Abstract
We describe the application of molecular biological techniques to estimate eukaryotic diversity (primarily fungi, algae, and protists) in Antarctic soils across a latitudinal and environmental gradient between approximately 60 and 87°S. The data were used to (i) test the hypothesis that diversity would decrease with increasing southerly latitude and environmental severity, as is generally claimed for “higher” faunal and plant groups, and (ii) investigate the level of endemicity displayed in different taxonomic groups. Only limited support was obtained for a systematic decrease in diversity with latitude, and then only at the level of a gross comparison between maritime (Antarctic Peninsula/Scotia Arc) and continental Antarctic sites. While the most southerly continental Antarctic site was three to four times less diverse than all maritime sites, there was no evidence for a trend of decreasing diversity across the entire range of the maritime Antarctic (60 to 72°S). Rather, we found the reverse pattern, with highest diversity at sites on Alexander Island (ca. 72°S), at the southern limit of the maritime Antarctic. The very limited overlap found between the eukaryotic biota of the different study sites, combined with their generally low relatedness to existing sequence databases, indicates a high level of Antarctic site isolation and possibly endemicity, a pattern not consistent with similar studies on other continents.
The Antarctic includes some of the most extreme terrestrial environments on Earth. However, not all habitats are hostile, and conditions range from moist eutrophic soils on maritime islands, through coastal “oases,” to remote inland nunataks and the frigid deserts of the McMurdo Dry Valleys (14, 61). According to general ecological principles, a more extreme (stressful) environment is expected to be less biodiverse (10, 23). Across Antarctica, many environmental stresses are severe relative to those at lower latitudes (15), while biodiversity is low and trophic systems are simple. Even within Antarctica, increasing environmental stress is linked to decreasing measures of diversity, with progressive loss of macrofaunal and macrofloral taxa, entire functional groups, and trophic levels (14, 29, 55, 59). Many studies have shown that many organisms, including higher plants, mosses, algae, fungi, arthropods, protozoa, and lichens, become less diverse as the latitude increases (6, 12, 14, 61).
The majority of previous studies of Antarctic eukaryotic microbiota in the terrestrial environment have been based on traditional morphological and/or culture approaches (6, 44, 61). However, the environmental range of some studies is relatively limited, and it is often difficult to compare results because of methodological variations (6). In recent years a selection of molecular methods which enable the identification of diverse community members in any given environment have become available. These methods do not require extraction of individuals or use of culture protocols and can be designed to target a very broad or very narrow range of organisms. To date, the majority of this molecular work has been directed towards prokaryote communities, often in extreme environments (38, 60, 66). Molecular studies have also been utilized to look at population differences in various environments or along stress gradients (10, 19, 20, 46). Recently, these methods have been applied to eukaryotic populations in a range of environments, although the majority have focused on aquatic microscopic eukaryotes (10, 18, 19, 31, 35, 37, 54, 65). These studies have identified unexpectedly high levels of diversity and suggest the possibility that a similarly hidden diversity of microscopic eukaryotes may exist in terrestrial environments.
The Antarctic forms a natural laboratory providing wide environmental gradients over which the effects of increasing stress on diversity and community structure can be observed. Conventionally, the region is separated into three terrestrial biogeographical zones (sub-Antarctic, maritime Antarctic, and continental Antarctic), based on consistent biological and climatic differences (14, 30, 48). The study described here utilizes molecular techniques to study diversity patterns across latitude, including material obtained across almost the entire latitudinal range of the maritime Antarctic, from the South Orkney Islands (ca. 60°S) to southern Alexander Island (ca. 72°S). Further data were obtained from the LaGorce Mountains (ca. 87°S), among the most southern terrestrial habitats existing in the continental Antarctic, and from Sky Hi Nunataks (ca. 75°S) in the currently undefined and largely unstudied “boundary region” between maritime and continental Antarctica. In addition to the obvious biogeographical significance of understanding processes across the boundary region between maritime and continental Antarctica, this study represents the first opportunity that biologists have had to work south of Alexander Island in the Antarctic Peninsula region.
The aim of this study was to utilize molecular methods to identify a wide range of eukaryotes from a selection of soils across the latitudinal and environmental gradient between the northern maritime and deep continental Antarctic, in order to test the hypothesis that diversity would decrease as latitude (and stress) increased. Prokaryote diversity in the soil community was not addressed in this study, due to the prohibitive number of clones that would have been required to perform an extensive survey. By using standard methods across all samples and study sites and targeting a specific type of soil habitat, we were able to assess the degree of similarity between the sites.
MATERIALS AND METHODS
Collection sites and sampling.
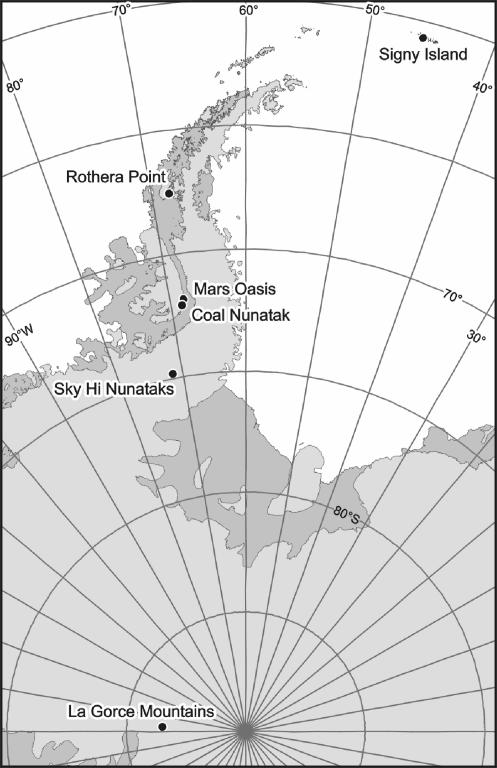
Soil samples were collected from six sites: Jane Col, Signy Island (South Orkney Islands) (60°42′S, 45°38′W); Rothera Point, Adelaide Island (western Antarctic Peninsula) (67°34′S, 68°08′W); Mars Oasis (71°52′S, 68°15′W) and Coal Nunatak (72°07′S, 68°32′W) (Alexander Island); Sky Hi Nunataks (74°50′S, 71°36′W) (Ellsworth Land); and the LaGorce Mountains (86°30′S, 147°W) (Fig. 1). Soils were collected over a number of Antarctic field seasons (1998 to 2001) (Table 1). They were stored initially at ambient field temperatures until returned to an Antarctic research station. They were then frozen at −20°C and returned to the United Kingdom for analysis. The biology of Signy Island, Rothera Point, Mars Oasis, and the La Gorce Mountains has received various levels of study (8, 16, 62), but little or no published biological information exists for the remaining two locations.
FIG. 1.
Map showing the study region and indicating locations of collection sites.
TABLE 1.
Summary of the physical and chemical properties of soils obtained from each study site
| Site | Sampling date | Organic (%) | H2O (%) | pH | C (mg/kg) | N (mg/kg) | Na (mg/kg) | K (mg/kg) | Ca (mg/kg) | Fe (mg/kg) | P (mg/kg) | Cd (mg/kg) | Cu (mg/kg) | Cr (mg/kg) | Ni (mg/kg) | Pb (mg/kg) | Zn (mg/kg) | V (mg/kg) | As (mg/kg) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Signy Island | February 2000 | 2.30 | 27.71 | 6.97 | 0.7 | 0.093 | 0.11 | 0.76 | 0.89 | 4.7 | 0.1 | 0.11 | 69 | 240 | 140 | 9.1 | 98 | 110 | <3 |
| Rothera Point | December 2000 | 5.94 | 24.74 | 5.47 | 2.3 | 0.31 | 0.4 | 0.27 | 17 | 1.8 | 9.2 | 15 | 220 | 23 | 4.5 | 17 | 1100 | 56 | 10 |
| Mars Oasis | December 1999 | 2.56 | 9.67 | 7.06 | 0.36 | 0.036 | 0.021 | 0.22 | 1.4 | 2.6 | 0.054 | 0.08 | 31 | 16 | 13 | 16 | 81 | 52 | 4 |
| Coal Nunatak | December 1999 | 4.34 | 12.36 | 7.67 | 1.5 | 0.047 | 0.015 | 0.76 | 1.4 | 1.7 | 0.044 | 0.1 | 16 | 8.1 | 6.9 | 23 | 61 | 36 | 5 |
| Sky Hi Nunataks | December 2000 | 1.83 | 10.51 | 5.42 | 0.42 | 0.05 | 0.01 | 0.11 | 0.91 | 2.88 | 0.07 | <0.07 | 5.56 | 19.60 | 10.80 | 10.6 | 38.40 | 50.80 | 12.80 |
| LaGorce Mts | December 1998 | 0.42 | 12.14 | 6.82 | 0.037 | 0.01 | 0.01 | 0.3 | 0.24 | 1.6 | 0.05 | <0.07 | 12 | 13 | 10 | 9.8 | 39 | 18 | <3 |
Maritime Antarctic soils are typically poorly developed with low organic content and physical stability. Repeated freeze-thaw action leads to large areas of patterned ground, with clearly defined areas of fine mineral particles surrounded by coarser material, stones, and rocks (11). The central mineral fines remain very unstable and are generally uncolonized by macroscopic vegetation,. To maximize comparability between the study sites, soil samples obtained from Signy Island, Rothera Point, Mars Oasis, and Coal Nunatak were taken from the central area of such frost-sorted polygons. Soil development is much more restricted on inland nunataks and mountain ranges. The geomorphology of the sites at Sky Hi Nunataks and the LaGorce Mountains does not lead to the formation of extensive areas of patterned ground. Rather, samples were obtained from small pockets of mineral fines concentrated by freeze-thaw action in spaces between rocks, particularly among boulder fields and areas of rock scree. None of the soils sampled had significant moss or lichen coverage, although traces of moss and lichen were visible in some samples.
Five separate soil samples (2 to 10 g) were collected from each site by one of two methods. If enough unfrozen soil was available, sterile monovette corers (Sarstedt Ltd., Leicester, United Kingdom) were plunged into soil fines to a depth of 10 to 15 mm. Alternatively, where little soil was available, larger particles precluded the use of monovettes, or the soil was frozen, samples were scraped into sterile whirlpack plastic bags with a sterile spatula. Soils were stored under ambient conditions in the field following collection before they could be returned to a research station, where they were then held at −20°C until use.
Physical and chemical analyses.
Physical analyses were undertaken to ascertain substratum similarities and any major differences. Soil samples (5 to 10 g) from each site were air dried at 80°C for 48 h, and the water content was estimated gravimetrically. A subsample of the air-dried soil was then incinerated at 400°C for 24 h to obtain organic matter content. The remainder of the air-dried sample was analyzed commercially (Centre for Ecology and Hydrology, Merlewood, United Kingdom) for pH and contents of C, N, P, K, Fe, As, Cd, Cr, Ni, Pb, V, and Zn. In order to provide sufficient material, soil from the five samples obtained at each site was combined, precluding statistical comparisons between sites.
Estimation of molecular diversity. (i) DNA extraction, amplification and clone library construction.
In order to provide comparable data across a wide range of organisms from all sites, a clone library-sequence approach was taken. Clone libraries were constructed by extracting total genomic DNA, amplifying the near-full-length eukaryotic small-subunit ribosomal DNA (SSU rRNA gene), and then cloning the products. Total genomic DNA was extracted separately from the five individual soil samples obtained from each site, and each of the five isolates was then used as a template in at least three amplification reactions. DNA was extracted from soil samples by using a combined physical and chemical treatment. Approximately 0.5 g of soil was placed in an Eppendorf tube with 500 μl of sterile 0.1-mm-diameter glass beads (Stratech Scientific, Soham, United Kingdom) and 700 μl of lysis buffer (0.1 M Tris [pH 7.4], 0.25 M EDTA, 2% [wt/vol] glucose, 0.5% [wt/vol] sodium dodecyl sulfate, and 2 mg of proteinase K [Sigma, Gillingham, United Kingdom] ml−1). Samples were vortexed at full speed for 5 min and then subjected to five cycles of freezing and thawing (−80°C for 2 min and +80°C for 2 min). Cell debris was then removed by centrifugation (12,000 × g, 2 min). The supernatant was phenol extracted, and the DNA was ethanol precipitated and resuspended in 400 μl of water. Aliquots from each sample (80 μl) were cleaned for PCR amplification with GFX DNA purification columns according to the instructions of the manufacturer (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom).
PCR amplifications were performed with 43 μl of ABgene 1.1× ReddyMix PCR master mix (ABgene, Epsom, United Kingdom), 20 pmol of each primer, and approximately 10 to 100 ng of template DNA in a final volume of 50 μl. The amplification program consisted of 1 cycle of 95°C for 5 min; 30 cycles of 94°C for 60 s, 55°C for 60 s, and 72°C for 70 s; and a final extension step of 72°C for 10 min. To amplify the near-full-length eukaryotic SSU rRNA gene, the primers E4F (5′-CTGGTTGATTCTGCCAGT-3′) and E1628R (5′-CGACGGGCGGTGTGTA-3′) (53) were used.
PCR products from the same sample were pooled and purified by using GFX DNA purification columns. Cleaned products originating from each sample from a single site were pooled and cloned into the pGEM-T Easy vector (Promega, Southampton, United Kingdom). Ligation products were transformed into JM109 (Promega) according to the manufacturer's instructions. Transformants were screened by color selection on Luria agar containing 100 μg of ampicillin ml−1, IPTG (isopropyl-β-d-thiogalactopyranoside), and either S-Gal or X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Sigma) as an indicator. White colonies were picked from the plates and resuspended in 50 of μl water. The cells were subjected to two freeze-thaw cycles (−80°C and +80°C), and an aliquot of 2 μl was used as the template in a screening PCR with M13 forward (TGTAAAACGACGGCCAGT) and reverse (CAGGAAACAGCTATGACC) universal primers. The amplification program consisted of 1 cycle of 95°C for 5 min; 30 cycles of 94°C for 45 s, 58.5°C for 45 s, and 72°C for 60 s; and a final extension step of 72°C for 10 min. Aliquots of the amplified products were electrophoresed in 1% (wt/vol) agarose gels, and products showing bands of the correct size were cleaned with GFX DNA purification columns.
(ii) Controls.
The methods described above are potentially open to contamination at a number of stages, and thus a series of controls were put in place. First, all samples were taken aseptically, and aliquots for DNA extraction were removed from the sampling containers in flow hoods. The hoods and balances were washed down with 70% ethanol and Virkon (main active ingredient, potassium monopersulfate; Antek International, Sudbury, United Kingdom) between samples. A control DNA extraction (containing no soil) was carried out in parallel with the soil DNA extractions and was used as the template in subsequent PCR amplification. Negative control reactions with mixtures containing water in place of genomic or plasmid DNA were also carried out at each step of the protocol. None of these control reactions generated positive results.
(iii) RFLP screening and sequencing.
Cleaned PCR products of the correct size were separately digested with HaeIII and RsaI for 2 to 3 h at 37°C. Digests were separated on 3% (wt/vol) high-resolution agarose (Appligene, Middlesex, United Kingdom) and stained with ethidium bromide, and images were captured and exported to the Gelcompar II software package (Applied Maths, Saint-Martens-Latem, Belgium). Patterns from all gels were normalized against a size standard, and all clones showing unique patterns were sequenced. Where groups of clones displayed the same restriction fragment length polymorphism (RFLP) patterns, two to four isolates were chosen, at random, for sequencing. Sequence reactions were carried out with the ET-Terminator sequence kit (Amersham Biosciences, Little Chalfont, United Kingdom) and M13 forward and reverse primers and were run on a Megabace 500 sequencer (Amersham Biosciences). For the purpose of this study, sequences corresponding to the first 700 bp of the eukaryotic SSU rRNA gene were used.
Operational taxonomic units (OTUs) were defined as clones sharing ≥97.5% sequence identity, regardless of whether or not they shared RFLP patterns. This value has been suggested for bacterial studies (3, 36). For eukaryotes no comparable cutoff has been proposed, and likely values appear to vary considerably depending on the DNA region used and the organisms studied. Hebert et al. (26) found that intraspecific divergences were rarely greater than 2% for the mitochondrial cytochrome c oxidase subunit I gene in animals, and a value of 0 to 3% divergence between species of basidiomycete fungi has been reported for internal transcribed spacer and 5.8S regions (64). Generally, the small-subunit regions of eukaryotes are very conserved, but given the potential for errors in both sequencing and amplification protocols from environmental samples (9), this value should ensure that differences are representative of biological differences and are not artifactual. Samples were taken until the sampling coverage (see below) for each clone library was greater than 85%.
(iv) Sequence identification.
All sequences were analyzed with the rRNA Database Project CHECK_CHIMERA program (32); six potential chimeric sequences were detected and removed from the analysis. All remaining clone sequences were compared with the GenBank database of genetic sequences by using gapped BLAST searches (1) to determine their approximate phylogenetic affiliations. Several sequences were most closely related to prokaryotes, and some sequences had no affiliations to any sequences in the database. These did not contain any of the SSU rRNA gene conserved regions and, along with the prokaryote sequences, were not included in further analyses. Sequences obtained from each site were aligned through the JalView utility of ClustalW run at the European Bioinformatics Institute (http://www.ebi.ac.uk), and a consensus phylogenetic tree was obtained from DNA distances and neighbor joining in PHYLIP. FASTA searches were then used to compare sequences against the appropriate EMBL subset database (e.g., EMBL-FUNGI) indicated by the position at which the sequence was recovered in the tree and the BLAST results. Sequences were identified to at least the subdivision level.
(v) Statistical analyses.
Library coverage, an estimate of the proportion of a clone library of infinite size that has been sampled, was calculated by using the relative distribution of OTUs and the equation described by Good (24). Collectors' curves, or species abundance curves, were constructed to compare the diversity of OTUs at each site. Curves were calculated by using the freeware program Analytic Rarefaction 1.3 (available at http://www.uga.edu/∼strata/software/). The program uses the rarefaction equations described by Hurlbert (28) and Heck et al. (27). The probability of drawing a new OTU (θn), on the next draw was estimated by using the method described by Clayton and Frees (13).
Between-site relationships based on numbers and types of taxa present were investigated by principal-coordinate analyses. Data in each group were transformed into presence-absence data and converted to a percentage of sequences recovered at each site. A distance matrix was constructed with the average taxonomic distance coefficient, and a principal-coordinate plot was made based on the first three axes. All computations used the MVSP package (Kovach Computing, Anglesea, United Kingdom).
Pairwise comparisons of sites were made by using Z scores, which indicate whether an estimated number of OTUs (E) at a given number of clones (n) is significantly different between pairs of sites. E was obtained by normalizing libraries at n = 77 by using rarefaction (as described above). The rarefaction calculations also gave the upper and lower 95% confidence levels, which were used to calculate the standard error for each site.
RESULTS
Physical and chemical analyses.
With the exception of the LaGorce Mountains, all soils had broadly similar levels of organic matter (Table 1). Water contents were not biologically limiting at any site, ranging from 10 to 28% (wt/wt). Soil pH varied around the neutral point, ranging from mildly acidic (Sky Hi Nunataks [pH 5.4]) to mildly basic (Coal Nunatak [pH 7.7]). Other parameters were similar across all soils, with the following exceptions: carbon showed a 30-fold variation across all of the sites, Signy Island soil had 10 to 30 times more Cr and Ni than other sites, and Rothera Point soil had higher levels of N, Na, Ca, Cd, Cu, and Zn (Table 1).
Clone libraries.
Clone library coverage ranged from 87 to 99%, and all libraries showed very low θn values. To reach this level of coverage, between 78 (Sky Hi Nunataks) and 169 (Signy Island) clones were required (Table 2). Across all sites, 712 clones produced 204 RFLP patterns that were divided into 111 OTUs. Several OTUs were comprised of more than one RFLP pattern, and in two cases the same RFLP pattern gave rise to different OTUs. Alexander Island sites (Mars Oasis and Coal Nunatak) had numbers of OTUs similar to or higher than those of northern maritime Antarctic Signy Island, while Rothera Point (central maritime Antarctic) generated fewer. Sky Hi Nunataks and LaGorce Mountains generated the lowest numbers of OTUs (Tables 2 and 3). The latter libraries also contained the lowest number of clones and the highest coverage values, supporting the conclusion that they are the least diverse of the sites studied.
TABLE 2.
Estimates of sampling coverage and θn (the unconditional probability of a new OTU discovery if another clone was taken) for each study sitea
| Site | Sampling coverage (%) | θn | No. of clones obtained | No. of OTUs identified |
|---|---|---|---|---|
| Signy | 92.31 | 0.0368 | 169 | 33 |
| Rothera Point | 89.74 | 0.0509 | 117 | 21 |
| Mars Oasis | 86.78 | 0.0673 | 121 | 32 |
| Coal Nunatak | 87.14 | 0.0714 | 140 | 48 |
| Sky Hi Nunataks | 93.59 | 0.0307 | 78 | 21 |
| La Gorce | 98.85 | 0.0135 | 87 | 10 |
See Materials and Methods for detail of calculation.
TABLE 3.
Identification and grouping of sequences. The table lists the major higher taxonomic groupings (phylum or division) into which sequences obtained were separated, the number (N) of separate operational taxonomic units (OTUs) obtained in each group from each study site, and the overall percentage contribution made by each kingdom or sub-kingdom to the biota of each study site.
| Kingdom or subkingdom | Phylum or divisiona | Signy Island
|
Rothera Point
|
Mars Oasis
|
Coal Nunatak
|
Sky Hi Nunatak
|
LaGorce Mountains
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nb | %c | N | % | N | % | N | % | N | % | N | % | ||
| Metazoa | Nematodes | 8 | 33 | 0 | 1 | 3 | 1 | 2 | 0 | 10 | 0 | ||
| Arthropods | 2 | 0 | 0 | 0 | 0 | 0 | |||||||
| Tardigrades | 1 | 0 | 0 | 0 | 2 | 0 | |||||||
| Fungi | Ascomycetes | 3 | 27 | 6 | 38 | 2 | 13 | 4 | 31 | 6 | 38 | 1 | 20 |
| Basidiomycetes | 0 | 2 | 0 | 1 | 0 | 1 | |||||||
| Zygosporic | 6 | 0 | 2 | 10 | 2 | 0 | |||||||
| Stramenopila | Bacillariophyta | 4 | 12 | 2 | 9 | 6 | 34 | 1 | 19 | 0 | 10 | 0 | |
| Xanthophyta | 0 | 0 | 4 | 8 | 2 | 0 | |||||||
| Chrysophyta | 0 | 0 | 1 | 0 | 0 | 0 | |||||||
| Chlorophyta | Chlorophyceae | 1 | 6 | 1 | 19 | 2 | 34 | 2 | 21 | 0 | 14 | 2 | 40 |
| Trebouxiaceae | 1 | 2 | 7 | 6 | 3 | 2 | |||||||
| Ulvophyceae | 0 | 1 | 2 | 2 | 0 | 0 | |||||||
| Streptophyta | Bryophytes | 0 | 1 | 5 | 0 | 1 | 2 | 0 | 0 | ||||
| Alveolata | Ciliophorans | 3 | 9 | 1 | 5 | 2 | 6 | 2 | 4 | 3 | 14 | 2 | 20 |
| Uncertain rank | Euglenazoa | 1 | 3 | 1 | 5 | 0 | 0 | 0 | 0 | ||||
| Cercozoa | 2 | 6 | 3 | 14 | 2 | 6 | 7 | 15 | 3 | 14 | 1 | 10 | |
| Plasmodiophora | 1 | 3 | 1 | 5 | 0 | 1 | 2 | 0 | 1 | 10 | |||
| Others | 0 | 0 | 1 | 3 | 2 | 4 | 0 | 0 | |||||
| Total from site | 33 | 21 | 32 | 48 | 21 | 10 | |||||||
Major taxonomic grouping into which sequences obtained were separated.
Number of separate OTUs.
Overall percent contribution made by kingdom or subkingdom to the biota.
Identification of sequences.
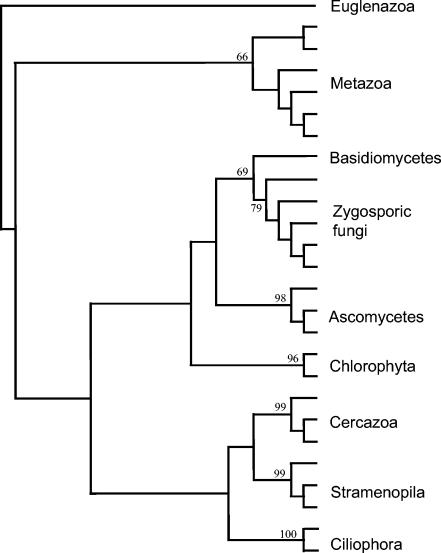
The consensus distance tree recovered the sequences in distinct, bootstrap-supported groups (Fig. 2), where sequences within each group showed greater than 85% homology to each other. The BLAST- and FASTA-derived identifications of the sequences indicated that each group of sequences belonged to a distinct kingdom or subkingdom, as listed in Table 3. The taxonomic level at which an individual sequence could be unequivocally identified varied both between and within taxonomic groups. In some cases sequences for individual species and genera were very distinctive, whereas in others sequences could only be loosely identified at the subdivision or phylum level. Comparison of sequences between sites showed that all sequences used were unique and that within each major taxonomic division there was no grouping according to site (results not shown).
FIG. 2.
Example of consensus tree of aligned complete sequences, showing major kingdom and subkingdom groups present in Signy Island samples. Bootsrap replications are shown for major groupings.
Community composition.
A detailed, class level analysis of OTU affiliations as determined by BLAST and FASTA matches is given in Table 4. All sites revealed a wide range of organisms, with fungal, algal, and nonalgal protist groups providing the majority of OTUs. Table 4 also includes a number of isolates showing greatest phylogenetic affiliation with “uncultured clones.” The published phylogenetic data referring to each of these uncultured clones suggest that they also belong to nonalgal protist groups (53, 65).
TABLE 4.
Taxonomic breakdown by class of OTUs identified at each study site by comparison with the GenBank sequence databasea
| Group | Number of OTUs at:
|
|||||
|---|---|---|---|---|---|---|
| Signy Island | Rothera Point | Mars Oasis | Coal Nunatak | Sky Hi Nunataks | LaGorce Mountains | |
| Fungi | ||||||
| Ascomycota indet | 2 | 1 | 2 | 1 | ||
| Chaetothyriomycetes | 1 | |||||
| Chytridiomycetes | 5 | 3 | 8 | |||
| Dothideomycetes | 1 | |||||
| Eurotiomycetes | 2 | 1 | ||||
| Fungi incertae sedis | 1 | |||||
| Heterobasidiomycetes | 1 | 1 | ||||
| Homobasidiomycetes | 1 | |||||
| Lecanoromycetes | 1 | 2 | 3 | |||
| Leotiomycetes | 1 | |||||
| Pezizomycetes | 1 | |||||
| Saccharomycetes | 1 | |||||
| Sordariomycetes | 1 | |||||
| Taphrinomycetes | 1 | |||||
| Zygomycetes | 1 | |||||
| Metazoa | ||||||
| Chromadorea | 1 | 1 | 1 | |||
| Enoplea | 1 | |||||
| Eutardigrada | 1 | 1 | ||||
| Heterotardigrada | 1 | |||||
| Collembola | 1 | |||||
| Uncultured clones | ||||||
| cln LKM30 | 1 | 1 | 2 | 1 | ||
| cln LKM45 | 1 | 1 | 1 | |||
| cln RT5iin25 | 1 | |||||
| Nonalgal protists | ||||||
| Nassophorea | 1 | |||||
| Colpodea | 2 | |||||
| Oligohymenophorea | 1 | |||||
| Phyllopharyngea | 1 | 1 | ||||
| Phytomastigophorea | 1 | |||||
| Prostomatea | 1 | |||||
| Spirotrichea | 1 | 1 | 1 | 1 | 1 | |
| Filosea | 1 | 1 | ||||
| Sarcomonadea | 2 | 2 | 1 | 3 | 1 | |
| Bodonea | 1 | 1 | 1 | |||
| Lobosea | 1 | 1 | ||||
| Protista incertae sedis | 1 | |||||
| Algae | ||||||
| Bacillariophyceae | 3 | 1 | 4 | 1 | ||
| Chrysophyceae | 2 | |||||
| Heterokonta incertae sedis | 1 | 1 | 1 | 1 | ||
| Hyphochytriaceae | 1 | |||||
| Xanthophyceae | 4 | 4 | ||||
| Chlorophyceae | 1 | 2 | 2 | 1 | 2 | |
| Trebouxiophyceae | 1 | 1 | 7 | 4 | 3 | 1 |
| Ulvophyceae | 1 | 2 | 2 | |||
| Bryopsida | 1 | 1 | ||||
See Materials and Methods for details.
Mars Oasis and Coal Nunatak soils contained representatives from all of the major eukaryotic taxonomic groups identified (Table 3). Chlorophyta and Stramenopila dominated Mars Oasis clones (68% of OTUs), while there was a fairly even spread of fungal (31%), chlorophyte (19%), and stramenopile (21%) classes at Coal Nunatak. Signy Island, Mars Oasis, Sky Hi Nunataks, and LaGorce Mountains clone libraries lacked plant clones, while Rothera Point and La Gorce Mountains also lacked metazoan clones. The limited occurrence of these macroscopic floral and faunal groups is likely to be a simple stochastic consequence of limited and/or spatially aggregated distributions or typically low representation in this soil habitat. Detailed phylogenetic analysis of the sequences obtained will be reported elsewhere.
Site comparisons.
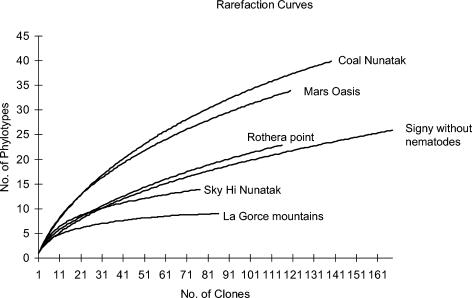
Rarefaction curves created for each of the sampling sites are shown in Fig. 3. A plateau, indicating close to full biodiversity coverage, was approached only for the LaGorce and Sky Hi Nunataks soils. While clone library coverage values were also high for the other sites (Table 2), the curves did not reach a plateau. The slopes of the Signy Island and Rothera Point curves were shallower than those of the curves obtained for Mars Oasis and Coal Nunatak, suggesting that a small number of OTUs were dominating these libraries. Indeed, the Signy Island clone library contained a single OTU corresponding to a nematode worm sequence that accounted for 55% of clones. No other library showed such a level of dominance by a single OTU; however, all had between one and three OTUs which each contributed 10% or more of the library. Removal of the dominating nematode OTU from the Signy Island data generated a rarefaction curve that was more similar to those of Mars Oasis and Coal Nunatak, although the number of OTUs found remained low in comparison (Fig. 3). Rarefaction was used to normalize each of the clone libraries, and comparisons were made between the sites based on the number of OTUs observed (Table 5). This comparison showed the two Alexander Island sites (Mars Oasis and Coal Nunatak) to be more diverse than the other maritime Antarctic sites, while the boundary region site at Sky Hi Nunataks was less diverse and the continental site at LaGorce Mountains had by far the lowest diversity.
FIG. 3.
Rarefaction curves constructed for eukaryotic clone libraries from each of the six collection sites. Clones were grouped into OTUs at a level of sequence similarity of >97.5%. The “Signy without nematodes” curve has had a dominating clone removed (see text).
TABLE 5.
Diversity comparisons between the six study sites
| Site |
Z value for comparison witha:
|
||||
|---|---|---|---|---|---|
| Rothera | Mars Oasis | Coal Nunatak | Sky Hi | LaGorce | |
| Signy | −0.476§ | −3.756* | −4.246* | 1.634§ | 4.191* |
| Rothera | −3.580* | −4.096* | 2.636* | 5.638* | |
| Mars Oasis | −0.768§ | 6.957* | 9.582* | ||
| Coal Nunatak | 7.062* | 9.347* | |||
| Sky Hi | 9.958* | ||||
Z values were calculated from data normalized by rarefaction (see Materials and Methods). §, not significant; *, significant (P < 0.01).
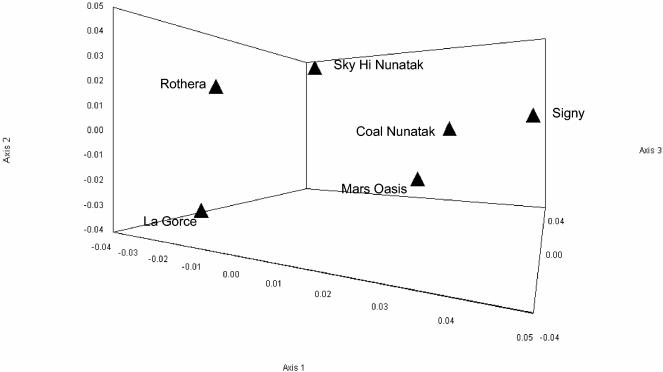
In order to test for similarities between the sites with regard to their basic community structure, the phylogenetic affiliations for each OTU were used to compare the sites at a high taxonomic level by using principal-coordinate analysis (Fig. 4). The principal-coordinate analysis representation showed that each site was distinct, with no obvious grouping of two or more sites together. The three dimensions shown in Fig. 4 accounted for 79.9% of the total variation, suggesting high correlation between variable characters. In this context, this finding can be taken as indicative of different combinations of taxa for each site.
FIG. 4.
Principal-coordinate analysis of sites based on the percentage of sequences obtained for each taxonomic group at each site (original data are in Table 3).
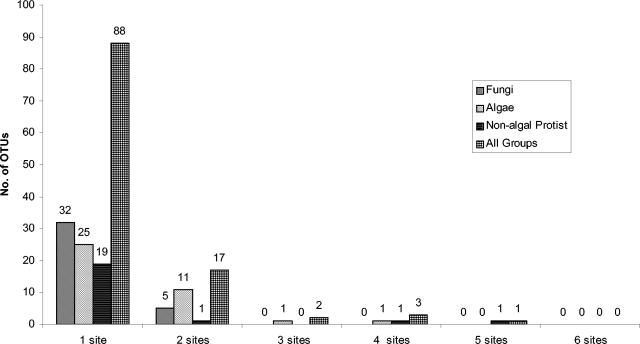
Finally, the number of sites at which each OTU occurred was quantified (Fig. 5) as an indication of whether the organisms detected are restricted to a single site or are present at multiple sites. The majority of OTUs were found at only one site, and no single OTU was present at all six sites. This pattern held for each of the various high-level taxonomic groupings when analyzed separately and also for a combined data set incorporating all OTUs.
FIG. 5.
Numbers of sites at which different OTUs were found. Fungal, algal, and nonalgal protist groups and a combination of all groups (including metazoans, plants, and uncultured clones) are shown. Numbers above the bars indicate the total number of OTUs in each group found at a given number of sites.
DISCUSSION
Methodological limitations.
There are well-documented limitations to the use of molecular methods in biodiversity studies (43, 57). Furthermore, clone library data are not quantitative and provide only presence-absence data, precluding the use of many diversity indices. To minimize biases, we used low template concentrations, and to minimize PCR drift, multiple PCR products were pooled prior to cloning (58). All samples have been treated in the same way, so results across sites should be fully comparable (36). The clone libraries generated in this study are representative of the eukaryote microbial biota of the immediate sampling environment but are not intended to form a comprehensive survey of all environments at each site.
Identification of sequences obtained is dependent on the quality of the sample data, the available reference data, and the discrimination accorded by the sequence used. In this study sample, sequence quality was good (sequences of ca. 1,000 bp were routinely recovered) and was clearly sufficient to separate OTUs into the major biological kingdoms and subkingdoms. Below this taxonomic level, discrimination was more variable, with some species and genera being very distinct, while other assignments could be made only at a family or order level. This illustrates a limitation in using a single sequence to screen and identify a wide diversity of organisms. Reference data quality remains problematic. Mislabeled and incorrect sequences appear in databases (5), while the proportion of organisms for which sequences are available is still relatively low, although coverage varies between taxonomic groups (see, e.g., reference 5). Our cutoffs of 97.5% homology for OTUs and identification to order level only are justified by the limitations of the reference information.
Latitudinal patterns.
It is generally accepted that Antarctic terrestrial diversity decreases as latitude (and stresses) increase (6, 29, 47). In contrast, the present data provide support for this pattern only when a large (regional)-scale comparison is made between the maritime and continental Antarctic. Instead, the two southern maritime Antarctic sites (Mars Oasis and Coal Nunatak) generated diversity measures comparable to or higher than those of the central and northern sites (Rothera Point and Signy Island). Greatest diversity was recorded for Coal Nunatak. While Coal Nunatak is only ca. 30 km from Mars Oasis and they are part of the same sedimentary geological formation, the two sites contrast markedly in their degree of colonization by macroscopic biota. Mars Oasis, which is ca. 30 m above sea level, has a relatively diverse fauna and flora (16), while those of Coal Nunatak, which is ca. 750 m above sea level, are extremely limited. The finding that the eukaryotic microbial diversity of Coal Nunatak soils is approximately 50% greater than that of Mars Oasis (Table 3) is, at least, surprising and may suggest a longer duration of exposure through recent glacial cycles.
The macroscopic biota of Alexander Island are closely related to those of the maritime Antarctic (16), as are the soil meiofauna as indicated by the very few published records (34). However, there are also endemic components, including a springtail and several currently undescribed nematode and tardigrade species (16, 34; N. R. Maslen and P. Convey, unpublished data; S. J. McInnes, personal communication). Although it is generally thought that contemporary terrestrial habitats of this island have been exposed for, at most, 5,000 to 6,000 years (16, 51), when combined with the current finding of high microeukaryotic diversity, these various observations suggest the existence of terrestrial habitats beyond the time scale allowed by the last glacial maximum.
Lower diversity was present at the two more southern sites examined, at Sky Hi Nunataks and the LaGorce Mountains. Given that the present study could include only these two sampling points, it is not possible to conclude whether their lower diversity is a function of increasing latitude or is simply representative of a different Antarctic biogeographical zone. The range of organisms isolated from the LaGorce soils is similar to that found by Broady and Weinstein (8), who used microscopy and culture-based methods to identify cyanobacteria, chlorophytes, and fungi in the same soils.
The organisms recovered are also likely to be influenced by the environmental conditions at each site. For instance, at the sites with the lowest availability of liquid water (LaGorce Mountains and Sky Hi Nunataks), largely water-associated stramenopilan taxa were not present or were poorly represented. Unlike in temperate and tropical solids, where basidiomycete and zygomycete fungi generally dominate, in this study ascomycetes predominated, a group that includes many lichenized taxa (Table 4). As the primarily lichen-forming chlorophyte Trebouxiaceae also contributed a high proportion of the sequences obtained, this suggests that the soils sampled were strongly lichen influenced (e.g., by propagules, early colonizing stages). Samples from four sites (Rothera Point, Signy Island, Coal Nunatak, and La Gorce Mountains) contained sequences that were conclusively identified as being plasmodiophoran. This group has not previously been reported from Antarctica.
Endemism or cosmopolitanism?
Comparison of molecular and traditional surveys is difficult. In studies of prokaryotes, they often show very little overlap, although greater similarity is seen in more extreme and less diverse environments (4, 40, 49). Some eukaryote groups (e.g., nematodes and arthropods) are relatively well documented in the Antarctic (2, 14). However, although sequences showed affiliation to nematodes or arthropods in the GenBank database, none of the latter species or their close relatives are present in Antarctica. Classical taxonomic data suggest that many metazoans are endemic in the continent (2, 25, 41, 42). Thus, while this study has most likely detected the presence of known Antarctic species, it is currently impossible to match the sequence data with known taxa. These difficulties are compounded for some groups (particularly the protists) by a lack of consistency within classical taxonomy.
Endemism, especially applied to microorganisms, is a controversial topic. The term generally implies that an organism is native or confined to a certain region, or, as expressed by Vincent (55) “genotypes of bacteria, protists or other microorganisms specific to a geographical region.” Vincent further postulates that if microbial endemism is possible, the Antarctic should be among the most likely places in which such organisms may be found. However, there is currently limited evidence for microbial endemism in Antarctic terrestrial, or indeed any, polar environments (6, 22, 50, 56).
The alternative to endemism is cosmopolitanism. It has long been suspected that microbial dispersal is not restricted by geographical boundaries (21, 63): “everything is everywhere, but the environment selects” (Baas-Becking [1934], cited in reference 50). Darling et al. (17) report evidence for bipolar genetic flow but do not confirm that cosmopolitan microorganisms exist in both polar regions. However, bipolar fungi, lichens, and mosses are known (48). Microorganisms may be transported into and around the Antarctic via various routes, including air currents (33), ocean currents (39), and animals (including birds [45] and humans [7]). Propagule banks in soil or snow are reported to contain cosmopolitan species of algae, fungi, protists, bacteria, and archaea (39, 52, 56, 61). However, while many of the organisms detected in our study show reasonable homology with isolates from other geographic locations, it remains impossible to categorically confirm that they are conspecific.
The clone library data from this study suggest that a minority of the organisms may be ubiquitous (Fig. 5), as a small number of OTUs were identified at multiple, widely separated sites and/or are closely related to sequences from cosmopolitan organisms (e.g., the alga Stichococcus bacillarus and the protists Heteromita globosa and Oxytricha granulifera). Many other isolates also show a high degree of similarity (>98% sequence identity) with non-Antarctic organisms, although 18S ribosomal gene sequence similarity does not necessarily imply functional similarity and species identity. Conversely, some sequences are considerably different from any others present in the GenBank database, an observation which may be indicative either of endemism or simply that these organisms are yet to be detected or sequenced in other environments. Although proof of endemism is not possible, our results suggest that there is very little effective transfer of biota between the study sites. Further insight will require isolation and genetic analysis of identified Antarctic organisms and comparison with known Antarctic and non-Antarctic relatives.
Acknowledgments
We gratefully acknowledge the considerable assistance of many British Antarctic Survey (BAS) staff in supporting the field operations at the various study sites. We thank Andrew Simms and Stephen Bromley for technical assistance and Andy Clarke, Lloyd Peck, Alex Rogers, David Pearce, and Tobias Garstecki for constructive comments on earlier manuscript versions.
This study forms part of the BAS “Biological Responses to Environmental Stress in Antarctica” core project and also contributes to the SCAR RiSCC (Regional Sensitivity to Climate Change in Antarctica) Program.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrássy, I. 1988. Nematodes in the sixth continent. J. Nematode Syst. Morphol. 1:107-186. [Google Scholar]
- 3.Bowman, J. P., S. M. Rea, S. A. McCammon, and T. A. McMeekin. 2000. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hills, Eastern Antarctica. Environ. Microbiol. 2:227-237. [DOI] [PubMed] [Google Scholar]
- 4.Brambilla, E., H. Hippe, A. Hagelstein, B. J. Tindall, and E. Stackebrandt. 2001. 16S rDNA diversity of cultured and uncultured prokaryotes of a mat sample from Lake Fryxell, McMurdo Dry Valleys, Antarctica. Extremophiles 5:23-33. [DOI] [PubMed] [Google Scholar]
- 5.Bridge, P. D., Roberts, P. J., Spooner, B. M., and Panchal, G. 2003. On the unreliability of published DNA sequences. New Phytol. 160:43-48. [DOI] [PubMed] [Google Scholar]
- 6.Broady, P. A. 1996. Diversity, distribution and dispersal of Antarctic terrestrial algae. Biodivers. Conserv. 5:1307-1335. [Google Scholar]
- 7.Broady, P. A., and R. A. Smith. 1994. A preliminary investigation of the diversity, survivability and dispersal of algae introduced into Antarctica by human activity. Proc. NIPR Symp. Polar Biol. 7:185-197. [Google Scholar]
- 8.Broady, P. A., and R. N. Weinstein. 1998. Algae, lichens and fungi in La Gorce Mountains, Antarctica. Antarct. Sci. 10:376-385. [Google Scholar]
- 9.Bruns, T. D., White, T. J., and Taylor, J. W. 1991. Fungal molecular systematics. Annu. Rev. Ecol. Syst. 22:525-564. [Google Scholar]
- 10.Casamayor, E. O., R. Massana, S. Benlloch, L. Ovreas, B. Diez, V. J. Goddard, J. M. Gasol, I. Joint, F. Rodriguez-Valera, and C. Pedros-Alio. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4:338-348. [DOI] [PubMed] [Google Scholar]
- 11.Chambers, M. G. 1967. Investigations of patterned ground at Signy Island, South Orkney Islands. III. Miniature patterns, frost heaving and general conclusions. Br. Antarct. Surv. Bull. 12:1-22. [Google Scholar]
- 12.Clarke, A. 2003. Evolution, adaptation and diversity: global ecology in an Antarctic context, p. 3-17. In A. H. L. Huiskes, W. W. C. Gieskes, J. Rozema, R. H. L. Schorno, S. M. van der Vries, and W. J. Wolff (ed.), Antarctic biology in a global context: proceedings of the VIII SCAR Biology Symposium. Backhuys, Leiden, The Netherlands.
- 13.Clayton, M. K., and E. W. Frees. 1987. Nonparametric-estimation of the probability of discovering a new species. J. Am. Stat. Assoc. 82:305-311. [Google Scholar]
- 14.Convey, P. 2001. Antarctic ecosystems, p. 171-184. In S. A. Levin (ed.), Encyclopedia of biodiversity, vol. 1. Academic Press, San Diego, Calif. [Google Scholar]
- 15.Convey, P. 1996. The influence of environmental characteristics on life history attributes of Antarctic terrestrial biota. Biol. Rev. 71:191-225. [Google Scholar]
- 16.Convey, P., and R. I. L. Smith. 1997. The terrestrial arthropod fauna and its habitats in northern Marguerite Bay and Alexander Island, maritime Antarctic. Antarct. Sci. 9:12-26. [Google Scholar]
- 17.Darling, K. F., C. M. Wade, I. A. Stewart, D. Kroon, R. Dingle, and A. J. L. Brown. 2000. Molecular evidence for genetic mixing of Arctic and Antarctic subpolar populations of planktonic foraminifers. Nature 405:43-47. [DOI] [PubMed] [Google Scholar]
- 18.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diez, B., C. Pedros-Alio, and R. Massana. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finlay, B. J. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061-1063. [DOI] [PubMed] [Google Scholar]
- 22.Franzmann, P. D. 1996. Examination of Antarctic prokaryotic diversity through molecular comparisons. Biodivers. Conserv. 5:1295-1305. [Google Scholar]
- 23.Frontier, S. 1985. Diversity and structure in aquatic ecosystems. Oceanogr. Mar. Biol. Annu. Rev. 23:253-312. [Google Scholar]
- 24.Good, I. J. 1953. The population frequencies of species and the estimation of the population parameters. Biometrika 40:237-264. [Google Scholar]
- 25.Greenslade, P. 1994. Collembola from the Scotia Arc and Antarctic Peninsula including descriptions of two new species and notes on biogeography. Polsk. Pism. Entomol. 64:305-319. [Google Scholar]
- 26.Hebert, P. D. N., S Ratnasingham, and J. R. deWaard. 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. London B 270(Suppl.):S96-S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heck, K. L. J., G. Van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1489-1491. [Google Scholar]
- 28.Hurlbert, S. H. 1971. The nonconcept of species diversity: a critique and alternative parameters. Ecology 52:577-586. [DOI] [PubMed] [Google Scholar]
- 29.Kennedy, A. D. 1995. Antarctic terrestrial ecosystem response to global environmental-change. Annu. Rev. Ecol. Syst. 26:683-704. [Google Scholar]
- 30.Longton, R. E. 1988. Biology of polar bryophytes and lichens. Cambridge University Press, Cambridge, Mass.
- 31.Lopez-Garcia, P., F. Rodriguez-Valera, C. Pedros-Alio, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 32.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall, W. A. 1996. Biological particles over Antarctica. Nature 383:680. [Google Scholar]
- 34.Maslen, N. R. 1979. Additions to the nematode fauna of the Antarctic region with keys to taxa. Br. Antarct. Surv. Bull. 49:207-230. [Google Scholar]
- 35.Massana, R., L. Guillou, B. Diez, and C. Pedros-Alio. 2002. Unveiling the organisms behind novel eukaryotic ribosomal DNA sequences from the ocean. Appl. Environ. Microbiol. 68:4554-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCaig, A. E., L. A. Glover, and J. I. Prosser. 1999. Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl. Environ. Microbiol. 65:1721-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 38.Moyer, C. L., F. C. Dobbs, and D. M. Karl. 1994. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S ribosomal RNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl. Environ. Microbiol. 60:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray, A. E., K. Y. Wu, C. L. Moyer, D. M. Karl, and E. F. DeLong. 1999. Evidence for circumpolar distribution of planktonic Archaea in the Southern Ocean. Aquat. Microb. Ecol. 18:263-273. [Google Scholar]
- 40.Ochsenreiter, T., F. Pfeifer, and C. Schleper. 2002. Diversity of Archaea in hypersaline environments characterized by molecular-phylogenetic and cultivation studies. Extremophiles 6:267-274. [DOI] [PubMed] [Google Scholar]
- 41.Øvstedal, D. O., and R. I. L. Smith. 2001. Lichens of Antarctica and South Georgia. Cambridge University Press, Cambridge, United Kingdom.
- 42.Pugh, P. J. A. 1993. A synonymic catalogue of the Acari from Antarctica, the sub-Antarctic islands and the Southern Ocean. J. Nat. Hist. 27:323-421. [Google Scholar]
- 43.Qiu, X. Y., L. Y. Wu, H. S. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Z. Zhou. 2001. Evaluation of PCR-generated chimeras: mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roser, D. J., R. D. Seppelt, and N. Ashbolt. 1993. Microbiology of ornithogenic soils from the Windmill Islands, Budd Coast, Continental Antarctica—microbial biomass distribution. Soil Biol. Biochem. 25:165-175. [Google Scholar]
- 45.Schlicting, H. E., B. J. Speziale, and R. M. Zink. 1978. Dispersal of algae and protozoa by Antarctic flying birds. Antarct. J. USA 13:147-149. [Google Scholar]
- 46.Sekiguchi, H., M. Watanabe, T. Nakahara, B. H. Xu, and H. Uchiyama. 2002. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 68:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, H. G. 1996. Diversity of Antarctic terrestrial protozoa. Biodivers. Conserv. 5:1379-1394. [Google Scholar]
- 48.Smith, R. I. L. 1984. Terrestrial plant biology of the sub-Antarctic and Antarctic, p. 61-162. In R. M. Laws (ed.), Antarctic ecology. Academic Press, London, United Kingdom.
- 49.Smith, Z., A. E. McCaig, J. R. Stephen, T. M. Embley, and J. I. Prosser. 2001. Species diversity of uncultured and cultured populations of soil and marine ammonia oxidizing bacteria. Microb. Ecol. 42:228-237. [DOI] [PubMed] [Google Scholar]
- 50.Staley, J. T., and J. J. Gosink. 1999. Poles apart: biodiversity and biogeography of sea ice bacteria. Annu. Rev. Microbiol. 53:189-215. [DOI] [PubMed] [Google Scholar]
- 51.Sugden, D. E., and C. M. Clapperton. 1977. The maximum ice extent on island groups in the Scotia Sea, Antarctica. Q. Res. 7:268-282. [Google Scholar]
- 52.Tong, S., N. Vors, and D. J. Patterson. 1997. Heterotrophic flagellates, centrohelid heliozoa and filose amoebae from marine and freshwater sites in the Antarctic. Polar Biol. 18:91-106. [Google Scholar]
- 53.van Hannen, E. J., W. Mooij, M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1999. Detritus-dependent development of the microbial community in an experimental system: qualitative analysis by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2478-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Hannen, E. J., M. P. van Agterveld, H. J. Gons, and H. J. Laanbroek. 1998. Revealing genetic diversity of eukaryotic microorganisms in aquatic environments by denaturing gradient gel electrophoresis. J. Phycol. 34:206-213. [Google Scholar]
- 55.Vincent, W. F. 2000. Evolutionary origins of Antarctic microbiota: invasion, selection and endemism. Antarct. Sci. 12:374-385. [Google Scholar]
- 56.Vincent, W. F. 1988. Microbial ecosystems of Antarctica. Cambridge University Press, Cambridge, United Kingdom.
- 57.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 58.Wagner, A., N. Blackstone, P. Cartwright, M. Dick, B. Misof, P. Snow, G. P. Wagner, J. Bartels, M. Murtha, and J. Pendleton. 1994. Surveys of gene families using polymerase chain-reaction-PCR selection and PCR drift. Syst. Biol. 43:250-261. [Google Scholar]
- 59.Wall, D. H., and R. A. Virginia. 1999. Controls on soil biodiversity: insights from extreme environments. Appl. Soil Ecol. 13:137-150. [Google Scholar]
- 60.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S ribosomal-RNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 61.Wynn-Williams, D. D. 1996. Antarctic microbial diversity: the basis of polar ecosystem processes. Biodivers. Conserv. 5:1271-1293. [Google Scholar]
- 62.Wynn-Williams, D. D. 1996. Response of pioneer soil microalgal colonists to environmental change in Antarctica. Microb. Ecol. 31:177-188. [DOI] [PubMed] [Google Scholar]
- 63.Zavarzin, G. A. 1993. An ecological approach to the systematics of prokaryotes, p. 555-558. In R. Guerrero and C. Pedros-Alio (ed.), Trends in microbial ecology. Spanish Society for Microbiology, Madrid, Spain.
- 64.Zervakis, G. I., J. M. Moncalvo, and R. Vilgalys. 2004. Molecular phylogeny, biogeography and speciation of the mushroom species Pleurotus cystidiosus and allied taxa. Microbiology 150:715-726. [DOI] [PubMed] [Google Scholar]
- 65.Zettler, L. A. A., F. Gomez, E. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Eukaryotic diversity in Spain's River of Fire—this ancient and hostile ecosystem hosts a surprising variety of microbial organisms. Nature 417:137. [DOI] [PubMed] [Google Scholar]
- 66.Zhou, J., M. E. Davey, J. B. Figueras, E. M. Rivkina, D. A. Gilichinsky, and J. M. Tiedje. 1997. Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913-3919. [DOI] [PubMed] [Google Scholar]