Abstract
Recent advances in mass spectrometry (MS)-based proteomics have greatly facilitated the robust identification and quantification of posttranslational modifications (PTMs), including those that are present at substoichiometric site occupancies. The abnormal posttranslational modification and accumulation of the microtubule-associated protein tau has been implicated in the pathogenesis of Alzheimer’s disease (AD), and it is thought that the primary mode of regulation of tau occurs through PTMs. Several studies have been published regarding tau phosphorylation; however, other tau PTMs such as ubiquitylation, acetylation, methylation, oxidation, sumoylation, nitration, and glycosylation have not been analyzed as extensively. The comprehensive detection and delineation of these PTMs is critical for drug target discovery and validation. Lysine-directed PTMs including ubiquitylation, acetylation, and methylation play key regulatory roles with respect to the rates of tau turnover and aggregation. MS-based analytical approaches have been used to gain insight into the tau lysine-directed PTM signature that is most closely associated with neurofibrillary lesion formation. This chapter provides details pertaining to the liquid chromatography tandem mass spectrometry (LC-MS/MS)-based analysis of the lysine-directed posttranslational modification of tau.
Keywords: Alzheimer’s disease (AD), Tau, Posttranslational modification, Methylation, Acetylation, Ubiquitylation, Mass spectrometry
1 Introduction
In Alzheimer’s disease (AD), the microtubule-associated protein tau dissociates from the neuronal cytoskeleton and aggregates to form paired helical filaments (PHFs) that comprise neurofibrillary tangles (NFTs) that are one of the prominent neuropathological hallmarks of AD [1, 2]. The molecular and cellular mechanisms that regulate tau aggregation remain unclear; however, it has been postulated that posttranslational modifications (PTMs) influence tau stability, function, and aggregation propensity [3]. Phosphorylation is the most well-established tau PTM, and a distinct hierarchical pattern of tau phosphorylation has been shown to correlate with the progression of AD neuropathology [4, 5]. PHFs of hyperphosphorylated tau aggregates are known to form the degradation-resistant core of NFTs in AD [4, 6, 7]. In addition to phosphorylation, tau is also subjected to several other PTMs that confer pro- or anti-aggregation effects on tau [8]. This chapter will provide details for the analysis of lysine-directed PTMs of tau, with particular emphasis on ubiquitylation, methylation, and acetylation.
Lysine residues can participate in electrostatic and hydrophobic interactions [9, 10], and they are known to play critical roles in tau assembly and toxicity [11]. The lysine-directed ubiquitylation of tau has been demonstrated to modulate tau accumulation and aggregation [12, 13]. In non-pathological conditions, tau has been shown to be ubiquitylated and proteolytically degraded by the ubiquitin-proteasome system [14–16]. The accumulation of degradation-resistant PHF-tau is linked to the impaired function of the ubiquitin proteasome system, and although PHF-tau is ubiquitylated, it is not subsequently degraded by the proteasome system [17–19]. Several ubiquitylation sites within filamentous tau isolated from AD brain have been identified, and they are all localized to the microtubule-binding repeat region [20–22].
Lysine methylation has recently been shown to contribute to the regulation of tau metabolism by directly competing with ubiquitylation and acetylation [22]. Lysine methylation has also been identified on tau purified from normal human brain [23], wild- type mice, and a mouse model of AD [24]. These data indicate that lysine methylation is part of the modification signature that is associated with the preservation of normal tau function. Acetylation is a relatively novel posttranslational modification of tau that has been shown to inhibit its degradation and contribute to tauopathy [25, 26]. Studies have shown that inhibiting histone deacetylase (HDAC)6—a tau deacetylase—has neuroprotective and beneficial consequences including microtubule stabilization [27, 28]. Hence, the pharmacological modulation of the levels of tau acetylation has been proposed as a novel therapeutic strategy. Because the co- occurrence of lysine-directed PTMs on tau has the potential to affect diverse cellular events, it is imperative to differentiate the lysine-directed tau PTM signature that is most closely associated with neuropathology in AD.
Recent advances in mass spectrometry (MS)-based proteomics have enabled the identification and quantification of thousands of PTMs in a single experiment [29–35]. This is due in large part to the development of highly specific and selective PTM-specific antibodies, new high performance liquid chromatography, and mass spectrometry instrumentation and bioinformatic algorithms for peptide and protein identification and PTM site-localization scoring that enable unambiguous PTM site assignments. Because many regulatory PTMs, including ubiquitylation and methylation, have low stoichiometry site occupancies of their target proteins, the use of antibody-based enrichment strategies is essential to achieve the requisite sensitivity in MS analysis for the comprehensive detection of these PTMs in complex matrices such as cell lysate and tissue homogenate [29, 31, 34, 36]. In this context, the stoichiometry refers to the fraction of proteins that are modified. For the majority of these antibody-based enrichment strategies, the enrichment is conducted at the peptide level after the protein mixture has been enzymatically digested. The amount of peptide input is typically on the order of a few milligrams. Accordingly, these strategies are optimized for the large-scale analysis of PTMs. The analysis of PTMs on a single protein of interest requires a different analytical approach, similar to what will be discussed in this chapter.
An inherent challenge in the mass spectrometry-based analysis of PTMs is that modified peptides can be low abundance and more difficult to identify from their fragmentation spectra compared to unmodified peptides. Unlike global proteome analysis, PTM analysis relies on the identification of single peptide species. Although database search engines are effective at identifying PTM-containing peptides with a defined measure of confidence, their localization of specific PTM sites can be arbitrary and unreliable [37]. Thus, in addition to the unambiguous identification of the modified peptides, the localization of modified amino acids within the peptide(s) of interest requires the calculation of a “localization score” [38].
The quantification of PTMs is important because PTM-based regulation in a biological context suggests that an amino acid site could be functional. Conducting quantitative MS-based analysis is essential for the elucidation of the stoichiometry of the specific tau lysine-directed PTMs that correlate with AD neuropathology. Multiple Reaction Monitoring (MRM) is a targeted mass spectrometry (MS)-based technology that is becoming increasingly utilized for protein quantification. MRM-based approaches have been used to determine the relative abundance of tau polyubiquitylation in human AD brain [20] and global tau in human CSF [39, 40]. In contrast to MS-based discovery proteomics experiments, MRM entails the targeted, simultaneous measurements of peptides that serve as surrogates for the protein targets of interest. MRM-based assays are considered to be the “gold standard” for MS-based targeted protein quantification because they are highly specific, precise, and accurate, and they can be multiplexed (hundreds of peptides can be quantified in a single assay), standardized, and readily reproduced. A targeted proteomics method that is similar to MRM is parallel reaction monitoring (PRM) wherein an accurate mass and high resolution mass spectrometer is utilized to permit the parallel detection of all target product ions [41–46].
A common fit-for-purpose quantification strategy entails the use of stable isotope-labeled internal peptide standards for relative quantification based on the establishment of calibration curves of dilutions of the peptide mixtures [47–49]. Because quantitative MS-based assays are negatively affected by a lack of proper procedures for storing and handling peptides, careful attention should be paid to the generation, quantification, storage, and handling of these peptides. Guidelines have been recently established for these practices [50]. To achieve reliable relative quantification of peptides in complex matrices, targeted proteomic assays must be analytically characterized with respect to their specificity, lower limit of quantification (LLOQ), linear range, precision, and repeatability. Various open source software tools are available to facilitate quantitative proteomic assay data analysis and sharing [51–54]. A detailed overview of targeted proteomic assays such as MRM and PRM is provided in several comprehensive reviews [55–60].
2 Materials
2.1 SDS-PAGE and In-Gel Trypsin Digestion
Pre-cast NuPAGE Novex 4–20 % Bis-Tris gels (ThermoFisher Scientific).
NuPAGE MES SDS running buffer (ThermoFisher Scientific).
NuPAGE LDS Sample buffer (ThermoFisher Scientific).
500 mM Dithiothreitol (DTT) in water; aliquot and store at −20 °C.
SeeBlue Pre-stained standard (ThermoFisher Scientific) (see Note 1).
Scalpels (see Note 2).
Gel-loading pipette tips.
Microcentrifuge tubes rinsed with 50 % methanol.
Coomassie blue stain or Silver stain (Silver Quest Silver Staining Kit; ThermoFisher Scientific) (see Note 3).
Optima LC/MS-grade Water (ThermoFisher Scientific).
1 M Ammonium bicarbonate (Sigma) stock in water; check to ensure pH is between 7.5 and 9.0.
Optima LC/MS-grade Acetonitrile (ThermoFisher Scientific).
Optima LC/MS-grade Methanol (ThermoFisher Scientific).
30 % methanol.
30 % acetonitrile in 100 mM Ammonium bicarbonate.
100 mM TCEP (“Bond-breaker neutral pH TCEP solution”; ThermoFisher Scientific): prepare 10 mM solution fresh in 100 mM Ammonium bicarbonate.
55 mM Iodoacetamide (Sigma): prepared in 100 mM Ammonium bicarbonate; Prepare fresh and keep in the dark—iodoacetamide is light sensitive.
Digestion buffer: 50 mM Ammonium bicarbonate.
Trypsin solution: 1.5 μg sequencing-grade modified trypsin (Promega) in 50 mM Ammonium bicarbonate.
Acetonitrile/5 % formic acid.
2.2 C18 StageTip Desalting
Empore C18 extraction disks.
Optima LC/MS-grade Methanol.
Optima LC/MS-grade Acetonitrile.
Optima LC/MS-grade Water.
Wash solvent 1: 0.1 % Formic acid in Water.
Elution solvent: 50 % Acetonitrile/0.1 % Formic acid.
2.3 In-Solution Trypsin Digestion
1 M Ammonium bicarbonate stock in water; ensure that the pH is between 7.5 and 9.0.
0.1 % RapiGest SF™ (Waters) in 50 mM Ammonium Bicarbonate.
TCEP (“Bond-breaker neutral pH TCEP solution”).
1 M Iodoacetamide stock prepared in 50 mM Ammonium bicarbonate; prepare fresh.
1 M DTT stock prepared in 50 mM Ammonium bicarbonate; 1 M DTT stock can be stored in aliquots at −20 °C.
Sequencing-grade modified trypsin.
Mass spectrometry-grade Trifluoroacetic acid (Sigma).
2.4 Liquid Chromatography
Mobile Phase “A” (aqueous solvent): 3 % Acetonitrile/0.1 % Formic acid in Water; Water and Acetonitrile: Optima, LC/ MS-grade; Formic acid: mass spectrometry-grade (Sigma).
Mobile Phase “B” (organic solvent): 90 % Acetonitrile/0.1 % Formic acid.
C18 trap column: 5 mm length × 300 μm inner diameter, 5 μm Acclaim PepMap 100 C18 (ThermoFisher Scientific).
C18 analytical column: 50 cm length × 75 μm inner diameter, 2 μm Acclaim PepMap RSLC C18 EASY-Spray column (ThermoFisher Scientific).
Dionex UltiMate 3000 RSLCnano LC system (ThermoFisher Scientific).
2.5 Mass Spectrometry and Data Analysis
Q Exactive hybrid quadrupole-Orbitrap mass spectrometer equipped with an EASY-Spray nanospray ionization source (ThermoFisher Scientific) for peptide and protein identification and parallel reaction monitoring (PRM)-based targeted proteomics analysis (see Note 4). A triple quadrupole or a QqTOF mass spectrometer is required for targeted analyses such as selected/multiple reaction monitoring (SRM/MRM).
MSConvert (freely available via ProteoWizard at http://proteowizard.sourceforge.net/tools.shtml).
2.6 Targeted MS Data Acquisition: Selected/Multiple Reaction Monitoring (SRM/MRM) or Parallel Reaction Monitoring (PRM)
3 Methods
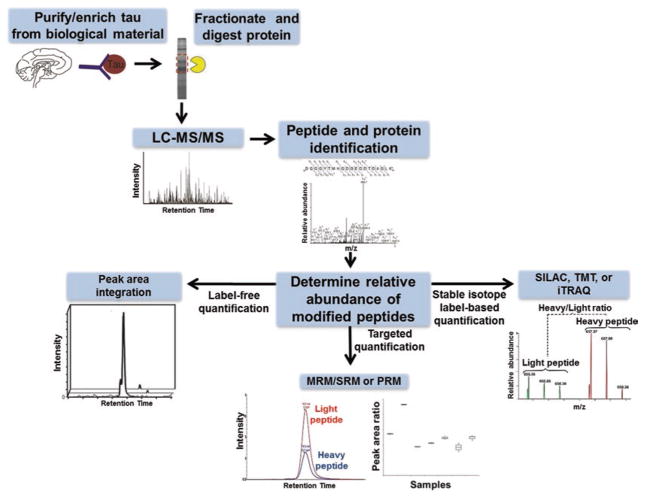
The methods detailed in this chapter are written based on the analysis of PHF-tau purified from AD brain tissue. As there are several biological material-based sources of tau (e.g., tissue, cerebrospinal fluid, serum), a specific tau purification protocol is not discussed here. The recommended minimum starting amount of purified tau for the methods described in this chapter is 20 μg. The protein concentration of the starting material used for the methods described below should be accurately determined using a BCA assay or similar protein quantification assay. A workflow entailing the methods that are described in this chapter is presented in Fig. 1.
Fig. 1.
Workflow for the mass spectrometry-based analysis of lysine-directed PTMs of tau. MRM/SRM multiple reaction monitoring/selected reaction monitoring, PRM parallel reaction monitoring, TMT tandem mass tags, iTRAQ isobaric tags for relative and absolute quantification, SILAC stable isotope labeling by amino acids in cell culture
3.1 In-Gel Trypsin Digestion
Perform SDS-PAGE to electrophoretically separate the tau- containing sample. Following electrophoresis, wash the gel three times for 10 min per wash with deionized water to remove any residual SDS.
Conduct Coomassie blue staining or Silver staining. After staining is complete, wash the gel for 10 min with deionized water.
Capture an image of the gel. Use a clean scalpel to excise a gel region corresponding to 50–75 kDa, divide the region into ~2–3 sections and cut into 1 × 1 mm cubes. Transfer gel pieces to a microcentrifuge tube washed with 50 % methanol (see Note 7). If using Silver stain, de-stain the gel pieces before proceeding to the next step.
Add 100 % methanol for 5 min to dehydrate the gel pieces. Add a sufficient volume of methanol to cover the gel pieces.
Remove 100 % methanol and add 30 % methanol for 5 min to rehydrate the gel pieces (see Note 8).
Remove 30 % methanol and wash the gel pieces twice for 10 min per wash with water.
Wash the gel pieces three times for 10 min per wash with 30 % acetonitrile in 100 mM Ammonium bicarbonate.
Dry gel pieces in a vacuum centrifuge (speed vacuum) for 15 min. Gel pieces will become opaque when dry.
Add 10 mM TCEP (enough volume to cover the gel pieces) and let incubate for 1 h at 60 °C to reduce protein disulfide bonds.
Briefly centrifuge the gel pieces and discard the liquid.
Add 55 mM iodoacetamide (enough volume to cover the gel pieces) and let incubate for 45 min at room temperature in the dark to alkylate cysteine residues.
Briefly centrifuge the gel pieces and discard the liquid.
Wash the gel pieces for 15 min with 100 mM Ammonium bicarbonate.
Discard the liquid. Shrink the gel pieces with 100 % acetonitrile.
Discard the liquid. Dry the gel pieces in a speed vacuum for 15 min. Ensure that the gel pieces are completely dry to facilitate the absorption of trypsin into the gel pieces in the next step.
Rehydrate the gel pieces in trypsin solution at 4 °C (on ice) for 45 min. Add an adequate volume of trypsin solution to completely cover the gel pieces (see Note 9).
Remove any remaining trypsin solution and add a sufficient volume of digestion buffer to completely cover the gel pieces.
Let incubate at 37 °C overnight with gentle shaking.
Briefly centrifuge the gel pieces and transfer the supernatant (containing tryptic peptides) to another microcentrifuge tube washed with 50 % methanol.
Add 50 mM Ammonium bicarbonate to cover the gel pieces and incubate at 37 °C with shaking for 15 min.
Add a volume of acetonitrile equal to the volume of 50 mM Ammonium bicarbonate utilized in step 20 and incubate at 37 °C with shaking for 15 min.
Centrifuge the gel pieces and add the supernatant to the supernatant collected in step 19.
Extract the peptides: add acetonitrile/5 % formic acid (50:50) to the gel pieces and incubate at 37 °C with shaking for 15 min.
Remove the supernatant and combine with the supernatant collected in steps 19 and 22.
Repeat step 23.
Remove the supernatant and combine with the supernatant collected in steps 19, 22, and 24.
Dry the combined supernatant in a speed vac.
Desalt the sample using C18 StageTips.
3.2 In-Solution Trypsin Digestion
Use at least 20 μg protein at a concentration of 0.2–1.0 μg/μL for in-solution digestion. If protein is in a pellet form, add 0.1 % RapiGest in 50 mM Ammonium Bicarbonate. If protein is in solution, the ideal buffer is 50 mM Ammonium bicarbonate. Conduct buffer exchange if necessary and add 1:1 (v/v) 0.2 % RapiGest:protein solution.
Denature protein by incubating at 90 °C for 3 min. Cool to room temperature.
Add TCEP to 5 mM final concentration to reduce protein disulfide bonds. Vortex and incubate for 30 min at 60 °C. Let cool to room temperature.
Alkylate cysteine residues by adding iodoacetamide to 10 mM final concentration. Vortex and incubate at room temperature in the dark for 30 min.
Add trypsin to sample at a trypsin:protein ratio of 1:50 (w/w). Let incubate overnight at 37 °C with gentle shaking (see Note 9).
Stop enzymatic digestion and hydrolyze RapiGest by adding trifluoroacetic acid to 1 % final concentration. Incubate for 45 min at 37 °C.
Centrifuge at 14,000 × g for 10 min at 4 °C and transfer the supernatant to a new microcentrifuge tube (see Note 10).
Desalt the sample using a C18-based spin column, cartridge, or tip (see Note 11).
3.3 C18 StageTip Desalting
Prepare C18 StageTips using two plugs of C18 material in each StageTip (see Notes 12 and 13).
Reconstitute the dried protein digest sample in 50 μL of Wash Solvent 1.
Wash the StageTip once with 50 μL of 100 % Methanol.
Equilibrate the StageTip twice with 50 μL of Wash Solvent 1.
Load the sample onto the equilibrated StageTip.
Wash and desalt the sample twice with 50 μL of Wash Solvent 1.
Elute the sample into a clean microcentrifuge tube using 50 μL of Elution Solvent.
Dry the desalted sample in a speed vac. Reconstitute in liquid chromatography Mobile Phase A. For in-gel digests, use ~1/3 of the sample for LC-MS/MS analysis. For in-solution digests, load ~1 μg of peptides.
3.4 Liquid Chromatography
Utilize an LC method with the following parameters:
Gradient pump flow rate = 0.3 μL/min; loading pump flow rate = 5.0 μL/min.
Column oven temperature = 35 °C.
Autosampler temperature = 6.0 °C.
Sample loading time = 2.5× sample loop volume.
Separation gradient: 60 min linear gradient from 3 to 35 % Mobile Phase B, followed by 35–90 % B in 5 min, 90 % B for 5 min, and column re-equilibration with 3 %B for 10 min.
3.5 Mass Spectrometry
Set spray voltage to 1.8 kV and heated capillary temperature to 180 °C.
-
Create a data-dependent MS/MS instrument method with the following parameters:
Full scan range: 400–1800 m/z; positive polarity
Ten most abundant ions selected for MS/MS fragmentation
Resolution = 70,000
Maximum injection time = 100 ms
Dynamic exclusion enabled—exclude ions for 20 s after being detected once within 20 s (see Note 14).
Automatic gain control target = 5e4
Activation time = 30 ms
Isolation width = 2.0 m/z
Normalized collision energy = 28 %
Charge state exclusion: z = unassigned, 1 and ≥6
3.6 Mass Spectrometry Data Analysis of Lysine PTMs
Use MSConvert to convert the acquired .raw files to .mzML format.
-
Conduct database search and peptide-spectrum match (PSM) validation using tools available in the Trans-Proteomic Pipeline [61]. Use the following database search parameters:
Database: UniProtKB human protein database with reviewed sequences. The data should also be searched against a targeted library of tau isoform sequences with the following UniProt accession numbers: P10637, P10637-2, P10637-3, P10637-4, P-10637-5, and P10637-6.
Fixed modification: Cys +57.02146 (carbamidomethy lation).
Variable Lys-directed modifications: Lys +14.01565 (monomethylation), Lys +28.03130 (dimethylation), Lys +42.04695 (trimethylation), Lys +42.01056 (acetylation), and Lys +114.042927 (di-Gly; tryptic ubiquitylation remnant). Other non-Lys-directed variable modifications to which tau is known to be subjected include the following: Met +15.99491 (oxidation); Ser, Thr, Tyr +79.96633 (phosphorylation); and Protein N-terminus +42.01056 (acetylation).
Precursor ion mass tolerance = 20 ppm.
Fragment ion (MS/MS) mass tolerance = 0.05 Da.
Maximum missed cleavages = 2.
Maximum modifications per peptide = 6.
Enzyme cleavage specificity = trypsin (or enzyme that was used for the in solution or in gel digestion).
Use PeptideProphet (available as part of the Trans-Proteomic Pipeline) to validate the peptide search results and set the false discovery rate (FDR) at 1 %.
Infer proteins from the identified peptides using ProteinProphet (available as part of the Trans-Proteomic Pipeline).
Manually inspect spectra of peptides passing the 1 % FDR filter to verify that the majority of the major fragment ions are assigned. Peptides with C-terminal ubiquitylation sites should be removed and considered false positive identifications because ubiquitylation commonly results in a missed trypsin cleavage site at the modified Lys residue on the ubiquitylation substrate protein [65].
Determine the PTM site localization probability (PTM score) using MaxQuant [62, 63]. When evaluating the quality of the data, take into consideration the delta score, which measures the difference between the best spectrum-match and the next best match with a different amino acid sequence. Also consider the score difference between the best and second-best positioning of a PTM within the same peptide sequence.
If the sample was isotopically labeled (e.g., chemical labeling with iTRAQ or TMT tags or metabolically labeled using SILAC), use a software package such as MaxQuant [62, 63] or QuantiMORE [66] to calculate the peptide and protein ratios (see Notes 15 and 16).
Researchers are encouraged to deposit their raw mass spectrometry data to repositories such as the ProteomeXchange Consortium via the PRIDE partner repository [67].
3.7 Label-Free Quantification of Mass Spectrometry Data Using Intensity-Based Peak Area Integration
Peptide features are quantified by measuring their corresponding spectral intensities or areas under the peaks in MS or MS/MS spectra (see Note 17).
Search the mass spectrometry data using MaxQuant with the default settings for label-free quantification wherein peptide intensity values are aggregated by charge state and modification status [68]. The peptide feature intensities can then be log2-transformed followed by conducting pairwise comparisons between the samples using an appropriate statistical test based on how the data are distributed. Intensity-based quantification can also be performed using other software such as SIEVE.
3.8 Targeted Mass Spectrometry-Based Quantitative Measurement of Peptides Using SRM or PRM
The main steps of targeted mass spectrometry method development are as follows: (1) generate a hypothesis that can be tested by quantitative protein measurements; (2) develop the SRM or PRM method using synthetic peptide standards; (3) refine the method; and (4) validate the method. These steps are outlined in further detail in [58].
Import tau protein sequence into Skyline to conduct in silico digestion, select peptides of interest, select all y-ion transitions (m/z > precursor m/z), select optimal collision energies predicted via mass spectrometer-specific linear equations, and export the instrument method that will be used for MS analysis. If the peptide of interest contains a posttranslational modification, ensure that at least one transition contains the modified residue.
Conduct transition refinement by manual inspection of data analyzed using Skyline.
Establish a scheduled instrument method using retention time prediction in Skyline.
Use the refined scheduled method to develop assay including response curve generation and determination of repeatability. An example of guidelines for assay development can be found at https://assays.cancer.gov/guidance-document/ [69].
Acknowledgments
This work was supported by National Institutes of Health grants MH59786 and AG25323 (A.Y.) and the Maryland Cigarette Restitution Fund.
Footnotes
Other pre-stained protein standards are suitable; however, the standard should include proteins with molecular weights close to those of tau isoforms (~45–65 kDa).
The use of scalpels as opposed to razor blades is preferred so as to minimize the chances of gloves making contact with the gel when excising gel bands and cutting the bands into smaller pieces. Avoid using latex gloves as these are a common source of keratin contamination.
There are alternative commercially available mass spectrometry compatible silver staining kits. Mass spectrometry compatible silver staining methods are those that do not involve the use of glutaraldehyde to fix the proteins in the gel.
Other types of mass spectrometers can be used to analyze the samples; however, for the best results, a mass spectrometer with high resolution and mass accuracy should be used. This is particularly important for differentiating among PTMs with similar masses such as trimethylation (42.046950 Da) and acetylation (42.010565 Da); Δ mass = 0.036385 Da.
Several other open source and freely available database search engines, algorithms, and software tools such as Mascot [70], MaxQuant [62, 63], Comet [71], MSGF+ [72], and ID Picker (MyriMatch) [73] can be used; however, be certain to adhere to the parameters that are outlined in Subheading 3.6.
Detailed guidelines for peptide procurement, characterization, storage, and handling, as well as approaches to the interpretation of the data generated by targeted MS-based assays have been developed [50].
All steps for in-gel digestion should be performed in a clean laminar flow hood to minimize keratin contamination.
For all steps of the in-gel digestion protocol that entail removing solutions from gel pieces, use a gel-loading pipette tip.
Enzymes other than trypsin such as LysC, ArgC, AspN, and GluC can be used to digest tau. The use of multiple proteases has been shown to improve the protein sequence coverage obtained from mass spectrometry-based analysis [74].
A cloudy pellet should appear after centrifugation. When pipetting the supernatant (containing peptides) after completion of the centrifugation, take caution to not disturb the pellet.
Consider the amount of peptides (measure using UV-based absorbance at 280 nm) and the binding capacity of the column, tip, or cartridge when selecting the format of the C18-based material used for desalting.
The original publication of the StageTip procedure provides detailed instructions and illustrations for how to prepare StageTips [75].
Use 2 mL microcentrifuge tubes and StageTip adapters so that the procedure can be completed using a microcentrifuge format. Conduct all centrifugation steps for the StageTip desalting at 3000–4000 × g for 3–5 min at room temperature or until all of the top-loaded solution has passed through the extraction disks in the StageTip.
The optimum duration for dynamic exclusion should be empirically determined based on the average chromatographic peak width. Using a dynamic exclusion duration of 20 s with an LC system that generates peak widths of ~40 s permits the acquisition of MS/MS data for a peptide as it begins to elute as a peak, at the apex of the peak and at the peak tail. The dynamic exclusion settings also affect the performance of spectral counting methods wherein low abundance proteins are under-sampled and high abundance proteins can be sampled to a degree such that near-maximum sequence coverage is achieved [76]. Optimization of the dynamic exclusion settings can increase the reproducibility of spectral counting and the quantification of low or high abundant proteins.
QuantiMORE (formerly named IsoQuant) is freely available for download at http://www.proteomeumb.org/MZw.html.
MaxQuant consists of several independent modules that enable the complete processing of raw mass spectrometry data files. It supports protein identification, quantification, recalibration, and the quality control of the raw and annotated mass spectra. A thorough step-by-step protocol explaining how MaxQuant can be used to analyze proteomics datasets was published in a recent edition of Methods in Molecular Biology [77].
Among the label-free methods for the quantification of mass spectrometry data, intensity-based quantification methods have been demonstrated to be more sensitive than spectral counting [78]. Although it is possible to conduct intensity-based quantification by summing the MS/MS-level feature intensities, quantification on the MS-level has been demonstrated to be more accurate [79].
References
- 1.Braak H, Braak E, Strothjohann M. Abnormally phosphorylated tau protein related to the formation of neurofibrillary tangles and neuropil threads in the cerebral cortex of sheep and goat. Neurosci Lett. 1994;171:1–4. doi: 10.1016/0304-3940(94)90589-4. [DOI] [PubMed] [Google Scholar]
- 2.Goedert M. Tau protein and the neurofibrillary pathology of Alzheimer’s disease. Trends Neurosci. 1993;16:460–465. doi: 10.1016/0166-2236(93)90078-z. [DOI] [PubMed] [Google Scholar]
- 3.Martin L, Latypova X, Terro F. Post-translational modifications of tau protein: implications for Alzheimer’s disease. Neurochem Int. 2011;58:458–471. doi: 10.1016/j.neuint.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 4.Augustinack JC, Schneider A, Mandelkow EM, et al. Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer’s disease. Acta Neuropathol. 2002;103:26–35. doi: 10.1007/s004010100423. [DOI] [PubMed] [Google Scholar]
- 5.Hampel H, Burger K, Pruessner JC, et al. Correlation of cerebrospinal fluid levels of tau protein phosphorylated at threonine 231 with rates of hippocampal atrophy in Alzheimer disease. Arch Neurol. 2005;62:770–773. doi: 10.1001/archneur.62.5.770. [DOI] [PubMed] [Google Scholar]
- 6.Buee L, Bussiere T, Buee-Scherrer V, et al. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev. 2000;33:95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 7.Holzer M, Holzapfel HP, Zedlick D, et al. Abnormally phosphorylated tau protein in Alzheimer’s disease: heterogeneity of individual regional distribution and relationship to clinical severity. Neuroscience. 1994;63:499–516. doi: 10.1016/0306-4522(94)90546-0. [DOI] [PubMed] [Google Scholar]
- 8.Gong CX, Liu F, Grundke-Iqbal I, et al. Post-translational modifications of tau protein in Alzheimer’s disease. J Neural Transm (Vienna) 2005;112:813–838. doi: 10.1007/s00702-004-0221-0. [DOI] [PubMed] [Google Scholar]
- 9.Marshall KE, Morris KL, Charlton D, et al. Hydrophobic, aromatic, and electrostatic interactions play a central role in amyloid fibril formation and stability. Biochemistry. 2011;50:2061–2071. doi: 10.1021/bi101936c. [DOI] [PubMed] [Google Scholar]
- 10.Sinha S, Lopes DH, Du Z, et al. Lysine-specific molecular tweezers are broad-spectrum inhibitors of assembly and toxicity of amyloid proteins. J Am Chem Soc. 2011;133:16958–16969. doi: 10.1021/ja206279b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Norlund MA, Lee JM, Zainelli GM, et al. Elevated transglutaminase-induced bonds in PHF tau in Alzheimer’s disease. Brain Res. 1999;851:154–163. doi: 10.1016/s0006-8993(99)02179-4. [DOI] [PubMed] [Google Scholar]
- 12.Petrucelli L, Dickson D, Kehoe K, et al. CHIP and Hsp70 regulate tau ubiquitination, degradation and aggregation. Hum Mol Genet. 2004;13:703–714. doi: 10.1093/hmg/ddh083. [DOI] [PubMed] [Google Scholar]
- 13.Shimura H, Schwartz D, Gygi SP, et al. CHIP-Hsc70 complex ubiquitinates phosphorylated tau and enhances cell survival. J Biol Chem. 2004;279:4869–4876. doi: 10.1074/jbc.M305838200. [DOI] [PubMed] [Google Scholar]
- 14.Arnaud LT, Myeku N, Figueiredo-Pereira ME. Proteasome-caspase-cathepsin sequence leading to tau pathology induced by prostaglandin J2 in neuronal cells. J Neurochem. 2009;110:328–342. doi: 10.1111/j.1471-4159.2009.06142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.David DC, Layfield R, Serpell L, et al. Proteasomal degradation of tau protein. J Neurochem. 2002;83:176–185. doi: 10.1046/j.1471-4159.2002.01137.x. [DOI] [PubMed] [Google Scholar]
- 16.Liu YH, Wei W, Yin J, et al. Proteasome inhibition increases tau accumulation independent of phosphorylation. Neurobiol Aging. 2009;30:1949–1961. doi: 10.1016/j.neurobiolaging.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 17.de Vrij FM, Fischer DF, van Leeuwen FW, et al. Protein quality control in Alzheimer’s disease by the ubiquitin proteasome system. Prog Neurobiol. 2004;74:249–270. doi: 10.1016/j.pneurobio.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Zhang JY, Liu SJ, Li HL, et al. Microtubule-associated protein tau is a substrate of ATP/Mg(2+)-dependent proteasome protease system. J Neural Transm (Vienna) 2005;112:547–555. doi: 10.1007/s00702-004-0196-x. [DOI] [PubMed] [Google Scholar]
- 19.Riederer IM, Schiffrin M, Kovari E, et al. Ubiquitination and cysteine nitrosylation during aging and Alzheimer’s disease. Brain Res Bull. 2009;80:233–241. doi: 10.1016/j.brainresbull.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 20.Cripps D, Thomas SN, Jeng Y, et al. Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament- Tau is polyubiquitinated through Lys-48, Lys- 11, and Lys-6 ubiquitin conjugation. J Biol Chem. 2006;281:10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
- 21.Morishima-Kawashima M, Hasegawa M, Takio K, et al. Ubiquitin is conjugated with aminoterminally processed tau in paired helical filaments. Neuron. 1993;10:1151–1160. doi: 10.1016/0896-6273(93)90063-w. [DOI] [PubMed] [Google Scholar]
- 22.Thomas SN, Funk KE, Wan Y, et al. Dual modification of Alzheimer’s disease PHF-tau protein by lysine methylation and ubiquitylation: a mass spectrometry approach. Acta Neuropathol. 2012;123:105–117. doi: 10.1007/s00401-011-0893-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Funk KE, Thomas SN, Schafer KN, et al. Lysine methylation is an endogenous post- translational modification of tau protein in human brain and a modulator of aggregation propensity. Biochem J. 2014;462:77–88. doi: 10.1042/BJ20140372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morris M, Knudsen GM, Maeda S, et al. Tau post-translational modifications in wild- type and human amyloid precursor protein transgenic mice. Nat Neurosci. 2015;18:1183–1189. doi: 10.1038/nn.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen TJ, Friedmann D, Hwang AW, et al. The microtubule-associated tau protein has intrinsic acetyltransferase activity. Nat Struct Mol Biol. 2013;20:756–762. doi: 10.1038/nsmb.2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min SW, Cho SH, Zhou Y, et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook C, Stankowski JN, Carlomagno Y, et al. Acetylation: a new key to unlock tau’s role in neurodegeneration. Alzheimers Res Ther. 2014;6:29. doi: 10.1186/alzrt259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irwin DJ, Cohen TJ, Grossman M, et al. Acetylated tau, a novel pathological signature in Alzheimer’s disease and other tauopathies. Brain. 2012;135:807–818. doi: 10.1093/brain/aws013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 30.Imamura H, Sugiyama N, Wakabayashi M, et al. Large-scale identification of phosphorylation sites for profiling protein kinase selectivity. J Proteome Res. 2014;13:3410–3419. doi: 10.1021/pr500319y. [DOI] [PubMed] [Google Scholar]
- 31.Kim W, Bennett EJ, Huttlin EL, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olsen JV, Vermeulen M, Santamaria A, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 33.Trinidad JC, Barkan DT, Gulledge BF, et al. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol Cell Proteomics. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Udeshi ND, Svinkina T, Mertins P, et al. Refined preparation and use of anti-diglycine remnant (K-epsilon-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol Cell Proteomics. 2013;12:825–831. doi: 10.1074/mcp.O112.027094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics. 2013;12:3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas SN, Zhang H, Cotter RJ. Application of quantitative proteomics to the integrated analysis of the ubiquitylated and global proteomes of xenograft tumor tissues. Clin Proteomics. 2015;12:14. doi: 10.1186/s12014-015-9086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chalkley RJ, Clauser KR. Modification site localization scoring: strategies and performance. Mol Cell Proteomics. 2012;11:3–14. doi: 10.1074/mcp.R111.015305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rigbolt KT, Prokhorova TA, Akimov V, et al. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal. 2011;4:rs3. doi: 10.1126/scisignal.2001570. [DOI] [PubMed] [Google Scholar]
- 39.Bros P, Vialaret J, Barthelemy N, et al. Antibody-free quantification of seven tau peptides in human CSF using targeted mass spectrometry. Front Neurosci. 2015;9:302. doi: 10.3389/fnins.2015.00302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McAvoy T, Lassman ME, Spellman DS, et al. Quantification of tau in cerebrospinal fluid by immunoaffinity enrichment and tandem mass spectrometry. Clin Chem. 2014;60:683–689. doi: 10.1373/clinchem.2013.216515. [DOI] [PubMed] [Google Scholar]
- 41.Peterson AC, Russell JD, Bailey DJ, et al. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics. 2012;11:1475–1488. doi: 10.1074/mcp.O112.020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsuchiya H, Tanaka K, Saeki Y. The parallel reaction monitoring method contributes to a highly sensitive polyubiquitin chain quantification. Biochem Biophys Res Commun. 2013;436:223–229. doi: 10.1016/j.bbrc.2013.05.080. [DOI] [PubMed] [Google Scholar]
- 43.Tang H, Fang H, Yin E, et al. Multiplexed parallel reaction monitoring targeting histone modifications on the QExactive mass spectrometer. Anal Chem. 2014;86:5526–5534. doi: 10.1021/ac500972x. [DOI] [PubMed] [Google Scholar]
- 44.Yu Q, Liu B, Ruan D, et al. A novel targeted proteomics method for identification and relative quantitation of difference in nitration degree of OGDH between healthy and diabetic mouse. Proteomics. 2014;14:2417–2426. doi: 10.1002/pmic.201400274. [DOI] [PubMed] [Google Scholar]
- 45.Thomas SN, Harlan R, Chen J, et al. Multiplexed targeted mass spectrometry-based assays for the quantification of N-linked glycosite- containing peptides in serum. Anal Chem. 2015;87:10830–10838. doi: 10.1021/acs.analchem.5b02063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim YJ, Gallien S, El-Khoury V, et al. Quantification of SAA1 and SAA2 in lung cancer plasma using the isotype-specific PRM assays. Proteomics. 2015;15:3116–3125. doi: 10.1002/pmic.201400382. [DOI] [PubMed] [Google Scholar]
- 47.Abbatiello SE, Schilling B, Mani DR, et al. Large-scale interlaboratory study to develop, analytically validate and apply highly multiplexed, quantitative peptide assays to measure cancer-relevant proteins in plasma. Mol Cell Proteomics. 2015;14:2357–2374. doi: 10.1074/mcp.M114.047050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carr SA, Abbatiello SE, Ackermann BL, et al. Targeted peptide measurements in biology and medicine: best practices for mass spectrometry-based assay development using a fit-for-purpose approach. Mol Cell Proteomics. 2014;13:907–917. doi: 10.1074/mcp.M113.036095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abbatiello SE, Mani DR, Schilling B, et al. Design, implementation and multisite evaluation of a system suitability protocol for the quantitative assessment of instrument performance in liquid chromatography-multiple reaction monitoring-MS (LC-MRM-MS) Mol Cell Proteomics. 2013;12:2623–2639. doi: 10.1074/mcp.M112.027078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoofnagle AN, Whiteaker JR, Carr SA, et al. Recommendations for the generation, quantification, storage, and handling of peptides used for mass spectrometry-based assays. Clin Chem. 2016;62:48–69. doi: 10.1373/clinchem.2015.250563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma V, Eckels J, Taylor GK, et al. Panorama: a targeted proteomics knowledge base. J Proteome Res. 2014;13:4205–4210. doi: 10.1021/pr5006636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reiter L, Rinner O, Picotti P, et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 53.Abbatiello SE, Mani DR, Keshishian H, et al. Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry. Clin Chem. 2010;56:291–305. doi: 10.1373/clinchem.2009.138420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aiyetan P, Thomas SN, Zhang Z, et al. MRMPlus: an open source quality control and assessment tool for SRM/MRM assay development. BMC Bioinformatics. 2015;16:411. doi: 10.1186/s12859-015-0838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ebhardt HA, Root A, Sander C, et al. Applications of targeted proteomics in systems biology and translational medicine. Proteomics. 2015;15:3193–3208. doi: 10.1002/pmic.201500004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Domon B. Considerations on selected reaction monitoring experiments: implications for the selectivity and accuracy of measurements. Proteomics Clin Appl. 2012;6:609–614. doi: 10.1002/prca.201200111. [DOI] [PubMed] [Google Scholar]
- 57.Picotti P, Aebersold R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nat Methods. 2012;9:555–566. doi: 10.1038/nmeth.2015. [DOI] [PubMed] [Google Scholar]
- 58.Bereman MS, MacLean B, Tomazela DM, et al. The development of selected reaction monitoring methods for targeted proteomics via empirical refinement. Proteomics. 2012;12:1134–1141. doi: 10.1002/pmic.201200042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gallien S, Duriez E, Domon B. Selected reaction monitoring applied to proteomics. J Mass Spectrom. 2011;46:298–312. doi: 10.1002/jms.1895. [DOI] [PubMed] [Google Scholar]
- 60.Khristenko N, Domon B. Quantification of proteins in urine samples using targeted mass spectrometry methods. Methods Mol Biol. 2015;1243:207–220. doi: 10.1007/978-1-4939-1872-0_12. [DOI] [PubMed] [Google Scholar]
- 61.Deutsch EW, Mendoza L, Shteynberg D, et al. Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics Clin Appl. 2015;9:745–754. doi: 10.1002/prca.201400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 63.Cox J, Neuhauser N, Michalski A, et al. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 64.MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peng J, Schwartz D, Elias JE, et al. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 66.Liao Z, Wan Y, Thomas SN, et al. IsoQuant: a software tool for stable isotope labeling by amino acids in cell culture-based mass spectrometry quantitation. Anal Chem. 2012;84:4535–4543. doi: 10.1021/ac300510t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vizcaino JA, Deutsch EW, Wang R, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Goeminne LJ, Argentini A, Martens L, et al. Summarization vs peptide-based models in label-free quantitative proteomics: performance, pitfalls, and data analysis guidelines. J Proteome Res. 2015;14:2457–2465. doi: 10.1021/pr501223t. [DOI] [PubMed] [Google Scholar]
- 69.Whiteaker JR, Halusa GN, Hoofnagle AN, et al. CPTAC assay portal: a repository of targeted proteomic assays. Nat Methods. 2014;11:703–704. doi: 10.1038/nmeth.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perkins DN, Pappin DJ, Creasy DM, et al. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 71.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 72.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tabb DL, Fernando CG, Chambers MC. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res. 2007;6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swaney DL, Wenger CD, Coon JJ. Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J Proteome Res. 2010;9:1323–1329. doi: 10.1021/pr900863u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rappsilber J, Mann M, Ishihama Y. Protocol for micropurification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 76.Hoehenwarter W, Wienkoop S. Spectral counting robust on high mass accuracy mass spectrometers. Rapid Commun Mass Spectrom. 2010;24:3609–3614. doi: 10.1002/rcm.4818. [DOI] [PubMed] [Google Scholar]
- 77.Tyanova S, Mann M, Cox J. MaxQuant for in-depth analysis of large SILAC datasets. Methods Mol Biol. 2014;1188:351–364. doi: 10.1007/978-1-4939-1142-4_24. [DOI] [PubMed] [Google Scholar]
- 78.Milac TI, Randolph TW, Wang P. Analyzing LC-MS/MS data by spectral count and ion abundance: two case studies. Stat Interface. 2012;5:75–87. doi: 10.4310/SII.2012.v5.n1.a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krey JF, Wilmarth PA, Shin JB, et al. Accurate label-free protein quantitation with high- and low-resolution mass spectrometers. J Proteome Res. 2014;13:1034–1044. doi: 10.1021/pr401017h. [DOI] [PMC free article] [PubMed] [Google Scholar]