Abstract
Acid mine drainage (AMD) microbial communities contain microbial eukaryotes (both fungi and protists) that confer a biofilm structure and impact the abundance of bacteria and archaea and the community composition via grazing and other mechanisms. Since prokaryotes impact iron oxidation rates and thus regulate AMD generation rates, it is important to analyze the fungal and protistan populations. We utilized 18S rRNA and beta-tubulin gene phylogenies and fluorescent rRNA-specific probes to characterize the eukaryotic diversity and distribution in extremely acidic (pHs 0.8 to 1.38), warm (30 to 50°C), metal-rich (up to 269 mM Fe2+, 16.8 mM Zn, 8.5 mM As, and 4.1 mM Cu) AMD solutions from the Richmond Mine at Iron Mountain, Calif. A Rhodophyta (red algae) lineage and organisms from the Vahlkampfiidae family were identified. The fungal 18S rRNA and tubulin gene sequences formed two distinct phylogenetic groups associated with the classes Dothideomycetes and Eurotiomycetes. Three fungal isolates that were closely related to the Dothideomycetes clones were obtained. We suggest the name “Acidomyces richmondensis” for these isolates. Since these ascomycete fungi were morphologically indistinguishable, rRNA-specific oligonucleotide probes were designed to target the Dothideomycetes and Eurotiomycetes via fluorescent in situ hybridization (FISH). FISH analyses indicated that Eurotiomycetes are generally more abundant than Dothideomycetes in all of the seven locations studied within the Richmond Mine system. This is the first study to combine the culture-independent detection of fungi with in situ detection and a demonstration of activity in an acidic environment. The results expand our understanding of the subsurface AMD microbial community structure.
The Richmond Mine at Iron Mountain, which is near the city of Redding in northern California, is a subsurface mine that contains chemolithotrophic microbial communities that are sustained by energy derived from pyrite (FeS2) oxidation (4). The dissolution of pyrite from the Richmond Mine ore body yields warm (35 to 57°C), extremely acidic (typically pHs 0.5 to 0.9), metal-rich (for a review, see reference 4) fluids referred to as acid mine drainage (AMD). At the Richmond Mine it is possible to access, via mining tunnels, several environments that harbor these extremely acidophilic communities.
Microbial eukaryotes likely play critical, but as yet only partially determined, roles in AMD communities. Fungal hyphae comprise a significant but variable portion of the total biomass in many biofilm communities in the Richmond Mine, particularly in those growing in flowing solutions. The hyphae may contribute to the anchoring of biofilms to pyrite sediments and may confer structure, especially to “slime streamers” (4). Relatively large fungal filaments also provide surfaces for the attachment of prokaryotes (see Fig. 3C in reference 4). In addition, they keep organic carbon levels low and produce dissolved carbonate ions, which are likely important for the growth of chemolithoautotrophic acidophilic prokaryotes.
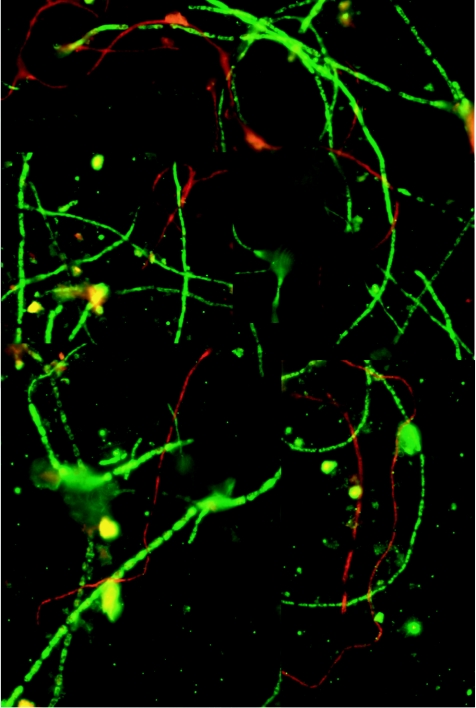
FIG. 3.
FISH analyses of five-way (August 2002) community, using Doh299 (Cy3 labeled, highlighting IM group II [Dothideomycetes] in red), and Eur1108 (fluorescein isothiocyanate labeled, highlighting IM group I [Eurotiomycetes] in green) rRNA probes. The image is a collage of four images to the same scale, and the filaments are all <5 μm thick. There is significant nonspecific fluorescence in these images from the minerals (rounded objects) in the sample. Note that the Eurotiomycetes (in green) are branched in several places and that the Eurotiomycetes are more abundant than the Dothideomycetes.
There have been few prior reports of eukaryotes in AMD (14, 18, 25, 34, 36, 38). Two extremely acidophilic (pH, <1) and metal-tolerant mitosporic fungi, Scytalidium acidophilium and Acontium velatum, were isolated by Starkey and Waksman (38). S. acidophilium is capable of growth down to pH 0 (18). The taxonomic characterization of these isolates was based on their morphology. McGinness and Johnson (34) and Johnson and Rang (25) cultured unidentified protists from an AMD site and showed that they were able to graze on acidophilic bacteria in culture.
Recently, a culture-independent study of eukaryotes of the AMD-impacted Tinto River in Spain identified small subunit rRNA (18S rRNA) gene sequences related to the Ascomycota phylum and a variety of protists (3). The Tinto River has a mean pH of 2.3, its temperature is <30°C, and it contains up to 41.2 mM Fe2+, 3.4 mM Zn, and 1.7 mM Cu metals (9, 29). The habitats at this site have higher pHs, are cooler, and are less enriched in metals than the environments examined in the present study.
Because high nucleotide substitution rates within the crown eukaryotes make it difficult to resolve variations at the species level, particularly in fungi (26), recent taxonomic studies have relied upon multigene phylogenies (6). The alpha-, beta-, and gamma-tubulin genes encode proteins whose heterodimers form the primary constituents of microtubules and other ultrastructural components in eukaryotes. Tubulin genes (particularly those for alpha- and beta-tubulin) are highly conserved, are ubiquitous in eukaryotes, have a significant representation in public databases, and thus have been shown to be reliable phylogenetic markers for the determination of evolutionary relationships among diverse eukaryotic groups (6, 27, 28).
For this study, we utilized both beta-tubulin and 18S rRNA gene sequence phylogenetic analyses to characterize the diversity of filamentous fungi and protists in the Richmond Mine at Iron Mountain, Calif. We also developed 18S rRNA probes to visualize the fungal filaments.
MATERIALS AND METHODS
Geochemical analyses.
Temperatures, pHs, and conductivities were measured directly in the mine with electrodes, as described elsewhere (14). Samples were collected in sterile 60-ml plastic syringes, filtered through Acrodisc (0.45- or 0.2-μm pore size) syringe filters, and stored on ice in sterile 15-ml Falcon tubes for transport back to the laboratory. Sulfate was analyzed by ion-exchange chromatography. Ferric and total iron concentrations were measured immediately after sample collection by the Ferrozine (39) and 1,10-phenanthroline (40) methods (HACH DR/2010 spectrophotometer methods 8147 and 8146). Other metal concentrations were determined via ion-exchange chromatography (12).
18S rRNA cloning and sequencing.
Environmental samples from the Richmond Mine were aseptically collected in October 1999 and on 19 January 2001 for the construction of two libraries. Samples were stored and transported back on wet ice to the laboratory, where they were frozen in 25% glycerol and stored at −20 and −80°C. After thawing, the samples were washed twice in phosphate-buffered saline or Tris-EDTA (TE) buffer to remove extracellular ions. DNAs were extracted from 0.75-ml aliquots of the samples by a modified phenol-chloroform extraction method as described by Bond et al. (8). DNA extraction products from the October 1999 sample were purified through Clontech Chroma Spin+TE-1000 columns. Several PCR amplifications of 18S rRNA genes were done by use of a variety of previously published primer combinations (82FE [5′-GAADCTGYGAAYGGCTC-3′], 378FE [5′-CGGAGARGGMGCMTGAGA-3′], 1391RE [5′-GGGCGGTGTGTACAARGRG-3′], and 1492RE [5′-ACCTTGTTACGRCTT-3′] [10]; EK-1F, EK-82F, and EK-1520R [30]; 28Fe [5′-TGGTTGATCCTGCCAGTAG-3′] and 514FU [5′-GTGCCAGCMGCCGCGG-3′] [13], and two primers designed for this study, EUKb518F [5′-GAGGRCMAGTCTGGTGC-3′] and EUKb1193R [5′-GGGCATMACDGACCTGTT-3′]) in an attempt to comprehensively amplify both the fungal and protistan populations. Most rRNA gene libraries were dominated by fungal or archaeal sequences. The library from the January 2001 sample reported here was constructed with primers 82FE, 378FE, 1391RE, and 1492RE and with AmpliTaq Gold (Perkin-Elmer) polymerase in a reaction mixture as specified previously (10). Eight replicate reactions were incubated on a gradient thermocycler at 94°C for 12 min, followed by 35 cycles of 94°C for 1 min, annealing at 45 to 65°C for 1 min, and extension at 72°C for 2 min, and then completed with a final 72°C extension for 10 min. Purified environmental DNAs from the October 1999 A drift upper sample were amplified with primers 28Fe and 1492RU; the reaction mixture and temperature conditions were different from those used for the January 2001 samples in the use of a 51°C annealing temperature and the addition of formamide to a concentration of 1.5% to increase the primer specificity (35). The PCR products were purified with a Qiagen Qiaquick PCR purification kit and were cloned by use of a Promega pGEM-T Vector System II cloning kit. The clones were PCR screened for inserts by use of the vector priming sites T7 and SP6. Amplification products from the 18 clones with inserts larger than 1.5 kb were digested with Msp1 and HinP1 for restriction fragment length polymorphism (RFLP) analysis, and eight novel phylotypes were identified for sequencing. Approximately 200 clones from the January 2001 library were analyzed via RFLP analysis. Plasmids from the January 2001 library were isolated from the clones with a Qiaprep Spin miniprep kit (Qiagen). End sequencing of the clones was done both on PCR products produced with T7 and SP6 primers and on the isolated plasmids. Nearly complete sequences of both strands were obtained by using combinations of broadly conserved primers (i.e., 28Fe, 514FU, 378FE, 515F, and 1492RU).
Beta-tubulin gene cloning and sequencing.
DNAs extracted from biofilm samples from the Richmond Mine on 19 January 2001 (see above) were used for the construction of a beta-tubulin library. The beta-tubulin genes were amplified by use of the primers BT107F (3′-AACAACTGGGCIAAGGTYACTACAC-5′) and BT261R (5′-ATGAAGAAGTGGAGICGIGGGAA-3′) under the same gradient PCR conditions described above. Screening and sequencing methods were performed as described above for the amplification of 18S rRNA genes.
Sequence analyses.
18S rRNA and tubulin gene sequences were compared to sequences available in the National Center for Biotechnology Information (NCBI) public database by BLAST searching (1). The rRNA genes from this study, as well as closely related gene sequences from the NCBI database, were imported into the ARB software package (32). One thousand beta-tubulin protein sequences from GenBank for various eukaryotic groups were used to create a beta-tubulin alignment with CLUSTALW (within ARB). The environmental beta-tubulin gene sequences were translated and aligned after the insertions were removed. Alignments were prepared in ARB by using the fast alignment tool, with subsequent manual refinements based on considerations of rRNA secondary structures. Ninety-six representative rRNA sequences incorporating 1,560 unambiguously homologous nucleotide positions were used for phylogenetic inference. One hundred sixty-eight homologous beta-tubulin amino acid positions were used for subsequent phylogenetic analyses. Initially, several tubulin trees were generated to determine the relative positions of the clones and to refine the large database for finer-scale phylogenetic analyses. Tree topologies from various phylogenetic inference techniques, including distance, quartet puzzling, maximum parsimony, and maximum likelihood (ML), were compared. The trees presented in Fig. 1 and 2 were generated with FastDNAMl, using the Felsenstein model (17), and with Protein_ML in ARB. The beta-tubulin amino acid trees were generated by using a Dayhoff substitution model.
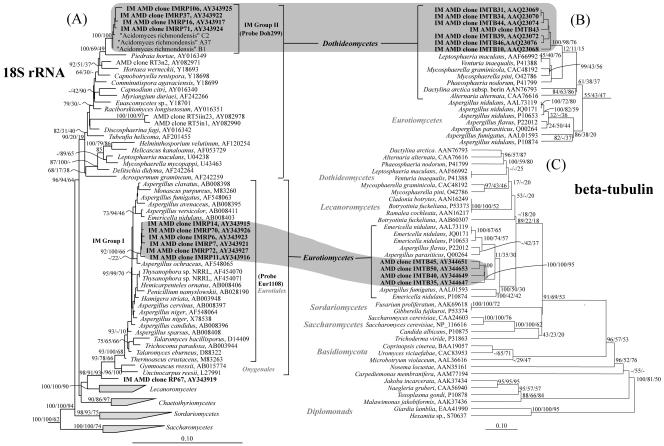
FIG. 1.
Phylogenetic correlations of AMD fungal 18S rRNA gene and beta-tubulin sequences belonging to the phylum Ascomycota. (A) Unrooted tree generated from a DNA alignment by the FastDNAMl ML method (22), with the Zygomycota and Basidiomycota fungal phyla as outgroups (not shown). The RT clones are from Rio Tinto AMD (8). Phylogenetic analyses of beta-tubulin protein sequences (generated with Protein_ML in ARB) associated with the Dothideomycetes (B) and Eurotiomycetes (C) classes from the mine are also shown. Tree B is a subset of taxa from tree C and was generated with the same sequences. In both B and C, the diplomonads Giardia intestinalis and Hexamita inflata were used as outgroups. Correlations between environmental sequences of the two molecules are shaded, and their classes are shown with brackets. Bayesian posterior probabilities, distance bootstrap values, and percentages of occurrence in the quartet-puzzling tree (in that order) are given at the corresponding nodes. The specificities of 18S rRNA probes are shown with brackets. Bar, 0.1 changes per site or 10% sequence difference.
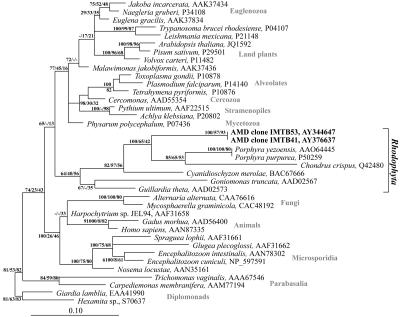
FIG. 2.
Phylogeny (ML) of protozoan beta-tubulin proteins, including the IMTB41 and IMTB53 (in bold) sequences from the Richmond Mine. The taxonomic affiliations of the reference sequences are labeled on the right. The mine clones were statistically supported to be within the Rhodophyta, or red algae, group. Bayesian posterior probabilities, distance bootstrap values, and percentages of occurrence in the quartet-puzzling tree (in that order) are shown at the corresponding nodes. The diplomonads Giardia lamblia and Hexamita were used as outgroups. Bar, 0.1 changes per site.
Bayesian posterior probabilities were approximated by using the Bayes 3.0 software package (22) and Markov chain Monte Carlo analysis (with a gamma rate correction). Based on comparisons of the means, variance values, and credible intervals for all generations, the first 25,000 generations (burn in) were removed. The chains were sampled every 1,000 generations, and inferences from each run were based on a total of 100,000 sampled trees. A majority rule consensus phylogeny was then generated with the PAUP* 4.0 software package. Bayesian inference has been shown to overestimate confidence values (41), and therefore distance and parsimony bootstrap values were also generated. Alternative topology full heuristic searches used 100 bootstrap replicates of random addition with a branch-swapping algorithm, tree bisection-reconnection. Quartet-puzzling trees were generated in PAUP* 4.0 with default parameters.
Whole-cell rRNA FISH analysis and probe design.
Samples were fixed in 4% paraformaldehyde on-site (at the time of collection), washed with phosphate-buffered saline, pH 1.2, and stored at −20°C within 8 h of collection. Oligonucleotide probes were designed to target the fungal sequences according to the methods reviewed by Hugenholtz et al. (24). Probe selection took into consideration the accessibility of the target region, as reported for Saccharomyces cerevisiae (7). Hybridizations were performed on fixed samples as previously reported, with incubation at 46°C and washing at 48°C for 15 min (8). Hybridizations were also counterstained with DAPI (4′,6′-diamidino-2-phenylindole dihydrochloride) DNA stain to estimate the total cell number. The optimal stringency for the probes was determined empirically by using fixed AMD samples and 5% formamide increments from 10 to 50%. For FISH analyses, S. cerevisiae, Morchella esculenta, and Debaryomyces hansenii var. hansenii were used as negative controls, and isolates were used as positive controls.
Fungal cultivation.
For the growth and isolation of fungi, a basal medium containing 0.8 g of (NH4)2SO4/liter, 0.4 g of KH2PO4/liter, 0.16 g of MgSO4 · 7H2O/liter, and trace salt solution was pH adjusted to 1.2 with H2SO4. This medium was supplemented with 0.2 g of yeast extract/liter and 6.6 mM glucose as carbon sources. Ampicillin was added to a concentration of 200 mg/liter to select for eukaryotes. Dilution series were used to obtain isolates A37, B1, and C2. The temperature of the mine fluctuates seasonally (15); therefore, all cultures were incubated at 37°C, which was roughly the mean of previous field measurements. Standard phenol-chloroform extraction (8) was done with 1.5-ml aliquots of the cultures lysed by either 2 min of bead beating, modified from a previously described method (8), or incubation with Zymolyase (modified from a previously described method [20]) followed by a freeze-thaw treatment (3). DNA extracts were purified through Clontech Chroma Spin+TE-1000 columns and were amplified with the 18S rRNA gene-specific primers 28Fe and 1492RU. Amplification products were purified by use of a Qiagen Qiaquick PCR purification kit. Purified amplification products were sequenced by using primers 28Fe, 515F, and 1492RU.
Nucleotide sequence accession numbers.
Completely sequenced clones were deposited in the NCBI GenBank under the following accession numbers: 18S rRNA sequences, AY343915 through AY343927, AY394431, and AY374298 through AY374306; and tubulin sequences, AY376637, AY376638, and AY344644 through AY344653. The oligonucleotide probe sequences (see Table 2) were deposited at probeBase (31).
TABLE 2.
FISH rRNA probes developed and used for this study
| Probe namea | Probe sequence (5′-3′) | Length (bp) | Tm (°C)b | GC% | Optimal amt (%) of formamide | Target |
|---|---|---|---|---|---|---|
| Doh299 | TCTCTCCGGTATCGTACCC | 19 | 58 | 58 | 10-25 | IM group II |
| Oil845 | TCGTACGGTGCCGATGGA | 18 | 61 | 65 | 20 | IM group II |
| Eur1108 | TTTAAGGGCCGAGGTCTC | 18 | 58 | 56 | 10 | Eurotiomycetes |
Probes were named according to their positions in Escherichia coli.
By nearest neighbor calculation (http://paris.chem.yale.edu/extinct.html) using 50 mM NaCl and 50 μM oligonucleotide.
RESULTS
Geochemistry of sample collection sites.
Table 1 lists the temperatures, pHs, and concentrations of some of the more prevalent ions in solution at the sample collection sites within the Richmond Mine (12). The A drift sites Red Pool and A Slump are both peripheral to the main drainage channels and are characterized by anomalously high pHs (>1.0). Red Pool is a remnant from a higher flow period, and A Slump is a pyrite accumulation associated with a sidewall mine opening. The solution in the stagnant Red Pool contains less Fe2+, Cu2+, and Zn2+ than the main drainage due to the oxidation of iron and the precipitation of reddish ferric iron oxyhydroxides that sorb metals (12). The A, B, and C drifts are tunnels in the actively dissolving Richmond ore body. The pH 0.8 solutions flowing along the A and B drifts have metal concentrations as follows: >261 mM Fe2+, 8.5 mM As, >2.2 mM Cu, and >14 mM Zn (12).
TABLE 1.
Physical conditions and solute concentrations of the Richmond Mine samples used for this studya
| Site | Temp (°C) | pH | Concn (mM) of solute
|
|||||
|---|---|---|---|---|---|---|---|---|
| Fe2+ | Fe3+ | SO42− | As | Cu | Zn | |||
| A drift | 42 | 0.83 | 269 | 50 | 665 | 8.5 | 4.1 | 14 |
| Red Pool (January 2001) | 30 | 1.38 | 10 | 25 | 142 | 0.1 | 0.3 | 2.7 |
| A Slump (June 2000) | 32 | 1.10 | 26 | 32 | 149 | 1.0 | 0.9 | 1.3 |
| B drift | 47 | 0.78 | 261 | 13 | 550 | NDb | 2.2 | 16.8 |
| C drift | 50 | 0.76 | 281 | 7 | 892 | 10.3 | 4.7 | 14.3 |
All measurements were from March 2002, except as indicated. Adapted with permission from Druschel et al. (12).
ND, not determined.
Phylogenetic analyses of AMD 18S rRNA genes.
All of the fungal sequences fell into three distinct phylogenetic clusters (Fig. 1A) within the phylum Ascomycota. Clones with IMRP designations are from the January 2001 library, and all others are from the October 1999 library. Ten clones grouped into Iron Mountain (IM) group I, and six of them were completely sequenced (IM AMD clones IMRP6, IMRP7, IMRP11, IMRP14, IMRP70, and IMRP72) and found to have >99% sequence similarity to one another. IM group II contains 17 clones, 7 of which were completely sequenced (IM AMD clones OPB24, IMRP16, IMRP37, IMRP56, IMRP61, IMRP63, IMRP71, and IMRP106) and found to have >99% sequence similarity to one another. IMRP67 represents a separate lineage that is basal to the Dothideomycetes class.
IM group I is in the fungal class Eurotiomycetes and the order Eurotiales. Its position in that class is strongly supported by multiple inference and statistical analyses (Fig. 1A) and by trees generated by several different inference methods. ML phylogenetic analyses suggested, with quite loose supporting bootstrap values, that this group shares a common ancestor with Aspergillus ochraceus. However, the bootstrap value was too low (22%) to say with certainty that this is its nearest neighbor. The phylogenetic association of group II with the class Dothideomycetes was statistically supported by a Bayesian inference of posterior probabilities and bootstrapping. Although some of the basal nodes to this class had little statistical support, this group was retained in several trees generated by using all inferences and variable substitution models. Trees generated by ML methods positioned it closest (≥96.7% sequence similarity) to Piedraia hortae (Fig. 1A). Clone IMRP67 also fell just outside of the Dothideomycetes class (Fig. 1). This position basal to that class was strongly supported by statistical analyses and the various trees generated. Based on these analyses, this clone may belong in a novel fungal class. Surprisingly, this clone's closest BLAST match is Rio Tinto RT3n2, with which it has 94% sequence similarity.
In addition to the fungal clones, two protistan clones were sequenced from the October 1999 library. A phylogenetic analysis of these clones revealed that they group within the Heterolobosea, of the order Schizopyrenida and the family Vahlkampfiidae, and have 94% similarity to Vahlkampfia lobospinosa. Several identical clone types to these were obtained from other libraries (not included in the analysis).
Analyses of beta-tubulin sequences and comparison to 18S rRNA gene phylogenies.
When compared to sequences in the NCBI database, approximately half of the beta-tubulin clones (including IM AMD clones IMTB35, IMTB40, IMTB45, and IMTB50) were closest to Aspergillus nidulans (81% sequence similarity), one of the Eurotiomycetes, and the others (IM AMD clones IMTB10, IMTB31, IMTB34, IMTB39, IMTB44, and IMTB46) had an 80% sequence similarity to Mycosphaerella pini, one of the Dothideomycetes.
The DNA sequences were aligned and translated to check the integrity of the alignments. This analysis revealed an insertion in four of the clones (IMTB35, IMTB40, IMTB45, and IMTB50). The insertions were 55 bp long (positions 329 to 384 in the clones) and were identical in all cases. This intron was in the same position in the gene as an insertion found in both Aspergillus nidulans (33) and Penicillium digitatum (33) and was nearly the same length (within 10 bp). However, there is no significant sequence similarity between the AMD clone introns and those reported for Aspergillus nidulans and Penicillium digitatum.
A phylogenetic analysis of all of the beta-tubulin sequences generated a tree with potential long-branch attraction artifacts and resulted in the grouping of the two clusters deep within the fungi. This placement was inconsistent with the 18S rRNA gene phylogeny. Thus, the sequences were subdivided based on these clusters, and trees were generated with identical alignments and reference organisms, using several inference methods (distance, parsimony, and ML). The results placed one clonal cluster into the Eurotiomycetes and the other into the Dothideomycetes. The ML trees are presented in Fig. 1B and C. Although these classes are only supported by a few statistical analyses, this topology was consistent in several trees generated by distance, ML, maximum parsimony, and quartet-puzzling methods.
Based on the relatedness of the Eurotiomycetes tubulin sequences (Fig. 1C) to Aspergillus sequences, we inferred that they share a common ancestor with Aspergillus (IM group I in Fig. 1). The 18S rRNA gene phylogeny of the Dothideomycetes lineage organisms was closest to that of Piedraia hortae (group II in Fig. 1A). The limited number of beta-tubulin sequences currently available for the Dothideomycetes and the lack of a Piedraia hortae sequence made it impossible to resolve their relationship in the tubulin phylogeny.
Two novel beta-tubulin protistan clones (IMTB41 and IMTB53) were fully sequenced. Their closest match (75% identity and an e value of 4 × 10−71) in GenBank was Cyanidioschyzon merolae, which is an acidophile. Multiple phylogenetic inferences placed these clones within the Rhodophyta (red algae) (Fig. 2). We presume that these sequences are from a heterotrophic acidophilic red alga, since other Rhodophyta organisms have been isolated from acidic environments (19).
Cultivation of fungi.
Three strains of fungi, A37, B1, and C2, were isolated from the A, B, and C drifts, respectively, by cultivation at 37°C. All three (tentatively named here “Acidomyces richmondensis” [see discussion]) were phylogenetically associated with IM group II (Fig. 1). Their placement within the Dothideomycetes was strongly supported by posterior probabilities of >80%. The isolates, like those in group II (see below), had ≥96.7% 18S rRNA gene sequence similarity to Piedraia hortae (Fig. 1A).
Microscopic characterization.
Fungal filaments have been noted in most biofilm samples collected from the Richmond Mine. In some cases, the hyphae constitute over 20% of the metabolically active cells that are visible by DNA and rRNA staining.
We designed rRNA-targeted oligonucleotide probes to confirm the presence and to estimate the relative abundance of organisms detected in the 18S rRNA libraries. Probes Doh299 and Oil845 were designed to target group II organisms only. Attempts to design a probe to target group organisms I only were unsuccessful due to the low sequence divergence within this group. Probe Eur1108 is a broader class-level probe for the Eurotiomycetes, including group I. A probe to solely target clone IMRP67 was also designed. The sequences, Tms, lengths, GC% values, and optimal stringencies of the probes are provided in Table 2.
The Eur1108 probe and the Oil845 and Doh299 probes were tested and found to be effective for visualizing groups I and II, respectively (Fig. 3). However, Doh299 also weakly bound to some protists. Given the clear morphological difference between the egg-shaped protists and the fungal hyphae, the probe can be used to evaluate the distribution of Dothideomycetes in these samples. Furthermore, probe Oil845 did not appear to have any nonspecific binding and targeted the same group effectively. Since the Oil845 and Doh299 probes targeted different regions of the gene, they could be used in concert to increase the brightness of the filaments, making them easier to visualize. Both Oil845 and Doh299 worked on the “Acidomyces richmondensis” isolates. Under identical hybridization conditions, none of these three probes hybridized significantly to the negative control, S. cerevisiae. The Eurotiomycetes were more abundant in all of the samples analyzed. Both types of fungal filaments were more abundant in the streamer biofilms in flowing AMD than in pools where the water was stagnant (Table 3). The probe designed for IMRP67 did not hybridize to filaments in any sample.
TABLE 3.
Relative abundance of the two fungal groups in several samples from the mine on 12 March and 18 September 2002a
| Mine site (date of sampling) | Abundance of fungal group
|
|
|---|---|---|
| Eurotiomycetes | Dothideomycetes | |
| A drift | ||
| Ten-foot streamers (September 2002) | +++ | − |
| Pool (March 2003) | − | − |
| Slime pool (September 2002) | − | − |
| Slump (March 2003) | ++ | + |
| Five-way streamers (September 2002) | +++ | + |
| B drift | ||
| Back (March 2003) | ++ | − |
| Back streamer (September 2002) | + | ++ |
The amount of filaments was estimated visually, with +++ being the largest amount and − indicating that none (or very few) were identified. The A drift 10-ft. streamer sample was collected ∼10 ft. downstream from the A drift weir (14).
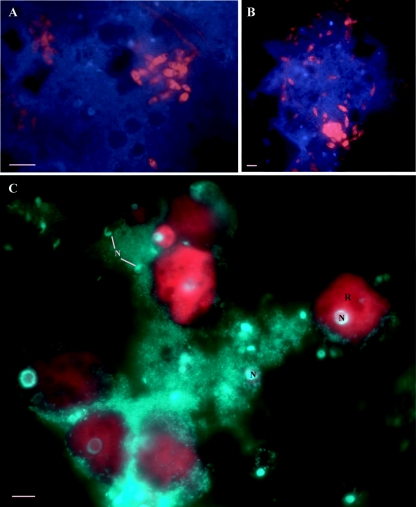
A variety of metabolically active morphologically distinct protists are shown in Fig. 4 (Eukb-mix probe); a subset contains bacterial endosymbionts (5). The most common shapes seen were ovoid (2 to 5 μm long) and larger spherical cells (<20 μm in diameter). Occasionally, groups of oblong (2 to 5 μm long) and amoeboid (<15 μm long) forms were observed.
FIG. 4.
Micrographs of protists from the Richmond Mine imaged with eukaryotic domain-level rRNA FISH probes. (A and B) The EUKb-mix probes (32) were labeled with Cy3 (red), and the DNAs were labeled with DAPI (blue). (C) The Euk502 probe labeled with Cy3 and DAPI is shown in green. Note that some of the protists were not targeted by this probe. N, nuclei of the eukaryotes; R, cytoplasmic staining of FISH probes corresponds to the cellular location of rRNA. Bars, ∼5 μm.
DISCUSSION
Although the power of the combination of culture-independent clone library analyses and FISH probe-based studies of prokaryotic populations in environmental samples is well established (11, 12, 23, 37), the method has not been adopted previously to study natural communities of fungi. For this paper, we used clone library analyses to design oligonucleotide probes to identify and study the distribution of microbial eukaryotes (both protists and fungi) in biofilms growing in pH 0.8 to 1.4, 24 to 50°C, subsurface, metal-contaminated AMD solutions. The FISH results confirmed that the Dothideomycetes and Eurotiomycetes fungal groups detected in the 18S rRNA gene and beta-tubulin libraries thrive in solutions with pHs of <1. Based on these genes, these taxa are phylogenetically closely related to neutrophilic lineages.
The discovery of lineages that are closely related to neutrophilic eukaryotes in extremely acidic environments is consistent with the results of Amaral Zettler et al. (3), who studied the pH 2, AMD-impacted Rio Tinto in Spain (2). Findings based only on an 18S rRNA gene library (e.g., not confirmed by whole-cell rRNA FISH) must be interpreted with caution because of the possibility of PCR contamination. The data presented here confirm the acid adaptation of some of the lineages detected by Amaral Zettler et al. and extend the ranges of these taxa to much lower pH (0.8) conditions.
Three clones from the Rio Tinto AMD site were linked to the Dothideomycetes (3). More recently, an acid-tolerant organism belonging to this class was isolated and named Hortaea acidophila (21). In addition to demonstrating the presence of Dothideomycetes (IM group II; Fig. 1) in environmental samples via FISH (Fig. 4), we were able to isolate representatives from the Richmond Mine. Although the IM group II strains clustered with Dothideomycetes, their lineages were distinct from those of previously isolated strains. On the basis of the 18S rRNA gene and beta-tubulin sequence divergence and the adaptation to extremes of the AMD habitat, we suggest a new genus and species with the name Acidomyces richmondensis (a.cid′myc.es rich.mon.den′sis. L. neut. N. acidum, acid; N.L. masc. adj. richmondensis, pertaining to Richmond Mine near Redding, Calif., where the species was first isolated). A preliminary description for this genus and species is as follows: member of the phylum Ascomycota, subphylum Pezizomycotina, and class Dothideomycetes, based on 18S rRNA gene sequences (accession numbers AY374298, AY374299, and AY374300) and hybridization with the oligonucleotide probe Doh299 (5′-CCTCTCCGGTATCGTACCC-3′); cultivated on cell-free medium, with branched filaments varying in diameter from 3 to 15 μm, acid-tolerant fungi native to an AMD.
Eurotiomycetes were detected via FISH, confirming that they are active in the AMD system. An Aspergillus sp. (strain P37), class Eurotiomycetes, isolated from an AMD site in Spain is capable of growth on acidic media containing ∼15,000 ppm of AsO43−, ∼3,100 ppm of Cu2+, and ∼2,600 ppm of Cr3+ (9).
In this study, we analyzed about 200 18S rRNA gene clones via RFLP and more than 30 via sequence-based phylogenetic analysis. The lack of deeply divergent fungal lineages in the AMD system is perhaps surprising when one considers the recent discovery of kingdom-level novelty in aquatic environments (10). There are two ways to interpret the close relatedness of these AMD lineages to neutrophilic lineages. Colonization may have occurred relatively recently so that rRNA genes have not had time to significantly diverge. Alternatively, these fungi may not be challenged by extreme acidity, high toxic metal concentrations, and moderately elevated temperatures (47°C), so there is no strong driving force for the evolution of specialized lineages to colonize AMD habitats. Since we know that other fungi found in acidic environments are capable of normal growth at a circumneutral pH, we believe that they have a selective advantage in surviving in acid that other closely related fungi do not (21).
The results indicated a low fungal diversity in the Richmond Mine biofilms. This finding parallels those of Bond et al. and Gross (8, 19) and of Druschel et al. (12), who demonstrated that the biofilms at this site are usually dominated by no more than about six 16S rRNA gene (prokaryote) phylotypes. Amaral Zettler et al. (2) reported a high eukaryal diversity in the AMD-impacted Rio Tinto. However, this river is colonized by phototrophic species, and only a few fungal lineages were detected. We attribute the difference between the two molecular biological studies of extremely acidic environments to the lack of phototrophic niches in the subsurface Richmond Mine system, not to its higher temperatures, lower pHs, and higher metal concentrations.
Only two protistan types were detected in the AMD system, despite many library construction attempts using various DNA extraction protocols and at least a dozen primer sets. However, morphological data (Fig. 4) indicated about five protistan types in the biofilms. New methods of DNA extraction, new primers, and/or the optimization of PCR conditions will be required to comprehensively characterize the AMD protistan community.
The only 18S rRNA gene sequences obtained from the protistan populations clustered with Vahlkampfia sequences. This clone type was found in both the October 1999 and January 2001 libraries. Furthermore, Vahlkampfia species were detected in the Rio Tinto study (3) and have been isolated from AMD (25). Thus, it is likely that Vahlkampfia species are metabolically active in the AMD biofilms. The only protistan clones detected via the analysis of beta-tubulin gene sequences were taxonomically associated with the Rhodophyta. We were not able to associate the clones with a previously identified acidophilic or heterotrophic red alga, which was likely due to the lack of representative beta-tubulin sequences available for this group.
Prior studies of the microbiology of AMD systems have focused almost exclusively on the prokaryotic populations because these are known to directly impact acid generation rates via the oxidation of iron and sulfur compounds (e.g., see references 7, 8, 12, and 16). Eukaryotes are almost ubiquitous in AMD biofilms, yet their identities and functions are very poorly understood. Protists such as Vahlkampfia impact prokaryote cell numbers (and thus oxidation rates and AMD generation rates) by grazing. They may also impact AMD biofilm membership when they graze selectively on certain community members (e.g., archaea). Fungal filaments, which were shown here to be Eurotiomycetes and Dothideomycetes, provide rigidity and organization to the biofilms and anchor them to pyrite sediment in flowing AMD solutions. Fungi also may impact the community structure and function by the consumption of organic waste products and the production of organic polymers, possibly including antibiotics.
Conclusions.
The development of 18S rRNA fungal probes allowed us to identify and study the distribution of lineages of morphologically indistinguishable fungi in environmental samples. We were able to phylogenetically correlate the beta-tubulin and 18S rRNA genes to indigenous phylotypes of the Eurotiomycetes and Dothideomycetes families belonging to the phylum Ascomycota. We obtained three isolates with 18S rRNA genes that are closely related to the Dothideomycetes clones. We suggest the name “Acidomyces richmondensis” for the isolates on the basis of their genus-level phylogenetic divergence and environmental adaptation. This study extends our understanding of the structure of AMD microbial communities and expands the range of geochemical adaptation for Vahlkampfiidae, Eurotiomycetes, Rhodophyta, and Dothideomycetes.
Acknowledgments
We thank Andreas Weber, Chuck Kaspar, and Philip Hugenholtz for their contributions to this work and Hans Trüper for advice on naming the isolates. We also thank Charles F. Wimpee, Gene W. Tyson, and John Taylor for helpful discussions. We thank Ted Arman, Iron Mountain Mines Ltd., for permission to work at the field site and Richard Sugarek, EPA, for facilitating access. We thank Don Dodds, North Pacific Research, and Rudy Carver for their assistance with field work.
This work was funded by the NSF Life in Extreme Environments (LExEn) and Biocomplexity Programs and by LDRD support from the Lawrence Berkeley National Laboratory.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaral Zettler, L. A., F. Gómez, E. R. Zettler, B. G. Keenan, R. Amils, and M. L. Sogin. 2002. Eukaryotic diversity in Spain's river of fire. Nature 417:137. [DOI] [PubMed] [Google Scholar]
- 3.Amaral Zettler, L. A., M. A. Messerli, A. D. Laatsch, P. J. S. Smith, and M. L. Sogin. 2003. From genes to genomes: beyond biodiversity in Spain's Rio Tinto. Biol. Bull. 204:205-209. [DOI] [PubMed] [Google Scholar]
- 4.Baker, B. J., and J. F. Banfield. 2003. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 44:139-152. [DOI] [PubMed] [Google Scholar]
- 5.Baker, B. J., P. Hugenholtz, S. C. Dawson, and J. F. Banfield. 2003. Extremely acidophilic protists host a Rickettsiales lineage with intervening sequence (IVS) in their 16S rRNA genes. Appl. Environ. Microbiol. 69:5512-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldauf, S. L., A. J. Roger, I. Wenk-Siefert, and W. F. Doolittle. 2000. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science 290:972-977. [DOI] [PubMed] [Google Scholar]
- 7.Behrens, S., C. Ruhland, J. Inacio, H. Huber, A. Fonseca, I. Spencer-Martins, B. M. Fuchs, and R. Amann. 2003. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probes. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bond, P. L., S. P. Smriga, and J. F. Banfield. 2000. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site. Appl. Environ. Microbiol. 66:3842-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cánovas, D., C. Durán, N. Rodriguez, R. Amils, and V. de Lorenzo. 2003. Testing the limits of biological tolerance to arsenic in a fungus isolated from the River Tinto. Environ. Microbiol. 5:133-138. [DOI] [PubMed] [Google Scholar]
- 10.Dawson, S. C., and N. R. Pace. 2002. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl. Acad. Sci. USA 99:8324-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains—ribosomal RNA-based for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 12.Druschel, G. K., B. J. Baker, T. H. Gihring, and J. F. Banfield. Biogeochemistry of acid mine drainage at Iron Mountain, California. Geochem. Trans. 5:13-32. [DOI] [PMC free article] [PubMed]
- 13.Edgcomb, V. P., A. J. Roger, A. G. B. Simpson, D. T. Kysela, and M. L. Sogin. 2001. Evolutionary relationships among “jakobid” flagellates as indicated by alpha- and beta-tubulin phylogenies. Mol. Biol. Evol. 18:514-522. [DOI] [PubMed] [Google Scholar]
- 14.Edwards, K. J., P. L. Bond, G. K. Druschel, M. M. McGuire, R. J. Hamers, and J. F. Banfield. 2000. Geochemical and biological aspects of sulfide mineral dissolution: lessons from Iron Mountain, California. Chem. Geol. 169:383-397. [Google Scholar]
- 15.Edwards, K. J., T. M. Gihring, and J. F. Banfield. 1999. Seasonal variations in microbial populations and environmental conditions in an extreme acid mine drainage environment. Appl. Environ. Microbiol. 65:3627-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards, K. J., B. M. Goebel, T. M. Rodgers, M. O. Schrenk, T. M. Gihring, M. M. Cardona, B. Hu, M. M. McGuire, R. J. Hamers, N. R. Pace, and J. F. Banfield. 1999. Geomicrobiology of pyrite (FeS2) dissolution: case study at Iron Mountain, California. Geomicrobiol. J. 16:155-179. [Google Scholar]
- 17.Felsenstein, J. 1993. Phylip (phylogeny inference package), version 3.5.1. Department of Genetics, University of Washington, Seattle.
- 18.Gould, W. D., J. I. Fujikawa, and F. D. Cook. 1974. A soil fungus tolerant to extreme acidity and high salt concentration. Can. J. Microbiol. 20:1023-1027. [DOI] [PubMed] [Google Scholar]
- 19.Gross, W. 2000. Ecophysiology of algae living in highly acidic environments. Hydrobiologia 433:31-37. [Google Scholar]
- 20.Guthrie, C., and G. R. Fink (ed.). 1991. Methods in enzymology, vol. 194. Guide to yeast genetics and molecular biology. Academic Press, Inc., San Diego, Calif. [PubMed]
- 21.Holker, U., J. Bend, R. Pracht, T. Muller, L. Tetsch, M. Hofer, and G. S. de Hoog. Hortaea acidophila, a new acid-tolerant black yeast from lignite. Antonie Leeuwenhoek, in press. [DOI] [PubMed]
- 22.Huelsenbeck, J. P., F. Ronquist, R. Nielsen, and J. P. Bollback. 2001. Evolution—Bayesian inference of phylogeny and its impact on evolutionary biology. Science 294:2310-2314. [DOI] [PubMed] [Google Scholar]
- 23.Hugenholtz, P., and N. R. Pace. 1996. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 14:190-197. [DOI] [PubMed] [Google Scholar]
- 24.Hugenholtz, P., G. W. Tyson, and L. L. Blackall. 2001. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridisation. Methods Mol. Biol. 179:29-42. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, D. B., and L. Rang. 1993. Effects of acidophilic protozoa on populations of metal-mobilizing bacteria during the leaching of pyritic coal. J. Gen. Microbiol. 139:1417-1423. [Google Scholar]
- 26.Kasuga, T., T. J. White, and J. W. Taylor. 2002. Estimation of nucleotide substitution rates in eurotiomycete fungi. Mol. Biol. Evol. 19:2318-2324. [DOI] [PubMed] [Google Scholar]
- 27.Katiyar, S. K., and T. D. Edlind. 1994. β-Tubulin genes of Trichomonas vaginalis. Mol. Biochem. Parasitol. 64:33-42. [DOI] [PubMed] [Google Scholar]
- 28.Keeling, P. J., and W. F. Doolittle. 1996. Alpha-tubulin from early eukaryotic lineages and the evolution of the tubulin family. Mol. Biol. Evol. 13:1297-1305. [DOI] [PubMed] [Google Scholar]
- 29.López-Archilla, A. I., and I. M. Amils. 2001. Microbial community composition and ecology of an acidic aquatic environment: the Tinto River, Spain. Microb. Ecol. 41:20-35. [DOI] [PubMed] [Google Scholar]
- 30.López-García, P., F. Rodríguez-Valera, C. Pedrós-Alió, and D. Moreria. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-606. [DOI] [PubMed] [Google Scholar]
- 31.Loy, A., M. Horn, and M. Wagner. 2003. ProbeBase—an online resource for rRNA-targeted oligonucleotide probes. Nucleic Acids Res. 31:514-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, W. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.May, G. S., M. L. Tsang, H. Smith, S. Fidel, and N. R. Morris. 1987. Aspergillus nidulans beta-tubulin genes are unusually divergent. Gene 55:231-243. [DOI] [PubMed] [Google Scholar]
- 34.McGinness, S., and D. B. Johnson 1992. Grazing of acidophilic bacteria by a flagellated protozoan. Microbiol. Ecol. 23:75-86. [DOI] [PubMed] [Google Scholar]
- 35.Sarkar, G., S. Kapelner, and S. S. Sommer. 1990. Formamide can drastically improve the specificity of PCR. Acids Res. 18:7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schleper, C., G. Puehler, I. Holz, A. Bambacorta, D. Janekovic, U. Santarius, H. P. Klenk, and W. Zillig. 1995. Picrophilus gen. nov., fam. nov.—a novel aerobic, heterotrophic, thermoacidophilic genus and family comprising archaea capable of growth around ph 0. J. Bacteriol. 177:7050-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schrenk, M. O., K. J. Edwards, R. M. Goodman, R. J. Hamers, and J. F. Banfield. 1998. Distribution of Thiobacillus ferrooxidans and Leptospirillum ferrooxideans: implications for generation of acid mine drainage. Science 279:1519-1522. [DOI] [PubMed] [Google Scholar]
- 38.Starkey, R. L., and S. A. Waksman. 1943. Fungi tolerant to extreme acidity and high concentrations of copper sulfate. J. Bacteriol. 45:509-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stookey, L. L. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42:779. [Google Scholar]
- 40.Stucki, J. W. 1981. The quantitative assay of minerals for Fe2+ and Fe3+ using 1,10-phenanthroline. II. A photochemical method. Soil Sci. Soc. Am. J. 45:638-641. [Google Scholar]
- 41.Suzuki, Y., G. V. Glazko, and M. Nei. 2002. Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc. Natl. Acad. Sci. USA 99:16138-16143. [DOI] [PMC free article] [PubMed] [Google Scholar]