Abstract
ITSF and ITSReub, constituting a new primer set designed for the amplification of the 16S-23S rRNA intergenic transcribed spacers, have been compared with primer sets consisting of 1406F and 23Sr (M. M. Fisher and E. W. Triplett, Appl. Environ. Microbiol. 65:4630-4636, 1999) and S-D-Bact-1522-b-S-20 and L-D-Bact-132-a-A-18 (L. Ranjard et al., Appl. Environ. Microbiol. 67:4479-4487, 2001), previously proposed for automated ribosomal intergenic spacer analysis (ARISA) of complex bacterial communities. An agricultural soil and a polluted soil, maize silage, goat milk, a small marble sample from the façade of the Certosa of Pavia (Pavia, Italy), and brine from a deep hypersaline anoxic basin in the Mediterranean Sea were analyzed with the three primer sets. The number of peaks in the ARISA profiles, the range of peak size (width of the profile), and the reproducibility of results were used as indices to evaluate the efficiency of the three primer sets. The overall data showed that ITSF and ITSReub generated the most informative (in term of peak number) and reproducible profiles and yielded a wider range of spacer sizes (134 to 1,387) than the other primer sets, which were limited in detecting long fragments. The minimum amount of DNA template and sensitivity in detection of minor DNA populations were evaluated with artificial mixtures of defined bacterial species. ITSF and ITSReub amplified all the bacteria at DNA template concentrations from 280 to 0.14 ng μl−1, while the other primer sets failed to detect the spacers of one or more bacterial strains. Although the primer set consisting of ITSF and ITSReub and that of S-D-Bact-1522-b-S-20 and L-D-Bact-132-a-A-18 showed similar sensitivities for the DNA of Allorhizobium undicula mixed with the DNA of other species, the S-D-Bact-1522-b-S-20 and L-D-Bact-132-a-A-18 primer set failed to detect the DNA of Pseudomonas stutzeri.
Automated ribosomal intergenic spacer analysis (ARISA) is an automated, culture-independent technique suitable for analyzing structures of microbial environmental communities. This PCR-based technique, developed by Fisher and Triplett (7), is based on the use of a fluorescent primer in the amplification of microbial ribosomal intergenic spacers, using DNA extracted from environmental samples as a template. The PCR products are analyzed by an automated capillary electrophoresis system that produces an electropherogram, the peaks of which correspond to discrete DNA fragments detected by a laser-based fluorescence detection system. The sensitivity of ARISA is very high (detection level, single-nucleotide difference), and reproducibility is guaranteed by instrumental automatism, as for length heterogeneity PCR (20, 24) and terminal restriction fragment length polymorphism (3, 15, 21), automated techniques commonly used in studying microbial community structure.
ARISA has been used to analyze the genetic structures of several bacterial and/or fungal communities from samples of freshwater (7), bacterioplankton (8, 10), and different soils (19) and has allowed an accurate estimation of community complexity, detecting from 38 to 232 peaks per profile (7, 19). In fact, the instrumental automatism of ARISA and the easy analysis of its output data make it a very suitable technique for analyzing and comparing large numbers of samples, the results obtained being both reliable and reproducible.
However, it is not easy to compare results, since different primer sets, exploring different annealing sites of the prokaryote ribosomal operon, have been used. Moreover, the use of different primers can result in a different amplification efficiency, depending on the primer sensitivity towards less-represented DNA populations in the mixture and primer specificity.
For these reasons, a primer set that could give the most-efficient DNA amplifications with a minimal amount of environmental DNA and that would reduce the possible PCR bias of preferential amplification of some templates in a mixture, for example, due to substrate substrate reannealing during PCR (23), would be most useful. In fact, the amount of DNA template available from precious samples that cannot be sampled easily and extensively, such as artworks and monuments, or that are expensive or collected with great difficulty could be the limiting factor for achieving a good description of microbial communities.
Our purpose was to select a suitable primer set for ARISA of environmental bacterial communities. We compared the efficiency of the previously published primer sets with that of the newly designed primer set consisting of ITSF and ITSReub (ITSF/ITSReub). The efficiencies of the three primer sets in generating ARISA profiles from bacterial communities were compared by using DNA extracted from six different environmental samples, including soils (natural and polluted), food and feed (goat milk and ensiled corn), a precious sample from an ancient monument (from the marble façade of the Certosa of Pavia), and a sample expensive to collect, anoxic brine from a deep hypersaline anoxic basin (DHAB) in the eastern Mediterranean Sea.
The three primer sets showed different amplification efficiencies, the electropherograms differing greatly in the number of peaks and peak size for all six of the environmental DNA samples. The efficiencies of the three primer sets were also tested by performing ARISA on different artificial model communities, prepared by mixing known amounts of DNA of various bacteria. These model communities allowed us to easily compare the efficiencies and the sensitivities of the three primer sets for amplification of DNA from specific strains in the mixture and for detection of minor components in the communities.
MATERIALS AND METHODS
Environmental samples and pure strains.
The polluted soil consisted of a 20-year-old anthropogenic soil generated in a municipal landfill (situated near Palermo in Sicily, Italy) that contained building refuse and municipal solid waste mixed with natural soil. The natural soil was a nutrient-rich soil, subjected to extensive agricultural practice, recovered from the nonpolluted area bordering the landfill. The brine sample was anoxic water taken from a DHAB in the eastern Mediterranean Sea. It was collected with a 10-liter Niskin bottle during the Urania R/V cruise in the Urania basin in the ambit of the Biodeep project, August 2001, Station Modus 12 (35°13′69N, 21°31′14E) at a depth of 3,535 m. The brine sample (1 liter) was filtered on board on a 0.2-μm-pore-size filter to recover the bacterial cells, and the filter was then used for DNA extraction. The ensiled maize sample was obtained by ensiling, for 30 days in anaerobic conditions at 30°C in sterile glass bottles, 1 kg of maize plants grown for 100 days in a greenhouse. The goat milk sample (25 ml) was collected in the summer of 2001 from a breeding goat fed with maize silage. The milk sample was mixed with an equal volume of 0.9% (wt/vol) NaCl and centrifuged at 5,000 × g for 10 min at 4°C. The pellet was washed with 10 ml of 0.9% (wt/vol) NaCl and used for DNA extraction. The monument sample consisted of 20 mg of marble powder collected, by using a sterile scalpel, from the greatly altered Carrara marble surface of the 16th-century façade of the Certosa of Pavia, Italy (25). All the samples were stored at −70°C until processing.
Allorhizobium undicula USDA 4903, Rhizobium leguminosarum USDA 2370, Sinorhizobium meliloti USDA 1002, Bacillus cereus DSMZ 31T, Streptococcus bovis JM 300, Escherichia coli DSMZ 50902, Pseudomonas stutzeri 1015, and Streptomyces coelicolor M145 were used as reference strains for testing the efficiency of ARISA. A. undicula, R. leguminosarum, and S. meliloti were grown in yeast extract-mannitol medium for 3 to 5 days at 28°C. B. cereus, E. coli, and P. stutzeri were grown in Luria-Bertani medium overnight at 37°C. S. bovis was grown in brain heart infusion medium overnight at 37°C. S. coelicolor was grown in R2Y medium for 3 days at 28°C. After growth, the cells were recovered by centrifugation and washed two times with phosphate-buffered saline.
Extraction and purification of DNA from pure bacterial strains and environmental samples.
Total DNA was extracted from the washed cells of pure strains by sodium dodecyl sulfate, proteinase K, and cethyltrimethyl-ammonium bromide (CTAB) treatment, as described by Ausubel et al. (2).
Soil DNA was extracted as described by Zhou et al. (26) and purified by fractionation on Sepharose 4B (Sigma, Milan, Italy) as previously described (11, 27). DNA was extracted from the filtered bacterial cells of the Urania brine with lysozyme, sodium dodecyl sulfate, and proteinase K, as previously described by Murray et al. (16). Five grams of ensiled corn silage were crushed with liquid nitrogen, and DNA was extracted in a CTAB-based buffer and purified by standard phenol-chloroform-isoamyl alcohol (25:24:1) treatment. The DNA from the milk pellet was extracted as previously described by Lipkin et al. (14), by treatment with dithiothreitol (70 mM) and proteinase K (0.2 mg ml−1) at 42°C overnight. Microbial DNA was extracted from the marble sample as described by Schabereiter-Gurtner et al. (22) by using lysozyme, sodium dodecyl sulfate, proteinase K, and CTAB.
The DNA extracted from each of the samples was quantified by agarose gel electrophoresis in Tris-borate-EDTA buffer, comparing it with known amounts of lambda DNA. The monument sample did not yield sufficient DNA to be visualized in the gel; thus, for that sample, 1 μl of extracted DNA was used for all the ARISA-PCR.
Artificial complex mixtures of DNA (model bacterial communities).
DNAs extracted from B. cereus DSMZ 31T, P. stutzeri 1015, S. bovis JM 300, E. coli DSMZ 50902, S. coelicolor M145, and A. undicula USDA 4903 were mixed to obtain an artificial community containing about 280 ng of total DNA μl−1, in which each strain was represented by the following DNA concentrations, B. cereus DSMZ 31T (22 ng μl−1), P. stutzeri 1015 (62 ng μl−1), S. bovis JM 300 (75 ng μl−1), E. coli DSMZ 50902 (50 ng μl−1), S. coelicolor M145 (38 ng μl−1), and A. undicula USDA 4903 (33 ng μl−1). This artificial community was used as the model to test primer efficiency in generating an ARISA profile that represented the real composition of the DNA mixture. The DNA mixture was serially diluted up to 1:2,000 (vol:vol) with sterile deionized water to obtain several solutions containing 28, 2.8, 1.4, and 0.14 ng of total DNA μl−1. One microliter of each dilution was used as the template in the ARISA-PCR in order to test the efficiency of the primer sets in amplifying small amounts of template. To calculate the expected ARISA profiles of the artificial mixed bacterial community, the theoretical fluorescence proportion for the peak of each bacterium was calculated, taking into account the genome size and the rRNA operon copy numbers (13). The considered genome sizes and rRNA operon copy numbers were the following: for B. cereus, 5.4 Mb, 12 copies; for P. stutzeri, 4.3 Mb, 4 copies; for S. bovis, 2 Mb, 5 copies; for E. coli, 5 Mb, 7 copies; for S. coelicolor, 8.67 Mb, 6 copies. In the case of A. undicula, for which information on genome size and rRNA operon copy number were not available, we have used these data from the related species belonging to the genera Rhizobium and Sinorhizobium.
In order to determine primer set sensitivity for detecting major and minor DNA populations in a mixture, model communities were prepared by making mixtures of comparable amounts of DNA (about 45 ng) from B. cereus 31T, E. coli DSM 50902, and S. coelicolor M145 with serial dilutions of DNA of P. stutzeri 1015 and A. undicula USDA 4903 separately to obtain final DNA mixtures in which P. stutzeri 1015 and A. undicula USDA 4903 DNA represented fractions of 50, 9.1, 4.8, 0.99, 0.5, and 0.1% of the total DNA. The expected relative fluorescences of the peaks corresponding to P. stutzeri 1015 and A. undicula USDA 4903 for each mixture were 39.1, 6.6, 3.4, 0.70, 0.35, and 0.07% for P. stutzeri 1015 and 36.3, 5.4, 2.8, 0.57, 0.28, and 0.06% for A. undicula USDA 4903, respectively.
Primers.
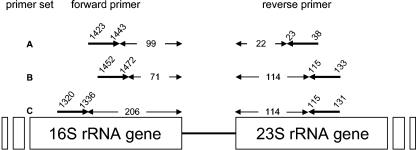
We tested the two universal primer sets 1406F and 23Sr (1406F/23Sr) (4) and S-D-Bact-1522-b-S-20 and L-D-Bact-132-a-A-18 (S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18) (17, 18). The 5′ and 3′ ends of primers 1406F (5′-TGYACACACCGCCCGT-3′) and 23Sr (5′-GGGTTBCCCCATTCRG-3′) were, respectively, complementary to positions 1320 and 1336 of the 16S rRNA and 131 and 115 of the 23S rRNA genes of E. coli. The 5′ and 3′ ends of primers S-D-Bact-1522-b-S-20 (5′-TGCGGCTGGATCCCCTCCTT-3′) and L-D-Bact-132-a-A-18 (5′-CCGGGTTTCCCCATTCGG-3′) were, respectively, complementary to positions 1452 and 1472 of the 16S rRNA and 133 and 115 of the 23S rRNA of E. coli. We also tested the new designed primer set ITSF/ITSReub. The 5′ and 3′ ends of primers ITSF (5′-GTCGTAACAAGGTAGCCGTA-3′) and ITSReub (5′-GCCAAGGCATCCACC-3′) were, respectively, complementary to positions 1423 and 1443 of the 16S rRNA and 38 and 23 of the 23S rRNA of E. coli. The positions of the six primers on the prokaryotic ribosomal operon and the numbers of amplified nucleotides belonging to the 16S and 23S rRNA genes are shown schematically in Fig. 1. The primers S-D-Bact-1522-b-S-20 and ITSReub were labeled with the phosphoramidite dye HEX (6-carboxy-1,4-dichloro-2′,4′,5′,7′-tetra-chlorofluorescein); the primer 1406F was labeled with the phosphoramidite dye 6-FAM (6-carboxyfluorescein) (7).
FIG. 1.
Positions of the six primers on the prokaryote ribosomal operon and numbers of nucleotides belonging to 16S and 23S rRNA genes amplified by PCR. The numbers of nucleotides amplified on 16S and 23S rRNA genes and the positions of the annealing sites by each primer set were calculated based on the E. coli O157:H7 ribosomal genes. (A) ITSF/ITSReub; (B) SD-Bact-1522-b-S-20/LD-Bact-132-a-A-18 (19); (C) 1406F/23Sr (4).
ARISA conditions.
PCR for ARISA was performed as previously described by Fisher and Triplett (7) for the 1406F/23Sr set and by Ranjard et al. (17, 18) for the S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer set. For the ITSF/ITSReub primer set, we amplified between 10 and 300 ng of environmental DNA (depending on the sample) in a reaction mixture containing 1× PCR buffer, 1.5 U of Taq DNA polymerase (Amersham-Pharmacia), 0.2 mM (each) deoxynucleoside triphosphate, and 0.25 μM (each) primer in a final volume of 25 μl. The mixture was held at 94°C for 3 min, followed by 30 cycles of 94°C for 45 s, 55°C for 1 min, 72°C for 2 min, and a final extension at 72°C for 7 min. The same protocols were followed to perform ARISA on artificial communities, using 0.14 to 280 ng of total DNA as a template.
A standardized amount (between 0.5 and 1 μl) of PCR product, along with 0.5 to 0.8 μl of an internal size standard, 1000 or 2500 ROX (Applied Biosystems), was added to 13 μl of deionized formamide, and the mixture was denatured at 95°C for 5 min, followed by 2 min on ice. The sample fragments were then discriminated by using the ABI 310 genetic analyzer (Perkin-Elmer), in which DNA was electrophoresed in a capillary tube filled with the electrophoresis polymer POP-4 (Applied Biosystems). The samples were run under standard ABI 310 denaturing electrophoresis conditions for 1 h each, and the data were analyzed using the GeneScan 3.1 software program (Perkin-Elmer). The program output is a series of peaks (an electropherogram), the sizes of which are estimated by comparison with fragments of the internal size standard. The GeneScan software calculates the height and the area of each peak, which are proportional to the quantity of DNA in the fragment.
All the ARISA experiments were replicated at least three times to confirm the results. For each environmental DNA, the reproducibility of the ARISA was measured as the average and the relative standard deviations of the peak number calculated from the three independent experiments.
Statistical analysis, database examination, and primer analysis.
The ARISA peak patterns were analyzed using GeneScan 3.1 software. The chi-square test was used to compare the relative distributions of peaks in the different artificial communities after normalization of the data by logarithmic transformation; chi-square values themselves were compared by using Student's t test. In order to determine which of the three primer sets described the complexities of the environmental bacterial communities in most detail, the peak patterns obtained by using the three primer sets with three independent ARISA experiment replicates were employed to calculate the ecological indices of Shannon-Weaver (H = −∑Pi logPi) and Simpson (1 − D = 1 − ∑Pi2), where Pi is the ratio between the intensity of fluorescence of each ith peak and the total intensity of fluorescence and D is the dominance Simpson index. For each environmental sample, the values of the ecological indices obtained by using the three primer sets were compared by using Student's t test.
We aligned the sequences of the six primers with the National Center for Biotechnology Information (NCBI) database using the BLAST program (1); this was done in order to verify both the theoretical capacity to match the sequence of all bacterial domains and the absence of alignment with sequences of nonbacterial organisms. For each phylogenetic group of the domain Bacteria, 1,000 alignments were requested, after setting the parameters of the searching to the following values: word size, 7; maximum e-value, 1,000. The alignments were evaluated for a total of 19,016 sequences belonging to the following phylogenetic groups: Actinomycetales (3,611 sequences examined); Proteobacteria (5,069 sequences examined); Firmicutes (4,169 sequences examined); Chlamydiae (575 sequences examined); Cyanobacteria (1,879 sequences examined); Spirochetales (1,127 sequences examined); Aquificales (59 sequences examined); Thermotogales (105 sequences examined); green nonsulfur bacteria (30 sequences examined); green sulfur bacteria (48 sequences examined); Termus-Deinococcus group (353 sequences examined); Cytophaga-Flexibacter-Bacteroides (CFB) group (1,588 sequences examined); planctomycetes (234 sequences examined); Synergistes (2 sequences examined); Holophaga (91 sequences examined); Thermodesulfobacterium group (8 sequences examined); and Verrucomicrobia (68 sequences examined).
Each primer set was analyzed for the secondary structures and the annealing temperatures using the oligoanalyzer 3.0 program (http://207.32.43.70/biotools/oligocalc/oligocalc.asp).
RESULTS
ARISA profiles of environmental bacterial communities.
In order to compare the efficiencies of the three primer sets in the ARISA, we used as the efficacy index the number of peaks generated via PCR, the ranges of peak size (width of the profile), the reproducibility of the results, the minimum amount of DNA required as a template to obtain reliable profiles, and the sensitivity for detection of minor DNA populations in the mixtures.
In the electropherograms we considered all the peaks corresponding to spacer sizes ranging between 96 and 1,400 nucleotides. For all the environmental communities, the electropherograms from the three primer sets showed differences in peak number, range, and intensity (Table 1). We were able to obtain ARISA profiles from all samples with the ITSF/ITSReub and S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer sets; however, no peak was observed in the electropherograms of the soil communities when the 1406F/23Sr primer set was used (Table 1). The other profiles obtained by the 1406F/23Sr primer set showed only one to four peaks per profile. This low amplification level was confirmed by two different primer batches from two different suppliers. The numbers of peaks per profile obtained with the ITSF/ITSReub primer set, with respect to that obtained with the S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer set, were comparable for the polluted soil and ensiled corn. Compared to S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18, the peak number obtained using primer set ITSF/ITSReub was double that of the natural soil community profile, triple that for the brine and marble community profiles, and seven times greater for the goat milk community profile (Table 1). Total fluorescence and peak height were greater in the profiles of all the environmental communities obtained by using the ITSF/ITSReub primer set (Table 1). The width of the profiles was between 218 (Urania brine profile) and 1,463 (goat milk profile) nucleotides for the ITSF/ITSReub primer set; between 285 (Urania brine) and 1,056 (ensiled corn) nucleotides for the S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer set; and between 444 (goat milk) and 1,088 (ensiled corn) nucleotides for the 1406F/23Sr primer set (Table 1). The smallest peak, corresponding to a hypothetical spacer of 96 nucleotides, was produced by the ITSF/ITSReub primer set in the ARISA of the brine DHAB community; the smallest peaks obtained by the S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 and 1406F/23Sr primer sets corresponded to spacers of 99 and 123 nucleotides, respectively. The largest peak, corresponding to a hypothetical spacer of 1,341 nucleotides, was produced by the ITSF/ITSReub primer set in the ARISA of the goat milk community; the larger peaks obtained by using S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 and 1406F/23Sr primer sets corresponded to spacers of 870 and 767 nucleotides, respectively, from the ensiled maize community. The mean similarities and standard deviations for all pairwise comparisons among three replicate amplifications of environmental samples were 89.4% ± 0.1%, 63.4% ± 5.6%, and 60.2% ± 8.0% for primer sets ITSF/ITSReub, S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18, and 1406F/23Sr, respectively.
TABLE 1.
Range of peak numbers and sizes in the ARISA profiles of six different environmental communities obtained with the three primer setsa
| Environmental sample | Value for primer set
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| ITSF/ITSReub
|
S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18
|
1406F/23Sr
|
|||||||
| No. of peaks ± SDb | Range of peak size (bp) | Range of spacer size (bp) | No. of peaks ± SDb | Range of peak size (bp) | Range of spacer size (bp) | No. of peaks ± SDb | Range of peak size (bp) | Range of spacer size (bp) | |
| Natural soil | 115 ± 6 | 231-1,087 | 109-965 | 55 ± 3 | 300-818 | 114-632 | 0 ± 0 | ||
| Polluted soil | 41 ± 16 | 245-698 | 123-576 | 42 ± 21 | 307-737 | 121-551 | 0 ± 0 | ||
| Urania brine | 36 ± 1 | 218-991 | 96-869 | 12 ± 4 | 285-724 | 99-538 | 3 ± 0 | 593-759 | 272-438 |
| Ensiled corn | 33 ± 1 | 294-1,030 | 172-908 | 29 ± 11 | 318-1,056 | 132-870 | 2 ± 1 | 570-1,088 | 249-767 |
| Goat milk | 34 ± 5 | 229-1,463 | 107-1,341 | 5 ± 2 | 293-603 | 107-417 | 4 ± 1 | 444-746 | 123-425 |
| Certosa of Pavia marble | 8 ± 0 | 228-894 | 106-772 | 3 ± 0 | 300-879 | 114-693 | 1 ± 1 | 579-608 | 258-287 |
The spacer sizes were calculated by subtracting the number of nucleotides belonging to 16S and 23S rRNA genes from the peak sizes.
Data were calculated from three independent ARISA experiments.
The standard deviations of the ARISA peak numbers were normalized, dividing for the respective average (Table 1), and expressed as a percentage. The average of the normalized standard deviations in the different environmental samples for each primer set was 10.8% for the ITSF/ITSReub primer set, 27.8% for S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18, and 43.7% for 1406F/23Sr. Shannon-Weaver and Simpson indices showed that for all the communities analyzed, primer set ITSF/ITSReub yielded the highest number of peaks and detected the highest level of diversity (P < 0.04) (Table 2). Only in the polluted soil, S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 showed higher values for the Shannon-Weaver and the Simpson indices, which, however were not significantly different (P = 0.28 and P = 0.11, respectively) from those of the ITSF/ITSReub primer set.
TABLE 2.
Values for ecological diversity indices of Shannon-Weaver and Simpson obtained by using the data of three independent ARISA experiments performed with each of the three primer sets for each environmental sample
| Primer set | Diversity index value (avg ± SD) for environmental samplea
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Polluted soil
|
Natural soil
|
Brines
|
Insiled maize
|
Monument
|
Goat milk
|
|||||||
| H | 1 − D | H | 1 − D | H | 1 − D | H | 1 − D | H | 1 − D | H | 1 − D | |
| ITSF/ITSReub | 3.48 ± 0.29 | 0.96 ± 0.01 | 4.52 ± 0.27 | 0.99 ± 0.01 | 2.50 ± 0.06 | 0.84 ± 0.02 | 2.86 ± 0.05 | 0.92 ± 0.01 | 1.85 ± 0.04 | 0.84 ± 0.07 | 2.81 ± 0.09 | 0.87 ± 0.01 |
| S-D-Bact-1522-b-S-20/ L-D-Bact-132-a-A-18 | 3.63 ± 0.33 | 0.97 ± 0.01 | 3.24 ± 0.28 | 0.96 ± 0.01 | 1.81 ± 0.12 | 0.81 ± 0.01 | 2.49 ± 0.18 | 0.86 ± 0.02 | 1.15 ± 0.18 | 0.66 ± 0.06 | 1.02 ± 0.29 | 0.51 ± 0.09 |
| 1406F/23Sr | 0 | 0 | 0 | 0 | 1.08 ± 0.01 | 0.66 ± 0.01 | 0.30 ± 0.52 | 0.18 ± 0.30 | 0.32 ± 0.35 | 0.22 ± 0.26 | 1.31 ± 0.24 | 0.72 ± 0.06 |
Data were calculated from three independent ARISA experiments. H, Shannon-Weaver index; 1 − D, Simpson index.
ARISA of DNA of pure strains.
The efficacies in amplifying the ribosomal spacers of the single DNAs of the various bacterial reference strains were not the same for the three primer sets. The ITSF/ITSReub and S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer sets were able to generate peaks of expected size from the eight reference strains belonging to four bacterial divisions. Primer set S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 generated peaks below 1,000 fluorescence units for R. leguminosarum, S. meliloti, and P. stutzeri, while the primer set ITSF/ITSReub produced peaks higher than 1,000 fluorescence units. The 1406F/23Sr primer set failed in spacer amplification for the α-proteobacteria R. leguminosarum and S. meliloti, while it generated peaks with a fluorescence intensity below 1,000 fluorescence units for A. undicola, P. stutzeri, and S. coelicolor (Table 3).
TABLE 3.
Results of ARISA of seven reference strains belonging to different bacterial groups performed with the three tested primer sets
| Strain | Phylogenetic group | Result with primer seta
|
||
|---|---|---|---|---|
| ITSF/ITSReub | S-D-Bact-1522-b-S-20/ L-D-Bact-132-a-A-18 | 1406F/23Sr | ||
| A. undicula USDA 4903 | α-Proteobacteria | ++ | ++ | + |
| R. leguminosarum USDA 2370 | α-Proteobacteria | ++ | + | − |
| S. meliloti USDA 1002 | α-Proteobacteria | ++ | + | − |
| E. coli DSMZ 50902 | γ-Proteobacteria | ++ | ++ | ++ |
| P. stutzeri 1015 | γ-Proteobacteria | ++ | + | + |
| B. cereus DSMZ 31T | Gram positive, low G + C | ++ | ++ | ++ |
| S. bovis JM 300 | Gram positive, low G + C | ++ | ++ | ++ |
| S. coelicolor M145 | Gram positive, high G + C | ++ | ++ | + |
++, peak height between 1,000 and 7,000 fluorescence units; +, peak height lower than 1,000 fluorescence units; −, no peak detected.
ARISA with different amounts of template.
The DNAs of the six strains successfully amplified by all the primer sets, i.e., A. undicola, B. cereus, S. bovis, E. coli, P. stutzeri, and S. coelicolor (Table 3), were mixed to create several model communities that were used to evaluate the sensitivities of the three primer sets for small amounts of template.
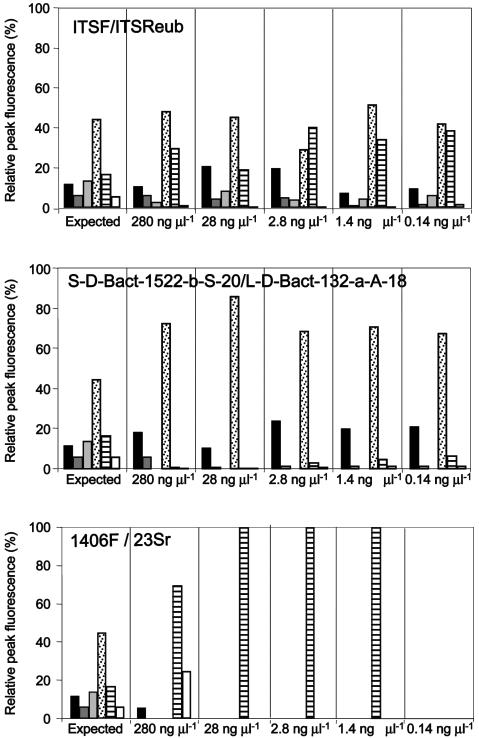
The 1406F/23Sr primer set produced ARISA profiles only from 280 to 1.4 ng of total DNA template. Moreover, these profiles lacked the peaks corresponding to the spacers of S. coelicolor, P. stutzeri, and S. bovis at 280 ng of total DNA template; with a smaller amount of DNA template, only E. coli was detected in the DNA mixture (Fig. 2). The other primer sets were able to produce complex ARISA profiles from all the amounts of template tested up to 0.14 ng of total DNA (Fig. 2). The profiles obtained with the ITSF/ITSReub primer set always showed all expected peaks from all the template amounts. All the profiles obtained from the S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer set lacked the peak corresponding to the spacer of P. stutzeri. The expected values of relative peak fluorescence for the different strains in the DNA mixture were calculated, taking into account the genome size and ribosomal operon copy number of each species included. The relative proportion of peak fluorescence for each species with the primer set ITSF/ITSReub fitted well the expected profile (Fig. 2). Chi-square and Student's t tests were used to compare the relative contribution of each strain in the artificial community to its expected fraction in the ARISA profiles. The results showed that the profiles obtained by using the ITSF/ITSReub primer set were significantly more similar to the expected profile of the artificial community structure than the 1406F/23Sr primer set (P = 0.0042) and S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer set (P = 0.0001). Moreover, the analysis showed that the S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer set was significantly more reliable than the 1406F/23Sr primer set (P = 0.0077).
FIG. 2.
Comparison of the relative areas of the peaks produced by the three primer sets in the ARISA of a model community containing 280, 28, 2.8, 1.4, or 0.14 ng of DNA of six bacterial reference strains. Black bar, B. cereus 31T; dark grey bar, S. coelicolor M145; light grey bar, P. stutzeri 1015; dotted bar, S. bovis JM300; lined bar, E. coli DSM 50902; white bar, A. undicula USDA 4903. The expected peak areas, corresponding to the DNA fractions in the model community, should range between 6.0 and 45.2% of the total fluorescence.
Sensitivity for detecting minor populations with ARISA.
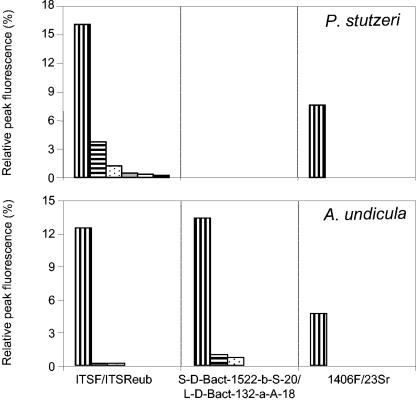
The 1406F/23Sr primer set failed to detect the DNA of A. undicula and P. stutzeri when they represented 9% or less of the mixture (Fig. 3). The S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer set failed to detect the DNA of P. stutzeri even when it represented 50% of the mixture. The ITSF/ITSReub and S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer sets were able to detect the presence of A. undicula to as low a level as 5% of the total DNA community. The ITSF/ITSReub primer set was able to detect the DNA of P. stutzeri even when it represented 0.1% of the mixture.
FIG. 3.
Relative areas of the peaks corresponding to the spacer of P. stutzeri 1015 and A. undicula USDA 4903 produced by the three primer sets when the strain DNA represented fractions of 50% (vertically lined bar), 9.1% (horizontally lined bar), 4.8% (dotted bar), 1.0% (grey bar), 0.5% (white bar), and 0.1% (black bar) of a model community DNA including three other bacterial species (B. cereus, E. coli, and S. coelicolor).
Database examination and primer analysis.
The sequences of the three primer sets were aligned in the NCBI database by using the BLASTN program, and they matched only bacterial organisms. By comparing the primer sequences with those of different phylogenetic groups (Table 4), we found that all six primers had some mismatches with some 16S or 23S rRNA gene sequences in the database. The ITSF, ITSReub, and 1406F primers showed significant alignment with 16S rRNA genes (23S rRNA genes in case of primer ITSReub) of all the explored phylogenetic groups. The S-D-Bact-1522-b-S-20 failed in the alignment with the 16S rRNA genes of the green nonsulfur bacteria, Synergistes, Thermodesulfotobacterium, and Verrucomicrobia. The L-D-Bact-132-a-A-18 and 23Sr primer failed in the alignment with the Verrucomicrobia 23S rRNA gene sequences. No 23S rRNA gene sequences are currently available in the NCBI database for the green nonsulfur bacteria, Synergistes, Thermodesulfobacterium group, and Holophaga phylogenetic group.
TABLE 4.
Mismatches between primers and bacterial gene sequences
| Bacterial division or parameter | No. of mismatches in sequence alignment with primera:
|
|||||
|---|---|---|---|---|---|---|
| ITSF | ITSReub | S-D-Bact-1522-b-S-20 | L-D-Bact-132-a-A-18 | 1406F | 23Sr | |
| Actinomycetales | 0/1,005 | 0/277 | 16/880 | 0/223 | 0/1001 | 0/225 |
| Chlamydiae | 0/108 | 0/76 | 0/93 | 0/75 | 0/153 | 0/70 |
| Cyanobacteria | 0/522 | 0/11 | 98/307 | 0/93 | 3/878 | 0/68 |
| Firmicutes | 0/1,000 | 0/571 | 0/1,082 | 0/247 | 0/1,014 | 1/255 |
| Proteobacteria | 0/1,008 | 0/697 | 1/1,008 | 2/663 | 0/1,006 | 4/687 |
| Spirochaetales | 0/273 | 0/5 | 4/184 | 0/13 | 2/637 | 0/15 |
| Aquificales | 0/12 | 0/1 | 0/1 | 0/1 | 0/43 | 0/1 |
| Thermotogales | 0/32 | 0/1 | 0/10 | 0/1 | 0/60 | 0/1 |
| Green nonsulfur bacteria | 0/1 | NSb | NMc | NS | 29/29 | NS |
| Green sulfur bacteria | 2/18 | 0/3 | 1/3 | 0/3 | 0/18 | 0/3 |
| Thermus | 0/91 | 0/3 | 0/56 | 0/3 | 1/121 | 0/3 |
| Deinococcus | 0/24 | 0/7 | 0/7 | 0/7 | 0/24 | 0/7 |
| CFB group | 2/514 | 0/3 | 2/75 | 0/3 | 0/990 | 0/3 |
| Planctomycetes | 0/87 | 0/2 | 0/19 | 0/1 | 0/124 | 0/1 |
| Synergistes | 0/1 | NS | NM | NS | 0/1 | NS |
| Holophaga | 0/31 | NS | 0/21 | NS | 0/39 | NS |
| Thermodesulfobacterium group | 0/1 | NS | NM | NS | 0/7 | NS |
| Verrucomicrobia | 1/9 | 3/7 | NM | NM | 1/52 | NM |
| Total no. of alignments | 4,737 | 1,664 | 3,746 | 1,333 | 6,197 | 1,339 |
| Total no. (%) of 3′-end mismatches | 5 (0.10) | 3 (0.18) | 122 (3.26) | 2 (0.15) | 7 (0.11) | 5 (0.37) |
| No. of phylogenetic groups showing 3′-end mismatches | 3 | 1 | 6 | 1 | 5 | 2 |
| No. of phylogenetic groups with no matches | 0 | 0 | 4 | 1 | 0 | 1 |
For each bacterial division, results are expressed as number of sequences containing a mismatch with the 3′-end position of the primers/total number of sequences showing a significant alignment, determined by using the BLAST program. BLAST parameters used were 1,000 matches as the maximum expected value, while the word size required was 7.
NS, no 23S rRNA gene sequence available in the NCBI database.
NM, no match with the available sequences.
All of the primers had mismatches at the 3′ ends with some sequences in some of the phylogenetic groups. In particular, primer ITSF had mismatches at the 3′ end with some 16S rRNA gene sequences of green sulfur bacteria, the CFB group, and Verrucomicrobia. Primer ITSReub had mismatches at the 3′ end with some 16S rRNA gene sequences of Verrucomicrobia. Primer S-D-Bact-1522-b-S-20 had mismatches at the 3′ end with some 16S rRNA gene sequences of the Actinomycetales, Cyanobacteria, Proteobacteria, Spirochetales, green sulfur bacteria, and CFB group; among Proteobacteria the nucleotide adjacent to the 3′ end of primer S-D-Bact-1522-b-S-20 had a mismatch with the 16S rRNA gene of P. stutzeri. Primer L-D-Bact-132-a-A-18 had mismatches at the 3′ end with some 23S rRNA gene sequences of the Proteobacteria. Primer 1406F had mismatches at the 3′ end with some 16S rRNA gene sequences of the Cyanobacteria, Spirochaetales, green nonsulfur bacteria, Thermus-Deinococcus group, and Verrucomicrobia. Primer 23Sr had mismatches at the 3′ end with some 23S rRNA gene sequences of the Firmicutes and Proteobacteria (Table 4).
The primers ITSF and ITSReub had very similar melting temperatures (only 0.3°C of difference), while primers S-D-Bact-1522-b-S-20 and L-D-Bact-132-a-A-18 had a difference of 4.8°C; the primers 1406F and 23Sr, because of degenerations, did not have two single melting temperatures but two ranges, with differences between 4.3 and 7.1°C.
The numbers of secondary structures produced by the three primer sets, including hairpins, homodimers, and heterodimers, were 33 for the ITSF/ITSReub, 39 for the 1406F/23Sr, and 42 for S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18.
DISCUSSION
Ribosomal intergenic spacer analysis (RISA) was used to analyze the structure of microbial populations in soil (4, 17, 18) by comparing the profiles after polyacrylamide gel electrophoresis. At most, only a few tens of bands were detected, suggesting a probable underestimation of diversity due to difficulty in identifying weak bands and resolving contiguous bands. The automated version of RISA, the ARISA, is a rapid and precise technique that allows microbial communities to be investigated and compared easily, highlighting the taxonomic diversity, evident from the marked variability in ribosomal spacer length, in the prokaryote genomes (7, 12, 19). ARISA explores microbial diversity at the intraspecific level (5, 6) and can be considered one of the most suitable techniques for rapidly analyzing and comparing great numbers of samples. Considering the very interesting potential of large-scale ARISA applications, we deemed it suitable for carrying out a standardization of the technique in order to permit an easy comparison of results from different studies.
An important step in standardizing PCR-based methods for the analysis of bacterial communities is to choose a primer set that can strongly achieve reliability in the results. We have compared the performance of two primer sets published for performing ARISA with a newly designed primer set. This new ITSF/ITSReub primer set yielded higher peaks than primer sets S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 and 1406F and 23Sr. Moreover, the primer set 1406F/23Sr was not able to amplify the internal transcribed spacers (ITS) of R. leguminosarum USDA 2370 and of S. meliloti 1002. Search for homology of the two primers for the 16S and 23S rRNA gene sequence of R. leguminosarum revealed that the forward primer has mismatches at the 3′ ends with 16S rRNA genes of R. leguminosarum. This defective annealing, coupled to the length of the ITS of these strains, which have ITS longer than 1 kb, did not allow the amplification of the ITS.
The S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 and 1406F/23Sr primer sets did not allow the amplification of all the spacers of a model community containing five different bacterial species. ARISA profiles obtained with primer set 1406F/23Sr lacked several peaks, while those successfully amplified showed a marked reduction in peak intensity. The S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer set generated a lower number of peaks than expected, even though it was able to amplify all the spacers when the template was purified DNA from single strains rather than a mixture of DNA from different species. The S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 set generated profiles that lacked the peaks of P. stutzeri even when it represented 50% of the community (Fig. 3). The ITSF/ITSReub primer set amplified all the strains in the DNA mixture when these DNA were mixed in comparable amounts (Fig. 2) but did not amplify the spacer of A. undicula when it represented less than 5% of the bacterial community DNA, like the S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 primer set (Fig. 3). Instead, the spacer of P. stutzeri was amplified by the ITSF/ITSReub primer set even when it represented only 0.1% of the community. This difference could have been due to different spacer sizes. In fact, the spacer of A. undicula is about 1,250 bp long, while that of P. stutzeri is about 530 bp long. The preferential amplification of the shorter templates is a bias of the PCR due to the kinetics of the reaction, possibly determining a false view of the real bacterial community structure. In this respect, the ITSF/ITSReub primer set would be more suitable than the other two primer sets because it reduces to about 120 the numbers of base pairs of 16S and 23S rRNA genes amplified by the PCR, while the S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 and 1406F/23Sr primer sets, respectively, amplify about 185 and 320 bp of the 16S and 23S rRNA genes (Fig. 1). The amplification of shorter stretches of the 16S and 23S rRNA genes would optimize the resolution power of the Gene Scan software, which tends to merge peaks with sizes over 500 bp, as observed by Suzuki et al. (24). Similar considerations could be made for the ARISA of S. meliloti that is known to have an ITS longer than 1 kb and did not yield any peak with primer set 1406F/23Sr, despite no primer mismatches were found with the 16S and 23S rRNA gene sequences.
The relative peak fluorescences obtained by the ITSF/ITSReub primer set for the different strains in the mixture when different amounts of total DNA were used in the PCR (Fig. 2) showed a good correspondence with the expected relative fluorescences calculated on the basis of the amount of DNA of each species added in the reaction, corrected for the factors genome size and ribosomal operon copy number. This indicated that with the DNA mixture used, the ITSF/ITSReub primer set did not suffer PCR biases like those due to substrate reannealing (23), known to potentially occur during the amplification of a complex DNA mixture. This was not the case for primer sets S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 and 1406F/23Sr, which yielded preferential amplification of S. bovis and E. coli ITS, respectively.
A higher number of peaks was found in the ARISA electropherograms of environmental DNA by using the primer set ITSF/ITSReub than with the other two primer sets (Table 1); this indicates a more informative capacity of this primer set with respect to the others. No peaks corresponding to spacers above 870 and 767 bp were observed in the environmental community profiles obtained by the S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 and 1406F/23Sr primer sets, respectively. From the same environmental samples, using primer set ITSF/ITSReub, we obtained peaks corresponding to spacers up to 1,341 bp (Table 1). Ranjard et al. (19) analyzed a soil bacterial community with the primer set S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 and observed peaks corresponding to spacers no longer than 800 bp, while Fisher and Triplett (7), using primer set 1406F/23Sr for the ARISA of freshwater samples, observed peaks up to 1,147 bp, corresponding to spacers of 826 bp. We speculate that these results were due to the preferential amplification of the shorter templates, as suggested by Fisher and Triplett (7), rather than to the biological absence of species possessing longer spacers (e.g., the α-Proteobacteria).
The 1406F/23Sr primer set generated the less-complex profiles, with no peak in many ARISA profiles, especially from soil samples (Table 1). Fisher and Triplett standardized ARISA with this primer set by using 150 to 300 ng of DNA as the template (7, 8). This amount is relatively large compared to what can be obtained by direct extraction from particular samples, such as monuments, artworks, sandy soils, etc. The ARISA results on the model communities allow us to argue that the 1406F/23Sr primer set does not efficiently amplify small amounts of a template, particularly when it consists of soil DNA. However, even when we set up ARISA using large amounts of template (about 300 ng of DNA) the peak profiles were not comparable with the real DNA composition of the model communities (Fig. 2).
The ITSF/ITSReub primer set allowed the amplification of only 27 and 22 bp of the 16S and 23S rRNA genes (calculated on the basis of the 16S and 23S rRNA gene sequences of E. coli O157:H7, excluding the primer sequences) and hence cannot be used for the taxonomic identification of the species by sequencing ARISA fragments and aligning the 16S or 23S rRNA gene stretches in the electronic database. When the principal research purpose includes the identification of species represented by ARISA fragments, the S-D-Bact-1522-b-S-20/L-D-Bact-132-a-A-18 set, which amplifies 115 bp of the 23S rRNA gene, or the 1406F/23Sr primer set, which amplifies 206 and 115 bp of the 16S and 23S rRNA gene, respectively (Fig. 1), would be more suitable. However, a specific database for the bacterial ITS, the Ribosomal Internal Spacer Sequence Collection (RISSC) database (9) is now available, and the number of sequences deposited (2,146 at the end of May 2003) is constantly increasing. This sequence database tool could be used for the taxonomic identification of ARISA fragment sequences independently of the 16S or 23S rRNA gene stretches.
The different efficiencies among the three tested primer sets in ARISA profiling of complex bacterial communities can be partially explained by the comparison of the primer sequences with the 16S and 23S rRNA gene database. All the primers were found to have some mismatches with some sequences in the database (Table 4). However, primers S-D-Bact-1522-b-S-20, L-D-Bact-132-a-A-18, and 23Sr did not match with four, one, and one bacterial division, respectively. Moreover the lowest percentages of sequences among those examined having mismatches at the 3′ end of the primer sequence, 0.1 and 0.18%, were found for primers ITSF and ITSReub, respectively. The in silico analysis of melting temperatures and putative secondary structures further indicated the ITSF/ITSReub primer set as the most effective in binding the target sequences. These observations could explain the different efficiencies of the three primer sets in amplifying the environmental samples and demonstrate the intrinsic difficulty in obtaining a primer set really universal for the bacterial domain.
This work presents a first tentative identification of a primer set suitable to reduce PCR biases (such as selective amplification of some templates in a mixture of DNA) during ARISA of complex bacterial communities. Our results have shown that the ITSF/ITSReub primer set emphasizes the capacity of ARISA to powerfully explore microbial diversity and to create complex, easy-to-analyze molecular fingerprintings. We observed that the ITSF/ITSReub primer set does not lead to a total avoidance of PCR biases, but it does reduce the effects of such bias in ARISA, allowing us to obtain a more reliable global view of communities than the other primer sets (Fig. 2). Hence, we suggest that the use of the ITSF/ITSReub primer set is appropriate for research where the purpose is to evaluate bacterial community structure by ARISA.
Acknowledgments
Support came from the EU projects BIODEEP (EVK3-2000-22057; Biotechnologies from the Deep) and TRANSBAC (QLK3-CT-2001-02242; Gene Flow from Transgenic Plants: Evaluation and Biotechnology). Partial support came from the Agenzia Regionale per la Protezione dell'Ambiente of Sicily, Italy.
We thank A. Tamburini for kindly giving us the goat milk sample. We thank two anonymous reviewers for helpful comments and suggestions.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1994. Current protocols of molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Blackwood, C. B., T. Marsh, S.-H. Kim, and E. A. Paul. 2003. Terminal restriction fragment length polymorphism data analysis for quantitative comparison of microbial communities. Appl. Environ. Microbiol. 69:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borneman, J., and E. W. Triplett. 1997. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and population shifts associated with deforestation. Appl. Environ. Microbiol. 63:2647-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cherif, A., S. Borin, A. Rizzi, H. Ouzari, A. Boudabous, and D. Daffonchio. 2003. Bacillus anthracis diverges from related clades of the Bacillus cereus group in 16S-23S ribosomal DNA intergenic transcribed spacers containing tRNA genes. Appl. Environ. Microbiol. 69:33-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daffonchio, D., A. Cherif, L. Brusetti, A. Rizzi, D. Mora, A. Boudabous, and S. Borin. 2003. Nature of polymorphisms in 16S-23S rRNA gene intergenic transcribed spacer fingerprinting of Bacillus and related genera. Appl. Environ. Microbiol. 69:5128-5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fisher, M. M., J. L. Klug, G. Lauster, M. Newton, and E. W. Triplett. 2000. Effects of resources and trophic interactions on freshwater bacterioplankton. Microb. Ecol. 40:125-138. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Martinez, J., I. Bescos, J. J. Rodriguez-Sala, and F. Rodriguez-Valera. 2001. RISSC: a novel database for ribosomal 16S-23S RNA genes spacer regions. Nucleic Acids Res. 29:178-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, L. E., J. M. Graham, G. H. Lauster, A. D. Kent, A. C. Yannarell, and D. E. Armstrong. 2001. Community dynamics of phytoplankton, protoplankton and bacterioplankton in selected Wisconsin lakes. J. Phycol. 37:20. [Google Scholar]
- 11.Jackson, C. R., J. P. Harper, D. Willoughby, E. E. Roden, and P. F. Churchill. 1997. A simple, efficient method for the separation of humic substances and DNA from environmental samples. Appl. Environ. Microbiol. 63:4993-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent, A. D., and E. W. Triplett. Microbial communities and their interactions in soil and rhizosphere ecosystems. 2002. Annu. Rev. Microbiol. 56:211-236. [DOI] [PubMed] [Google Scholar]
- 13.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. rrndb: the Ribosomal RNA Operon Copy Number Database. 2001. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lipkin, E., A. Shalom, H. Khatib, M. Soller, and A. Friedmann. 1993. Milk as a source of deoxyribonucleic acid and as a substrate for the polymerase chain reaction. J. Dairy Sci. 76:2025-2032. [DOI] [PubMed] [Google Scholar]
- 15.Liu, W.-T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 1S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murray, A. E., C. M. Preston, R. Massana, L. T. Taylor, A. Blakis, K. Wu, and E. F. Delong. 1998. Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl. Environ. Microbiol. 64:2585-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ranjard, L., S. Nazaret, F. Gourbière, J. Thioulouse, P. Linet, and A. Richaume. 2000. A soil microscale study to reveal the heterogeneity of Hg(II) impact on indigenous bacteria by quantification of adapted phenotypes and analysis of community DNA fingerprints. FEMS Microbiol. Ecol. 31:107-115. [DOI] [PubMed] [Google Scholar]
- 18.Ranjard, L., E. Brothier, and S. Nazaret. 2000. Sequencing bands of ribosomal intergenic spacer analysis fingerprints for characterization and microscale distribution of soil bacterium populations responding to mercury spiking. Appl. Environ. Microbiol. 66:5334-5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ranjard, L., F. Poly, J.-C. Lata, C. Mougel, J. Thioulouse, and S. Nazaret. 2001. Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl. Environ. Microbiol. 67:4479-4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie, N. J., M. E. Schutter, P. D. Richard, and D. D. Myrold. 2000. Use of length heterogeneity PCR and fatty acid methyl ester profiles to characterize microbial communities in soil. Appl. Environ. Microbiol. 66:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scala, D. J., and L. J. Kerkhof. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schabereiter-Gurtner, C., G. Piñar, W. Lubitz, and S. Rölleke. 2001. An advanced molecular strategy to identify bacterial communities on art objects. J. Microbiol. Methods 45:77-87. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki, M., M. S. Rappé, and S. J. Giovannoni. 1998. Kinetic bias in estimates of coastal picoplankton community structure obtained by measurements of small-subunit rRNA gene PCR amplicon length heterogeneity. Appl. Environ. Microbiol. 64:4522-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zanardini, E., V. Andreoni, S. Borin, F. Cappitelli, D. Daffonchio, P. Talotta, C. Sorlini, G. Ranalli, S. Bruni, and F. Cariati. 1997. Lead-resistant microorganisms from red stains of marble of the Certosa of Pavia, Italy and use of nucleic acid-based techniques for their detection. Int. Biodeterior. Biodegrad. 40:171-182. [Google Scholar]
- 26.Zhou, J., M. A. Bruns, and J. M. Tiedje. 1996. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62:316-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zucchi, M., L. Angiolini, S. Borin, L. Brusetti, N. Dietrich, C. Gigliotti, P. Barbieri, C. Sorlini, and D. Daffonchio. 2003. Response of bacterial community during bioremediation of an oil-polluted soil. J. Appl. Microbiol. 94:248-257. [DOI] [PubMed] [Google Scholar]