mutant and reporter plasmids containing HO-1 3′UTR and quantified as in (B). (D) Immunoblot analysis of the extracts from A549 and NCI-H1975 cells transfected with miR-1304 or its mutant for 48 h. Left: Representative blots. Right: Quantification of HO-1 protein levels normalized to β-actin protein levels and plotted as fold changes relative to the levels in cells transfected with nc. (E) HO-1 mRNA levels in A549 and NCI-H1975 cells transfected with miR-1304 or its mutant for 48 h were detected by RT-qPCR. Data are presented as the mean±SEM of three independent experiments. **P<0.01; #P<0.05, ##P<0.01.
HO-1 plays a critical role in miR-1304-mediated growth suppression of NSCLC cells
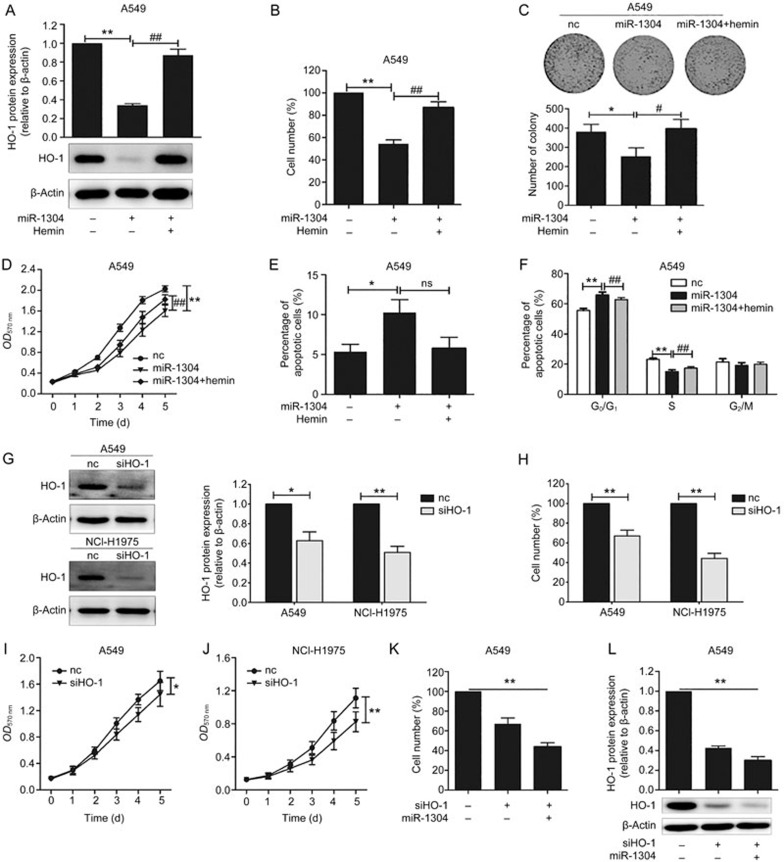
The data presented above suggest that miR-1304 negatively regulates the expression of HO-1 protein and mRNA by targeting its 3′UTR. Most mammalian cells induce HO-1 upon stimulation with hemin. To explore whether HO-1 is involved in modulating NSCLC cell survival, we restored the expression of HO-1 in miR-1304-transfected A549 cells via treatment of the cells with 20 μmol/L hemin, as previously described14. We evaluated the levels of HO-1 protein in A549 cells transfected with miR-1304 alone or in the presence of hemin, and we found that the reduction of HO-1 expression by miR-1304 was abolished by hemin (Figure 3A). Hemin-mediated HO-1 re-expression attenuated miR-1304-promoted growth of NSCLC cells (Figure 3B–3D). MiR-1304-induced cell apoptosis and G0/G1 phase cell cycle arrest were also abolished in hemin-treated cells (Figure 3E, 3F). We used RNAi to study the effect of HO-1 loss of function on NSCLC cell growth. The results showed that in A549 and NCI-H1975 cells transfected with small-interfering RNA against HO-1 (siHO-1), HO-1 protein expression was dramatically decreased compared with cells transfected with negative control oligonucleotides (nc), thus confirming the success of HO-1 knockdown by siHO-1 (Figure 3G). Upon downregulation of HO-1 by siHO-1, the number of NSCLC cells was significantly reduced (Figure 3H), and cell viability was inhibited (Figure 3I, 3J). These results of cell viability upon HO-1 knockdown were consistent with that of miR-1304 transfection in NSCLC cells. To further demonstrate that miR-1304 directly targets HO-1, we co-transfected siHO-1 with miR-1304 mimic into A549 cells. Strikingly, transfection with siHO-1 and miR-1304 together led to a 23% greater reduction in cell numbers compared with siHO-1 transfection alone (Figure 3K). We also analyzed HO-1 protein expression in these cells and found that a greater HO-1 suppression effect was observed in cells co-transfected with siHO-1 and miR-1304, which was in accordance with the cell number inhibition effect (Figure 3L).
Figure 3.
HO-1 plays a critical role in miR-1304-mediated cell growth suppression of NSCLC cells. (A) Immunoblot analysis of A549 cells treated with or without 20 μmol/L hemin after transfection with nc or miR-1304. Top: Quantification of HO-1 protein levels normalized to β-actin protein levels and plotted as fold changes relative to the levels in cells transfected with nc. Bottom: Representative blots. (B–D) Number, colony-forming efficiency and viability of A549 cells treated as in (A). (E, F) Flow cytometry analysis of apoptosis and cell cycle in A549 cells treated as in (A). (G) Immunoblot analysis of the extracts from A549 and NCI-H1975 cells transfected with chemically synthesized siHO-1 or nc for 48 h. Left: Representative blots. Right: Quantification of HO-1 protein levels normalized to β-actin protein levels and plotted as fold changes relative to the levels in cells transfected with nc. (H) Number of A549 and NCI-H1975 cells transfected with siHO-1 or nc. (I, J) MTT analysis of A549 and NCI-H1975 cells transfected with siHO-1 or nc. (K, L) Number and western blot analysis of A549 cells transfected with siHO-1 alone or siHO-1 together with miR-1304. Data are presented as the mean±SEM of three independent experiments. *P<0.05, **P<0.01; nsP>0.05, ##P<0.01.
These results supported the explanation that miR-1304 suppresses growth of NSCLC cells by targeting HO-1.
Endogenous miR-1304 suppresses NSCLC cell growth
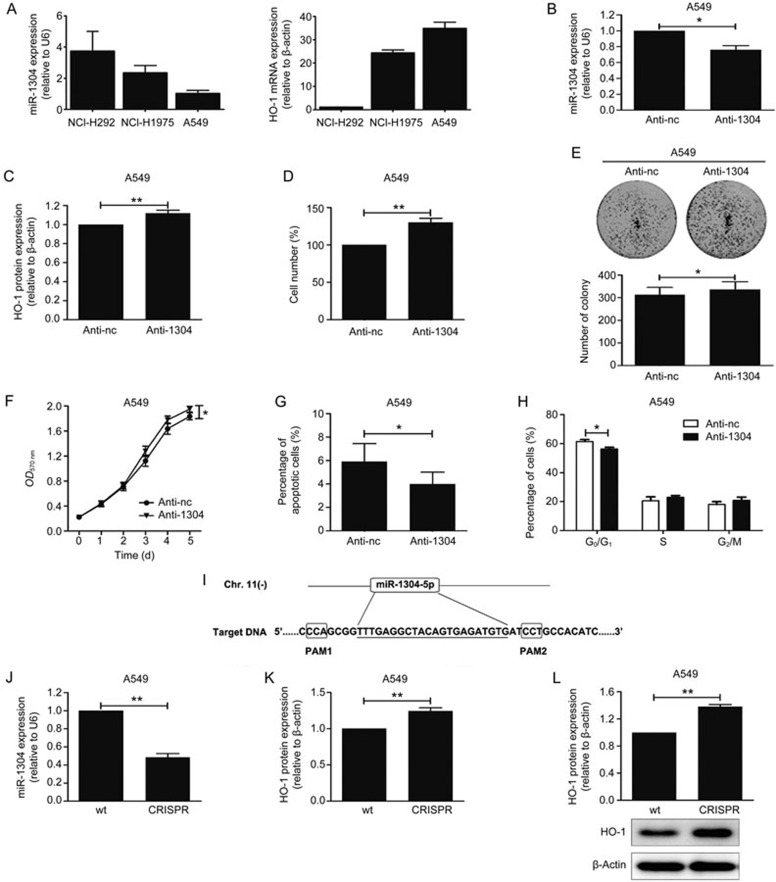
We were also interested in whether endogenous miR-1304 plays a pathophysiological role in NSCLC cells. First, RT-qPCR was used to assess endogenous miR-1304 and HO-1 expression in NSCLS cell lines, as shown in Figure 4A, A549 cells have a relatively low level of miR-1304 and a high level of HO-1. Transfection of A549 cells with anti-sense oligonucleotides against miR-1304 (Anti-1304) resulted in a 24% decrease in miR-1304 expression compared with cells transfected with negative control oligonucleotides (Anti-nc) (Figure 4B). Accordingly, HO-1 expression, cell numbers, colony-forming efficiency and cell viability were slightly enhanced in A549 cells transfected with Anti-1304 (Figure 4C-4F). Flow cytometry analysis revealed that transfection with anti-1304 in A549 cells decreased the percentage of apoptotic and G0/G1 phase cells (Figure 4G, 4H). To further determine the relationship between miR-1304 and HO-1, we used CRISPR/Cas9 to knockdown endogenous miR-1304. The results showed that miR-1304 was dramatically reduced in cells with miR-1304 knocked down by CRISPR/Cas9 (Figure 4J), and the mRNA and protein expression levels of HO-1 increased significantly in these cells (Figure 4K, 4L).
Figure 4.
Endogenous miR-1304 suppresses NSCLC cell growth. (A) RT-qPCR analysis of endogenous miR-1304 (left) and HO-1 (right) expression in NSCLC cell lines. (B) miR-1304 expression in A549 cells transfected with antisense oligonucleotides against miR-1304 (Anti-1304) or negative control antisense oligonucleotides (Anti-nc). (C) Immunoblot analysis of the extracts from A549 cells transfected with Anti-1304 or Anti-nc. (D–F) Number, colony-forming efficiency and viability analyses of A549 cells transfected with Anti-1304 or Anti-nc. (G, H) Flow cytometry analysis of apoptosis and cell cycle in A549 cells transfected with Anti-1304 or Anti-nc. (I) Schematic representation of the CRISPR/Cas9 strategy mediated deletion of miR-1304-5p. The mature miR-1304-5p sequence is underlined. (J) RT-qPCR measurement of the expression level of miR-1304 in miR-1304 knockdown cells. (K, L) HO-1 mRNA (K) or protein (L) levels in miR1304 knockdown cells. Data are presented as the mean±SEM of three independent experiments. *P<0.05, **P<0.01.
These results provided further evidence that miR-1304 suppresses growth of NSCLC cells by targeting HO-1.
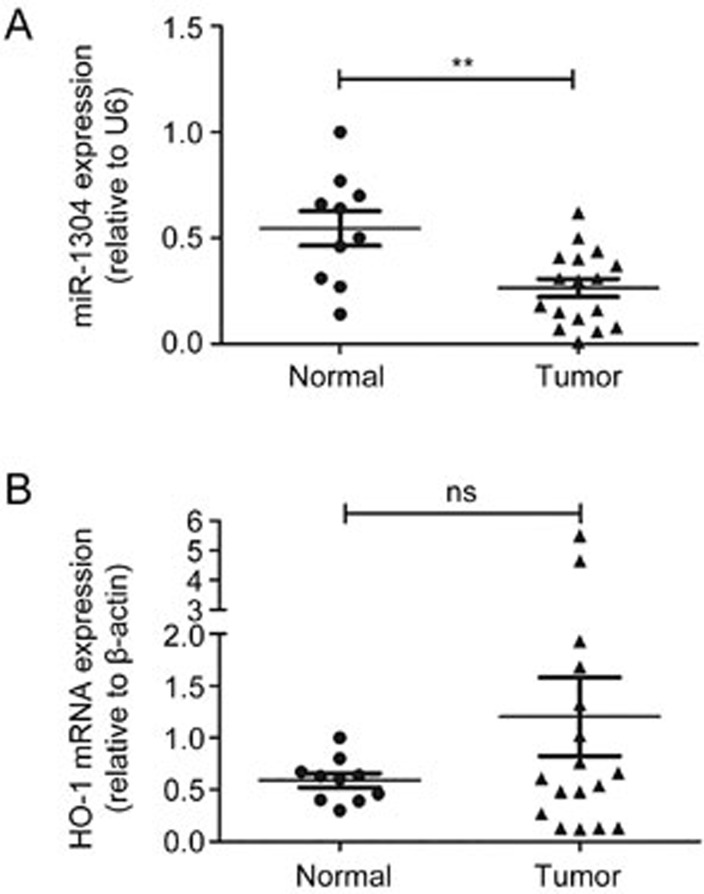
MiR-1304 is down-regulated in human NSCLC tissues
To investigate the pathophysiological role of miR-1304 in human lung carcinogenesis, we evaluated the expression of miR-1304 and HO-1 in seventeen frozen human NSCLC tissues and ten normal human lung tissues using RT-qPCR. The expression level of miR-1304 in tumor tissues was significantly lower than in normal lung tissues (Figure 5A), whereas HO-1 was up-regulated in tumor tissues compared with normal lung tissues, although no statistically significant difference was found (Figure 5B), probably due to the small sample size.
Figure 5.
MiR-1304 is down-regulated in human NSCLC tissues. (A, B) RT-qPCR analysis of miR-1304 (A) and HO-1 (B) expression in NSCLC tissues compared with normal lung tissues. Data are presented as the mean±SEM. nsP>0.05, **P<0.01.
These findings suggested that decreased miR-1304 and increased HO-1 expression might be responsible for NSCLC growth.
Discussion
MicroRNA dysregulation has been observed in various pathological conditions, such as cancer. It plays an important role in cancer development and progression by suppressing the expression of genes involved in cell proliferation, apoptosis, differentiation, invasion and metastasis. MicroRNAs are also considered potential targets for cancer treatment. Therefore, identification of tumor-related miRNAs and their direct target genes is critical to understanding the biological function of miRNA in cancer and might offer novel biomarkers or therapeutic targets for effective treatment of cancer.
miR-1304 is a primate-specific miRNA that has low expression in human embryonic stem cells, peripheral blood, brain cortex, and melanoma15,16,17,18. Recently, two studies indicated that miR-1304 might influence the cell viability in different cancer types. In paclitaxel-treated hypopharynx cancer cells, miR-1304 was found to be dramatically up-regulated coupled with decreased cell viability10. In contrast, miR-1304 was down-regulated in Bcl-xl silenced lung adenocarcinoma, and cell viability was reduced in the tumor11. These results suggest that miR-1304 might exert different roles via different pathways in response to different stimuli. Although certain targets have been reported and predicted10,12, the direct target of miR-1304, by which it influences cancer cell viability, is still unclear. In our study, we found that miR-1304 decreased the number of NSCLC cells and suppressed cell viability and colony-formation ability. Furthermore, ectopic expression of miR-1304 led to a higher percentage of apoptotic cell and cells in G0/G1 phase in vitro.
HO-1 was further identified as a new, direct and functional target of miR-1304 using miRNA target prediction software and experimental validation. Using dual-luciferase reporter assays, we found that miR-1304 could target the 3′UTR sequence of HO-1. In addition, miR-1304 but not its mutant form can inhibit HO-1 mRNA and protein expression. Moreover, we validated the importance of miR-1304/HO-1 axis via rescue experiments. Restoration of HO-1 expression abolished miR-1304-mediated cell growth suppression of NSCLC cells. Consistent with miR-1304 transfection, knockdown of HO-1 decreased the number of NSCLC cells and suppressed cell viability. In addition, we found that endogenous miR-1304 suppressed HO-1 expression. In miR-1304-inhibited cells, HO-1 expression, cell growth, colony-forming efficiency and cell viability were increased. Probably owing to the low level of endogenous miR-1304 in A549 cells and insufficient suppression efficiency of Anti-1304 antisense oligonucleotides, Anti-1304-induced changes in cell growth and colony formation were not significant. However, cell apoptosis was significantly suppressed by Anti-1304, which indicated that these cells were more sensitive to miR-1304 with respect to apoptosis. Decreased miR-1304 and increased HO-1 were detected in NSCLC tissues compared with related normal tissues.
Heme oxygenase-1 (HO-1), also known as Hsp32, catalyzes the rate-limiting step in heme degradation. HO-1 converts heme to carbon monoxide (CO), free iron and biliverdin, which is quickly reduced to bilirubin. Numerous studies have demonstrated the function of HO-1 in many types of cancer, including thyroid cancer19, acute myeloid leukemia20, breast cancer21, prostate cancer22, melanoma23, renal cancer24 and non-small cell lung cancer25. Compared with surrounding healthy tissue, HO-1 is extensively expressed in tumor tissues, and HO-1 is believed to exert a cancer cell growth-promoting effect that is dependent on its three metabolites26 and itself27. Consequently, several anticancer strategies aimed at HO-1 have been developed, and it has been clearly demonstrated that specific HO inhibitors such as metalloporphyrins present tumor inhibitory effects. However, clinical use of HO-1 inhibitors is limited because of various side effects28. Therefore, the development of novel HO-1 inhibitors is in great demand. MicroRNA, which suppresses gene expression at post-transcriptional level, offers a potential strategy. Several reports have found that miR-217, miR-377, miR-200c and miR-24-3p can directly interact with HO-1 3′UTR in hemin chloride-treated HEK293, human umbilical vein endothelial cells, clear-cell renal cell carcinoma cells and MARC-145 cells14,29,30. In this work, we found that microRNA-miR-1304 could significantly suppress the expression of HO-1 in NSCLC cells by targeting the HO-1 3′UTR.
In conclusion, we demonstrated that miR-1304 inhibited cell growth and decreased cell viability and colony-forming activity in NSCLC cells. Furthermore, miR-1304 most likely exerts its biological functions on NSCLC cell growth and viability by targeting HO-1. Our findings suggest that modulation of miR-1304/HO-1 expression offers a potential therapeutic strategy for NSCLC.
Author contribution
Jin REN, Ling-ling MIAO, and Cheng-gang LI designed the project and wrote the manuscript; Cheng-gang LI performed the research; Meng-fan PU, Chun-zhu LI, Man GAO, Ming-xia LIU, Cun-zhi YU, Hong YAN, Chun PENG, Yang ZHAO, Yu LI, Ze-long MA, and Xin-ming QI helped to design the overall study and analyze the data; and all authors contributed to manuscript preparation.
Acknowledgments
We are grateful to Shanghai Pulmonary Hospital for providing lung cancer tissues.
This work was supported by grants from the National Science and Technology Major Project (2012ZX09302-003, 2012ZX09301-001-006 and 2015ZX09102005).
References
- Wood SL, Pernemalm M, Crosbie PA, Whetton AD. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev 2014; 40: 558–66. [DOI] [PubMed] [Google Scholar]
- Ye L, Wang H, Liu B. miR-211 promotes non-small cell lung cancer proliferation by targeting SRCIN1. Tumour Biol 2016; 37: 1151–7. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010; 79: 351–79. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136: 215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. Getting to the root of miRNA-mediated gene silencing. Cell 2008; 132: 9–14. [DOI] [PubMed] [Google Scholar]
- Liu K, Li X, Cao Y, Ge Y, Wang J, Shi B. miR-132 inhibits cell proliferation, invasion and migration of hepatocellular carcinoma by targeting PIK3R3. Int J Oncol 2015; 47: 1585–93. [DOI] [PubMed] [Google Scholar]
- Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell 2010; 18: 510–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Huang F, Yu D, Zhang Y, Xu H, Zhang L, et al. Autoregulatory loop between TGF-beta1/miR-411-5p/SPRY4 and MAPK pathway in rhabdomyosarcoma modulates proliferation and differentiation. Cell Death Dis 2015; 6: e1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay D, Leblanc R, Grunewald TG, Ambatipudi S, Ribeiro J, Clezardin P, et al. The LPA1/ZEB1/miR-21-activation pathway regulates metastasis in basal breast cancer. Oncotarget 2015; 6: 20604–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu CZ, Shi RJ, Chen D, Sun YY, Wu QW, Wang T, et al. Potential biomarkers for paclitaxel sensitivity in hypopharynx cancer cell. Int J Clin Exp Pathol 2013; 6: 2745–56. [PMC free article] [PubMed] [Google Scholar]
- Othman N, In LL, Harikrishna JA, Hasima N. Bcl-xL silencing induces alterations in hsa-miR-608 expression and subsequent cell death in A549 and SK-LU1 human lung adenocarcinoma cells. PLoS One 2013; 8: e 81735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Valenzuela M, Ramirez O, Rosas A, Garcia-Vargas S, de la Rasilla M, Lalueza-Fox C, et al. An ancestral miR-1304 allele present in Neanderthals regulates genes involved in enamel formation and could explain dental differences with modern humans. Mol Biol Evol 2012; 29: 1797–806. [DOI] [PubMed] [Google Scholar]
- Miao L, Yao H, Li C, Pu M, Yao X, Yang H, et al. A dual inhibition: microRNA-552 suppresses both transcription and translation of cytochrome P450 2E1. Biochim Biophys Acta 2016; 1859: 650–62. [DOI] [PubMed] [Google Scholar]
- Beckman JD, Chen C, Nguyen J, Thayanithy V, Subramanian S, Steer CJ, et al. Regulation of heme oxygenase-1 protein expression by miR-377 in combination with miR-217. J Biol Chem 2011; 286: 3194–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu AL, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 2008; 18: 610–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaz C, Ahmad HM, Sharma P, Gupta R, Kumar L, Kulshreshtha R, et al. Analysis of microRNA transcriptome by deep sequencing of small RNA libraries of peripheral blood. BMC Genomics 2010; 11: 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti E, Pantano L, Banez-Coronel M, Llorens F, Minones-Moyano E, Porta S, et al. A myriad of miRNA variants in control and Huntington's disease brain regions detected by massively parallel sequencing. Nucleic Acids Res 2010; 38: 7219–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MS, Tyagi S, Nancarrow DJ, Boyle GM, Cook AL, Whiteman DC, et al. Characterization of the melanoma miRNAome by deep sequencing. PLoS One 2010; 5: e9685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TY, Liu CL, Chen MJ, Lee JJ, Pun PC, Cheng SP. Expression of haem oxygenase-1 correlates with tumour aggressiveness and BRAF V600E expression in thyroid cancer. Histopathology 2015; 66: 447–56. [DOI] [PubMed] [Google Scholar]
- Lin X, Fang Q, Chen S, Zhe N, Chai Q, Yu M, et al. Heme oxygenase-1 suppresses the apoptosis of acute myeloid leukemia cells via the JNK/c-JUN signaling pathway. Leuk Res 2015; 39: 544–52. [DOI] [PubMed] [Google Scholar]
- Lee HN, Jin HO, Park JA, Kim JH, Kim JY, Kim B, et al. Heme oxygenase-1 determines the differential response of breast cancer and normal cells to piperlongumine. Mol Cells 2015; 38: 327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labanca E, De Luca P, Gueron G, Paez A, Moiola CP, Massillo C, et al. Association of HO-1 and BRCA1 is critical for the maintenance of cellular homeostasis in prostate cancer. Mol Cancer Res 2015; 13: 1455–64. [DOI] [PubMed] [Google Scholar]
- Barbagallo I, Parenti R, Zappala A, Vanella L, Tibullo D, Pepe F, et al. Combined inhibition of Hsp90 and heme oxygenase-1 induces apoptosis and endoplasmic reticulum stress in melanoma. Acta Histochem 2015; 117: 705–11. [DOI] [PubMed] [Google Scholar]
- Balan M, Mier y Teran E, Waaga-Gasser AM, Gasser M, Choueiri TK, Freeman G, et al. Novel roles of c-Met in the survival of renal cancer cells through the regulation of HO-1 and PD-L1 expression. J Biol Chem 2015; 290: 8110–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JR, Wang HM, Liu PL, Chen YH, Yang MC, Chou SH, et al. High expression of heme oxygenase-1 is associated with tumor invasiveness and poor clinical outcome in non-small cell lung cancer patients. Cell Oncol (Dordr) 2012; 35: 461–71. [DOI] [PubMed] [Google Scholar]
- Fang J, Akaike T, Maeda H. Antiapoptotic role of heme oxygenase (HO) and the potential of HO as a target in anticancer treatment. Apoptosis 2004; 9: 27–35. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Yeh CT, Sun YJ, Chiang MT, Lan WM, Li FA, et al. Signal peptide peptidase-mediated nuclear localization of heme oxygenase-1 promotes cancer cell proliferation and invasion independent of its enzymatic activity. Oncogene 2015; 34: 2360–70. [DOI] [PubMed] [Google Scholar]
- Pittala V, Salerno L, Romeo G, Modica MN, Siracusa MA. A focus on heme oxygenase-1 (HO-1) inhibitors. Curr Med Chem 2013; 20: 3711–32. [DOI] [PubMed] [Google Scholar]
- Gao C, Peng FH, Peng LK. MiR-200c sensitizes clear-cell renal cell carcinoma cells to sorafenib and imatinib by targeting heme oxygenase-1. Neoplasma 2014; 61: 680–9. [DOI] [PubMed] [Google Scholar]
- Xiao S, Wang X, Ni H, Li N, Zhang A, Liu H, et al. MicroRNA miR-24-3p promotes porcine reproductive and respiratory syndrome virus replication through suppression of heme oxygenase-1 expression. J Virol 2015; 89: 4494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]