Abstract
This study aimed to evaluate the biocontrol potential of bacteria isolated from different plant species and soils. The production of compounds related to phytopathogen biocontrol and/or promotion of plant growth in bacterial isolates was evaluated by measuring the production of antimicrobial compounds (ammonia and antibiosis) and hydrolytic enzymes (amylases, lipases, proteases, and chitinases) and phosphate solubilization. Of the 1219 bacterial isolates, 92% produced one or more of the eight compounds evaluated, but only 1% of the isolates produced all the compounds. Proteolytic activity was most frequently observed among the bacterial isolates. Among the compounds which often determine the success of biocontrol, 43% produced compounds which inhibit mycelial growth of Monilinia fructicola, but only 11% hydrolyzed chitin. Bacteria from different plant species (rhizosphere or phylloplane) exhibited differences in the ability to produce the compounds evaluated. Most bacterial isolates with biocontrol potential were isolated from rhizospheric soil. The most efficient bacteria (producing at least five compounds related to phytopathogen biocontrol and/or plant growth), 86 in total, were evaluated for their biocontrol potential by observing their ability to kill juvenile Mesocriconema xenoplax. Thus, we clearly observed that bacteria that produced more compounds related to phytopathogen biocontrol and/or plant growth had a higher efficacy for nematode biocontrol, which validated the selection strategy used.
Keywords: Rhizobacteria, Antibiosis, Mesocriconema xenoplax, Obligate parasites, Perennial plants
Introduction
Concerns regarding food safety and the environment have led to reduced use of agrochemicals and the development of sustainable agriculture. In this context, the focus of biological control studies reflects the desire of several sectors to develop sustainable methods for plant disease control.1 However, efficient antagonists must be obtained for biological control to become a reality.
Soil microorganisms coexist in association with plant roots and interfere with plant performance and microbial community structure. Bacteria are estimated to occupy between 7% and 15% of the total root surface area.2 Of these, some bacteria positively affect plants and have been designated as plant growth-promoting rhizobacteria (PGPR).3
Rhizobacteria can indirectly or directly promote positive effects on plants. Indirectly, they suppress pathogens mediated by competition and the production of antimicrobial compounds and lytic enzymes. Directly, they solubilize minerals and cause a wide range of changes in the rhizosphere, which promotes higher efficiency in the absorption of water and macro- and micronutrients by plants and changes in phytohormone concentrations, nitrogen fixation, and siderophore production.4, 5, 6
In addition to the rhizosphere, the phylloplane is a source of antagonists; however, bacteria from this area are still underused as biological control agents, especially compared to rhizobacteria.7 Kishore et al.,8 found that both rhizoplane and phylloplane bacteria promote peanut seedling growth. In addition, bacteria that colonize the shoots have a better chance of surviving and multiplying in a nutrition-rich environment such as the soil, whereas in the phylloplane, they are exposed to high temperatures, moisture content fluctuations, and limited nutrient availability.
In vivo biocontrol agent selection is not a simple task due to the diversity of agents and interactions with the host plant, and therefore, efficient search methods are required. Thus, it is necessary to develop efficient selection strategies to reduce costs and increase the possibility of selecting organisms that can be produced in a large scale at low cost and that maintain their viability and efficiency for long periods. In 1997, Schisler and Slininger9 divided the selection process into three categories: (i) choosing the appropriate pathosystem, (ii) choosing the adequate method, and (iii) characterizing the isolates and evaluating efficiency. In recent years, this concept has evolved, and other groups of researchers have proposed initial selection criteria based on evaluations in the absence of the host.10, 11
In this sense, in vitro tests are appropriate during the initial selection steps due to the large number of microorganisms that can be evaluated and, especially, their low cost. Thus, due to the need for alternative management strategies for difficult-to-control pathogens, and considering that the initial steps for biocontrol agent selection should be performed in the absence of the host, the present study aimed to (i) characterize bacteria to determine their in vitro potential for the production of compounds related to phytopathogen biocontrol and/or promotion of plant growth (CRBPGs); (ii) select bacterial isolates with the highest number of CRBPGs; and (iii) validate the selection process by studying the effect of the bacteria selected on the ringed peach nematode.
Material and methods
Origin of the bacteria
A total of 1219 bacteria that belonged to the collection of the Plant Bacteriology Laboratory (Laboratório de Bacteriologia Vegetal – LBV) at the Universidade Federal de Pelotas, Brazil, were used. The bacteria were obtained from different niches (phylloplane, rhizosphere, and soil) and grouped according to their isolation source: fig tree (Ficus carica L. – 55), Gramineae (72), Leguminoseae (151), Liliaceae (219), peach tree (Prunus persica L. – 297), Tagetes sp. (51), non-rhizospheric soil (309), and others (65 – tomato plant (Solanum lycopersicum L.), Brassicae, and culture medium contaminants).
Evaluation of CRBPG production
The abilities to produce antimicrobial compounds (antibiosis and ammonia production) and hydrolytic enzymes (amylases, lipases, proteases, and chitinases) and to solubilize phosphates were evaluated. Bacteria that previously exhibited or did not exhibit the ability to produce each compound studied were used as positive and negative controls, respectively.
The ability of bacteria to produce antibiotic compounds against Monilinia fructicola (Winter) Honey, the causal agent of peach brown rot was evaluated. Four bacteria arranged equidistant at the edges were streaked on each Petri dish containing 523 medium12 and in the center, a mycelial disk containing the fungus previously grown in a potato dextrose agar (PDA). After seven days at 22 ± 2 °C, scores were assigned according to the inhibition zone of the fungal growth as follows: 0 – no inhibition; 1 – ≤10 mm; 2 – ≥11 and ≤20 mm; and 3 – ≥21 mm).
Ammonia production was observed by the presence of a yellow-orange precipitate after five days of incubation at 28 °C.13
Starch hydrolysis was evaluated according to the method of Schaad (1988). After four days of incubation at 28 °C, the addition of Lugol's Iodine allowed the visualization of clear zones around the bacterial colony, indicative of starch hydrolysis, and the bacteria were classified using the scale described for antibiotic compounds.
Lipid hydrolysis was evaluated in medium containing 1% Tween 80, according to the method used by Fahy and Persley,14 after four and seven days of incubation at 28 °C and verified by the presence of a milky white precipitate surrounding the colonies.
Two substrates were used to evaluate the ability of the bacteria to hydrolyze proteins, Litmus® milk (Difco) and 5% gelatin medium, as described by Schaad.15 After four and ten days of incubation at 28 °C, the medium changed from milky to translucent (Litmus) or became liquefied after being refrigerated at 4 °C for 1 h (gelatin). Chitin hydrolysis was evaluated using 0.5% chitin medium as the sole carbon source16 and calcium phosphate solubilization was evaluated in NBRIP culture medium,17 both activities were observed at seven, 14, and 21 days of incubation at 28 °C to verify the degradation zone of each compound.
The hydrolysis of chitin, lipid and proteins (Litmus® milk and gelatin) was determined in a semiquantitative way, using different incubation times for evaluating. The intensity of production was low for positive reaction after 10 (lipids and proteins) or 21 days (chitin) of incubation; middle if the positive reaction happened after 14 days of incubation (chitin); and high when the positive reaction occurred after four (lipids and proteins) or seven days of incubation (chitin).
Validation of biocontrol bacterial selection – evaluation of nematicide activity
At this step, the bacteria that produced at least five of the eight CRBPGs analyzed were used, as well as DFs1985 (which did not produce any of the compounds evaluated) and DFs2180 (which produced one compound), for contrast effects. The ringed nematode (Mesocriconema xenoplax (Raski) Loof and de Grisse) species associated with Peach Tree Short Life disease was chosen as the target species to evaluate the efficiency of bacterial biocontrol.
Mobile forms (juvenile and adult) of the nematode (25 per plot) were treated with a bacterial suspension (A540 = 0.5) or with saline solution (control) at a 1:1 ratio (v/v). The microtiter plates containing the treatments were incubated at 25 °C for 24 h and arranged in a completely randomized design (86 bacterial treatments in four replicates). The number of dead nematodes was evaluated by adding 10 μL of 1 N NaOH, according to Chen and Dickson.18 The values were sin acsc transformed, subjected to analysis of variance, and grouped by the method of Scott and Knott.19
Bacterial identification
The bacteria with best control were identificated by their 16S rRNA sequences. Total DNA was extracted with Wizard genomic DNA purification Kit (Promega Corporation, USA). Partial amplicom of 16S rRNA of samples were produced by PCR with primers 27F and 1492R.20 The amplicons were cleansed with Wizard SV Gel and PCR Clean-up kit (Promega) and sent for sequencing by Ludwig Sequenciamento.
Results
Of the 1219 bacterial isolates studied, 92% were able to produce one or more of the eight compounds evaluated. However, only 1% of the isolates produced all the compounds. The ability of the bacteria to produce compounds was called ‘production of compounds related to biocontrol and/or promoting growth (CRBPG)’, and most isolates (17%) produced three CRBPGs (Table 1).
Table 1.
Percentage of bacteria producing compounds related to biocontrol and/or promoting growth according to number of produced compounds and total isolated by grouping as the origin of the bacterial isolates (Ficus carica, Gramineae, Leguminoseae, Liliaceae, Prunus persica, soil, Tagetes and others – total = 1219 isolates).
| Source groups/(no. isolates) | Number of the compounds produced |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| Ficus carica | 10.9* | 18.2 | 23.6 | 14.5 | 9.1 | 10.9 | 9.1 | 3.6 | 0.0 |
| Gramineae | 8.3 | 11.1 | 16.7 | 15.3 | 11.1 | 6.9 | 20.8 | 8.3 | 1.4 |
| Leguminoseae | 7.9 | 13.9 | 17.9 | 7.9 | 12.6 | 15.9 | 17.9 | 6.0 | 0.0 |
| Liliaceae | 2.3 | 6.4 | 8.2 | 16.9 | 13.7 | 19.2 | 17.8 | 15.5 | 0.0 |
| Prunus persica | 10.8 | 16.8 | 18.2 | 20.5 | 12.5 | 12.1 | 6.1 | 2.7 | 0.3 |
| Soil | 11.0 | 12.0 | 11.0 | 17.5 | 12.9 | 14.6 | 13.9 | 5.5 | 1.6 |
| Tagetes | 0.0 | 2.0 | 23.5 | 21.6 | 19.6 | 15.7 | 13.7 | 3.9 | 0.0 |
| Others** | 6.2 | 10.8 | 18.5 | 20.0 | 12.3 | 10.8 | 13.8 | 7.7 | 0.0 |
Percentage values for 1219 bacterial isolates tested.
Others = Solanum lycopersicum, Brassicae, and culture medium contaminants.
Upon analysis of the source groups and the number of compounds produced (Table 1), three groups exhibited bacterial isolates that produced all the compounds evaluated (soils – 1.6%, grasses – 1.4%, and peach tree – 0.3%). Additionally, a large number of isolates from Liliaceae (15.5%) were able to produce at least seven CRBPGs. The isolates from Tagetes comprised the only group in which 100% of the isolates exhibited production of at least one of the compounds. In contrast, the group of isolates from soils exhibited the highest percentage of isolates, without producing any of the compounds, followed by isolates from the fig tree and peach tree.
In general, the groups exhibited different percentage distributions among the numbers of compounds produced. The highest percentage for bacteria isolated from fig tree, leguminous plants and Tagetes was observed for two compounds and for the bacteria isolated from peach tree, soil and others, took place in three compounds. The isolates obtained from the soil had a more homogeneous distribution, ranging from 11% to 17.5% for zero to six compounds produced, in contrast to Tagetes, which concentrated the distribution between two and six compounds produced (85.9%).
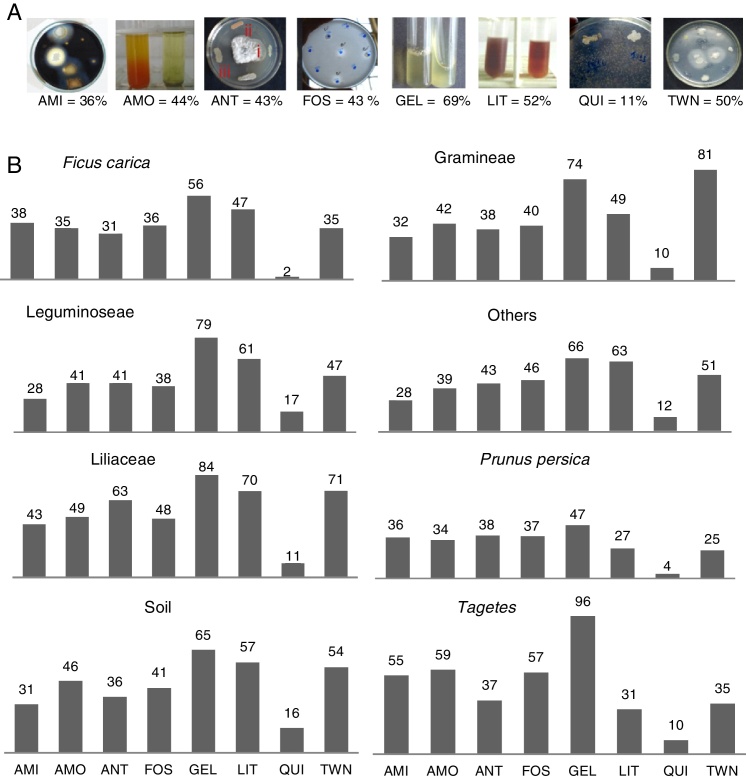
When observing each of the evaluated compounds (Fig. 1A), it is noteworthy that proteolytic activity was most frequently observed for both substrates, and chitin hydrolysis was least frequently observed. In the other tests, the quantity of isolates that produced compounds varied between 36% (amylolytic) and 50% (lipolytic). However, when comparing the eight groups originating from the bacterial isolates (Fig. 1B), it is possible to distinguish which isolates have higher production potential for each of the compounds evaluated. The Tagetes group exhibited the highest percentage of phosphate solubilizers, protease-producers in gelatin, ammonia-producers, and amylase-producers. The bacterial isolates from Liliaceae were notable as protease-producers and for their antibiosis activity against M. fructicola. The bacterial isolates from grasses proved to be the best lipase-producers, and those from legumes were the best chitinase-producers.
Fig. 1.
Percent bacteria producing compounds related to biocontrol and/or promoting growth depending on the total isolates number. (A) AMI, amylase; AMO, ammonia; ANT, antibiotics (halos type I – ≤10 mm; II – ≥11 and ≤20 mm; and III – ≥21 mm); FOS, phosphate solubilization; GEL, proteases medium gelatin; LIT, proteinase medium Litmus®; CHI, chitinase; TWN, lipases medium Tween 80. (B) Groups formed as the isolation origin (Ficus carica, Gramineae, Leguminoseae, others, Liliaceae, Prunus persica, soil, Tagetes).
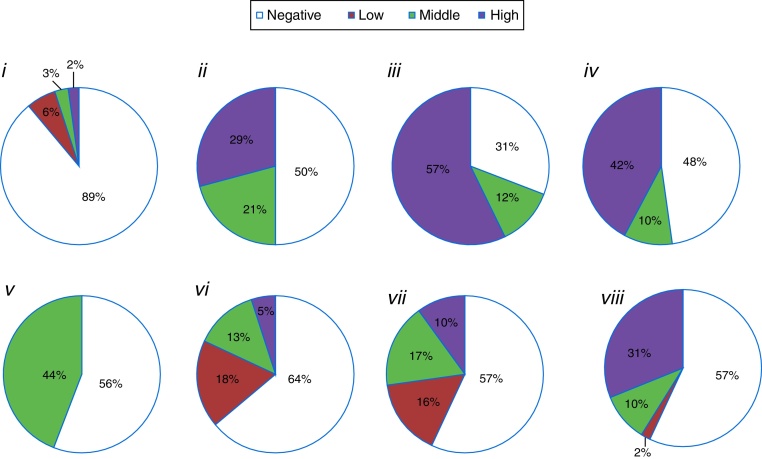
The semiquantitative component of the evaluations, through the assignment of grades or evaluations during distinct incubation periods (Fig. 2), allowed us to summarize the results of compound production intensity into negative, low (type I zone or slowest isolates), medium (type II zone or isolates with positive reaction after intermediate incubation time), and high (type III zone or fast isolates) categories. Thus, protease-producing isolates (both substrates) predominated upon the negative ones. Among the positive results, the high intensity was more frequent to isolates that hydrolyzed lipids, gelatin and milk or solubilized phosphate (Fig. 2).
Fig. 2.
Percentage of isolates (total = 1219) grouped as reaction intensity: (i) chitinolytic; (ii) lipolytic; (iii) proteolytic on gelatin medium; (iv) proteolytic Litmus® milk medium; (v) ammonia-production; (vi) amylase-production; (vii) antibiotic against Monilinia fructicola; (viii) phosphate-solubilization.
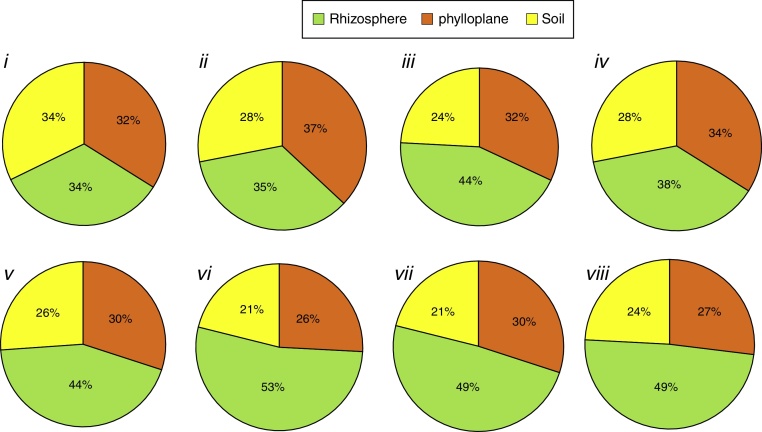
In contrast, considering only the isolates that produced each compound and their isolation niches (Fig. 3), rhizospheric isolates predominated for all the compounds (ranging from 53 to 38%), except for chitinase (Fig. 3i) and lipase production (Fig. 3ii), where the three niches exhibited practically the same percentage.
Fig. 3.
Percentage of isolates with positive reaction grouped as isolation niche (phylloplane, rhizosphere and soil): (i) individually chitinolytic (135); (ii) lipolytic (604); (iii) proteolytic on gelatin medium (839); (iv) proteolytic medium Litmus® milk (630); (v) ammonia-producers (531); (vi) amylolytic (443); (vii) antibiotics producing (529); (viii) phosphate solubilizers (530). Values between brackets represent the number of isolates with positive results.
Thus, upon determination of the isolates ability to produce CRBPGs, it was possible to select 86 bacteria that produced from five to eight compounds evaluated (three bacteria that produced eight, 32 that produced seven, 23 that produced six, and 28 that produced five compounds).
Evaluation of ringed nematode mortality upon exposure to a bacterial suspension for 24 h allowed us to group the isolates into 10 groups with different efficiency levels (a–j, Table 2). Eight of these groups positively differed from the control, i.e., comprised bacteria that caused mortality in juvenile M. xenoplax (groups a–h); one was equal to the control (group i); and another reduced their mortality (group j). Of these, 89.4% of bacteria were effective, and most bacteria (42.5%) resulted in at least 96.7% mortality (group a).
Table 2.
Mortality percent of Mesocriconema xenoplax due the action of bacteria producing at least five compounds related to biocontrol and/or promotion growth (CRBPGs) after 24 h incubation at 25 °C.
| Bacterial treatment | Mortality (%) | Bacterial treatment | Mortality (%) | Bacterial treatment | Mortality (%) |
|---|---|---|---|---|---|
| Producers eight compounds CRBPGs | |||||
| DFs1391 | 100.00a* | DFs1486 | 100.00a | DFs19866 | 100.0a |
| Producers seven compounds CRBPGs | |||||
| DFs0066 | 100.00a | DFs2247 | 100.00a | DFs0081 | 55.67f |
| DFs0390 | 100.00a | DFs2251 | 100.00a | DFs1021 | 49.08g |
| DFs0695 | 100.00a | DFs0439 | 97.95a | DFs0582 | 46.87g |
| DFs0886 | 100.00a | DFs1887 | 93.15b | DFs1985 | 31.56h |
| DFs1256 | 100.00a | DFs1039 | 86.87b | DFs1421 | 29.63h |
| DFs1258 | 100.00a | DFs0419 | 80.64c | DFs0706 | 22.57i |
| DFs1341 | 100.00a | DFs0023 | 78.33c | DFs2137 | 19.25i |
| DFs1947 | 100.00a | DFs1219 | 72.15d | DFs1967 | 17.77i |
| DFs2049 | 100.00a | DFs0756 | 69.56e | DFs1320 | 12.65j |
| DFs2108 | 100.00a | DFs0377 | 59.51f | DFs1344 | 9.38j |
| DFs2124 | 100.00a | DFs2121 | 58.73f | ||
| Producers six compounds CRBPGs | |||||
| DFs1983 | 100.00a | DFs2191 | 90.23b | DFs2172 | 50.00g |
| DFs2007 | 100.00a | DFs2122 | 85.42b | DFs1934 | 28.37h |
| DFs2126 | 100.00a | DFs2175 | 82.89c | DFs1923 | 22.55i |
| DFs2156 | 100.00a | DFs0032 | 81.68c | DFs2162 | 22.33i |
| DFs2227 | 100.00a | DFs1691 | 71.94d | DFs2048 | 16.11i |
| DFs2229 | 100.00a | DFs2265 | 67.09e | DFs2062 | 13.29j |
| DFs2236 | 100.00a | DFs2052 | 65.06e | DFs0422 | 7.09j |
| DFs2110 | 96.74a | DFs2246 | 62.75e | ||
| Producers five compounds CRBPGs | |||||
| DFs2008 | 100.00a | DFs2148 | 82.55c | DFs2155 | 54.17f |
| DFs2099 | 100.00a | DFs2113 | 81.03c | DFs1955 | 45.71g |
| DFs2109 | 100.00a | DFs0418 | 79.02c | DFs1970 | 40.91g |
| DFs2133 | 100.00a | DFs2168 | 77.11d | DFs1932 | 37.29h |
| DFs2147 | 100.00a | DFs1650 | 76.06d | DFs2193 | 37.08h |
| DFs1928 | 98.04a | DFs2060 | 74.54d | DFs1935 | 19.46i |
| DFs1899 | 91.92b | DFs1909 | 69.88e | DFs1950 | 13.92i |
| DFs2250 | 90.77b | DFs1902 | 69.61e | DFs2051 | 9.17j |
| DFs2136 | 83.94c | DFs2027 | 68.60e | DFs1985** | 31.56h |
| DFs1958 | 83.90c | DFs2089 | 64.12e | DFs2180** | 6.01j |
| CV (%) | 10.02 | Control 16.08i | |||
Original values were converted to arc sen whose original means followed by the same letter in the column do not differ by Scott–Knott test at 5% probability.
Isolated used as contrast: DFs1985, not produces evaluated compounds and DFs2180, producing one compound.
In contrast, when considering the bacteria grouped by number of CRBPGs produced, we clearly observed that bacteria that produced more CRBPGs had a higher nematicide percentage (Table 2). Thus, all isolates evaluated that produced eight CRBPGs resulted in 100% nematode mortality; producers of seven CRBPGs had 43.7% of their isolates in the superior group; producers of six CRBPGs had 34.8% of their isolates in group a; and among those that produced five CRBPGs, only 21.4% exhibited high efficiency.
The partial 16S rRNA gene sequencing (Table 3) resulted 85–100% identity and 76% isolates identified belonging to the genus Bacillus.
Table 3.
Bacterial identification by homology of partial sequence of 16S rRNA using 27F and 1492R primers.
| Bacterial Treatment | Description | Identity | Accession |
|---|---|---|---|
| DFs066 | Bacillus subtilis subsp. subtilis strain KISR-1 16S ribosomal RNA gene, PSa | 98% | KM068047.1 |
| DFs390 | Bacillus thuringiensis strain BT-EM14 16S ribosomal RNA gene, PS | 99% | KT588443.1 |
| DFs439 | Bacillus subtilis strain CI1 16S ribosomal RNA gene, PS | 99% | KU557409.1 |
| DFs695 | Bacillus sp. AR493 partial 16S rRNA gene, strain AR493 | 100% | LN829576.1 |
| DFs886 | Bacillus thuringiensis Bt407 16S ribosomal RNA, CSb | 98% | NR_102506.1 |
| DFs1256 | Bacillus subtilis subsp. subtilis strain DSM 10 16S ribosomal RNA, PS | 98% | NR_027552.1 |
| DFs1258 | Bacillus subtilis strain 1-3 16S ribosomal RNA gene, PS | 97% | HQ693082.1 |
| DFs1341 | Bacillus subtilis subsp. subtilis str. 168 strain 168 16S ribosomal RNA, CS | 99% | NR_102783.1 |
| DFs1928 | Paenobacillus polymyxa gene 16 S RNA, PS | 99% | AB689755.1 |
| DFs1947 | Bacillus cereus strain CF-S30 16S ribosomal RNA gene, PS | 97% | KJ781400.1 |
| DFs1983 | Bacillus thuringiensis Bt407 16S ribosomal RNA, CS | 99% | NR_102506.1 |
| DFs2008 | Pseudomonas poae RE*1-1-14 strain RE*1-1-14 16S ribosomal RNA, CS | 97% | NR_102514 |
| DFs2049 | Pseudochrobactrum saccharolyticum strain ALK626 16S ribosomal RNA gene, PS | 96% | KC456591.1 |
| DFs2099 | Bacillus cereus strain YLB-P5 16S ribosomal RNA gene, PS | 99% | KF376341.1 |
| DFs2108 | Pseudomonas sp. VS-2 16S ribosomal RNA gene, PS | 98% | JF699699.1 |
| DFs2109 | Pseudomonas fluorescens strain M5 16S ribosomal RNA gene, PS | 99% | KT767962.1 |
| DFs2110 | Bacillus cereus strain NCIM 2106 16S ribosomal RNA gene, PS | 100% | KU218541.1 |
| DFs2124 | Bacillus cereus ATCC 14579 strain ATCC 14579 16S ribosomal RNA, CS | 99% | NR_074540.1 |
| DFs2126 | Bacillus sp. | 85% | |
| DFs2133 | Bacillus sp. SNG-3 16S ribosomal RNA gene, PS | 99% | KP992135.1 |
| DFs2147 | Pseudomonas putida strain BB3 16S ribosomal RNA gene, PS | 98% | KP314235.1 |
| DFs2229 | Bacillus cereus ATCC 14579 strain ATCC 14579 16S ribosomal RNA, CS | 100% | NR_074540.1 |
| DFs2236 | Bacillus cereus strain FT9, CGc | 97% | CP008712.1 |
| DFs2247 | Bacillus amyloliquefaciens FZB42 strain FZB42 16S ribosomal RNA, CS | 99% | NR_075005.1 |
| DFs2251 | Bacillus cereus strain KCd1 16S ribosomal RNA gene, PS | 99% | FJ613630.1 |
(PS) Partial sequence.
(CS) Complete sequence.
(CG) Complete genome.
Discussion
In general, all the plant species and soils used to isolate bacteria were host candidates for pathogen biocontrol. Each source group exhibited different percentages for each number of CRBPGs produced. The isolates that originated from grasses, legumes, and Liliaceae exhibited higher percentages for production of six or seven CRBPGs, although the two first groups comprised approximately 20% of the isolates that did not produce any or one of these compounds.
However, the group of isolates obtained from Tagetes is noteworthy because different species from these plants are considered antagonists to species of Pratylenchus and Meloidogyne.21 There are no reports of antagonism of Tagetes bacterial isolates against species of Mesocriconema. Additionally, studies in the 1990s showed that using isolates from roots of antagonistic plants increased the chances of finding good candidates for biocontrol agents by up to six times in in vivo tests.22, 23 Thus, the high percentages of CRBPG producers (98% between 2 and 7 CRBPGs) isolated from the Tagetes rhizosphere observed in the present study reflect this premise.
In contrast, approximately 90% of the isolates obtained from peach trees produced some CRBPGs. Although these isolates are not among those with highest CRBPG production, they should receive special attention because peach trees are affected by the ringed nematode, and biocontrol agents from the hosts are desirable because they have been adapted to survive in this habitat. This aspect was successfully exploited in the control of the ringed nematode.24, 25, 26 For most of the source groups, the isolates from the rhizosphere exhibited higher production of the evaluated compounds, as well as higher intensities of CRBPG production. These results reflect the richness of the rhizospheric environment, which is responsible for the occurrence and preference of several organisms, including those that exhibit antagonistic effects against several pathogens, such as nematodes.27
It has been previously established that enzymes, especially lytic enzymes, are synthesized by microorganisms and also act as an antagonistic mechanism, which, in many situations, may partially explain the biological control of plant disease.28 The isolates studied here exhibited a higher frequency for protein and lipid hydrolysis, which is an important result, as these compounds are present in the membranes surrounding the eggs and in the cuticle of mobile forms of different nematode species.26 Thus, these lytic activities have been associated with the biological control of nematodes.29
Chitinases are also important in controlling phytopathogens, as chitin is the main constituent of nematode eggs26, 30 and the fungal wall.31 Thus, chitinolytic bacteria are potential biocontrol agents for both pathogen groups.32, 33, 34 However, just over 10% of the isolates studied produced chitinases, which indicates that this ability is more common in actinomycetes and less common in bacteria and fungi present in the soil.35
Several researchers consider antimicrobial compound production one of the most important antagonist mechanisms. Antibiotics and toxic compounds produced by the bacteria can act on both the hatching and development of nematodes.26, 36 The isolates studied here (43%) inhibited mycelial growth of M. fructicola, which is used as an indicator of antibiotic production because M. xenoplax is an obligate pathogen and thus cannot be cultured in vitro. Therefore, it cannot be affirmed that the antibiotic activity is the same for the nematode.
In addition, 44% of the isolates produced ammonia, which is a compound that has previously been associated with pathogen control26, 37 and has the additional advantage of being volatile and the ability to expand through soil pores, reaching a higher volume around the bacterial colonies producing the compound. Furthermore, ammonia is a compound that acts during organic matter decomposition, which improves soil structure and plant nutrition, resulting in higher productivity by the crop and higher tolerance to phytoparasites.38 The phosphate solubilization observed among the isolates may improve plant nutrition. This aspect is especially important, as the damage to peach trees by M. xenoplax is aggravated under conditions of low soil fertility.39
A high percentage (85%) of bacteria selected in vitro exhibited high mortality in mobile forms of the ringed nematode. The potency of this ability may be related to different mechanisms of action, which is indicated by the number of bacteria-produced CRBPGs. Other studies have associated the individual production of some compounds with the control of different nematodes.26, 40, 41 However, none of these studies examined the diversity in compounds produced for the selection of biocontrol isolates.
Sequencing of 16S rRNA allows the accurate identification of bacterial genera from various plant species. Bacillus is an important bacteria genus because of the synthesis of a wide range of metabolites with diverse properties.42, 43 However, for the precise definition of species and subspecies analysis of other genes is needed, for example gyrA.44
It is very important to develop fast and efficient methods to select biocontrol microorganisms, especially when evaluating a large number of candidates. In vitro evaluations allow for a reduction in the number of isolates, which makes further in vivo tests viable. This aspect is crucial when working with perennial plants and long-cycle, obligate pathogens, such as the peach tree-M. xenoplax pathosystem.
Thus, the results obtained in the present study indicate the efficacy of the selection strategy and validate the use of the CRBPG production profile in reducing the number of isolates for in vivo evaluation.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgments
The authors would like to thank the Rio Grande de Sul State Research Foundation (Fundação de Amparo a Pesquisa do Estado do Rio Grande do Sul–FAPERGS) for financial support (10.10360), the Brazilian Federal Agency for the Support and Evaluation of Graduate Education (Conselho Nacional de Desenvolvimento Científico e Tecnológico – CAPES) for the doctoral scholarship given to the first author, and the Brazilian National Council for Scientific and Technological Developmentarship given to the last author.
Associate Editor: Jerri Édson Zilli
References
- 1.Hyakumachi M. Research on biological control of plant diseases: present state and perspectives. J Gen Plant Pathol. 2013;79(6):435–440. [Google Scholar]
- 2.Gray E.J., Smith D.L. Intracellular and extracellular PGPR: commonalities and distinctions in the plant–bacterium signaling processes. Soil Biol Biochem. 2005;37:395–412. [Google Scholar]
- 3.Kloepper J.W., Schroth M.N. Proceedings of the 4th International Conference on Plant Pathogenic Bacterial. INRA; Angers: 1978. Plant growth promoting rhizobacteria on radishes; pp. 879–882. [Google Scholar]
- 4.Van Loon L.C. Plant responses to plant growth promoting bacteria. Plant Pathol. 2007;119:243–254. [Google Scholar]
- 5.Lugtenberg B., Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 6.Glick B.R. Hindawi Publishing Corporation, Scientifica; 2012. Plant Growth-Promoting Bacteria: Mechanisms and Applications; p. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindow S.E., Leveau J.H.J. Phyllosphere microbiology. Curr Opin Biotechnol. 2002;13:238–243. doi: 10.1016/s0958-1669(02)00313-0. [DOI] [PubMed] [Google Scholar]
- 8.Kishore K., Pande S., Podile A.R. Phylloplane bacteria increase seedling emergence, growth and yield of field-grown groundnut (Arachis hypogaea L.) Appl Microbiol. 2005;40:260–268. doi: 10.1111/j.1472-765X.2005.01664.x. [DOI] [PubMed] [Google Scholar]
- 9.Schisler D.A., Slininger P.J. Microbial selection strategies that enhance the likelihood of developing commercial biological control products. J Ind Microbiol Biotechnol. 1997;19:172–179. [Google Scholar]
- 10.Validov S., Kamilova F., QI S. Selection of bacteria able to control Fusarium oxysporum f. sp. Radices lycopersici in stonewool substrate. J Appl Microbiol. 2007;102:461–471. doi: 10.1111/j.1365-2672.2006.03083.x. [DOI] [PubMed] [Google Scholar]
- 11.Köhl J., Postma J., Nicot P., Ruocco M., Blum B. Stepwise screening of microorganisms for commercial use in biological control of plant-pathogenic fungi and bacteria. Bio Control. 2011;57:1–12. [Google Scholar]
- 12.Kado C.I., Heskett M.G. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas and Xanthomonas. Phytopathology. 1970;60:969–976. doi: 10.1094/phyto-60-969. [DOI] [PubMed] [Google Scholar]
- 13.Cappuccino J.G., Sherman N. Benjamin Cummings Science Publishing; Menlo Park: 1998. Microbiology – A Laboratory Guide; p. 477. [Google Scholar]
- 14.Fahy P.C., Persle G.J. Academic Press; Sydney: 1983. Plant Bacterial Diseases: A Diagnostic Guide; p. 393. [Google Scholar]
- 15.Schaad N.W. 2nd ed. The American Phytopathology Society; St. Paul: 1988. Laboratory Guide for Identification of Plant Pathogenic Bacteria; p. 158. [Google Scholar]
- 16.Renwick A., Campbell R., COE S. Assessment of in vitro screening systems for potential biocontrol agents of Gaeumannomyces graminis. Plant Pathol. 1991;40:524–532. [Google Scholar]
- 17.Nautiyal C.S. An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol Lett. 1999;170:265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x. [DOI] [PubMed] [Google Scholar]
- 18.Chen S.Y., Dickson D.W. A technique for determining live second-stage juveniles of Heterodera glycines. J Nematol. 2000;32(1):117–121. [PMC free article] [PubMed] [Google Scholar]
- 19.Scott A.J., Knott M. A cluster analysis method for grouping means in the analysis of variance. Biometrics. 1974;30(2):507–512. [Google Scholar]
- 20.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M.N., editors. Nucleic Acid Techniques in Bacterial Systematics. Wiley; Chichester: 1991. pp. 115–147. [Google Scholar]
- 21.Fabry C.F.S., Freitas L.G., Neves W.S. Obtenção de bactérias para o biocontrole de Meloidogyne javanica por meio de aquecimento de solo e tratamento com filtrado de raízes de plantas antagonistas a fitonematoides. Fitopatol Bras. 2007;32:079–082. [Google Scholar]
- 22.Kloepper J.W., Rodriguez-Kábana R., Mcinroy J.A., Collins D.J. Analysis of populations and physiological characterization of microorganisms. Plant Soil. 1991;136:95–102. [Google Scholar]
- 23.Kloepper J.W., Rodriguez-Kábana R., McInroy J.A., Young R.W. Rhizosphere bacteria antagonistic to soybean cyst (Heterodera glycines) and root-knot (Meloidyne incognita) nematodes: identification by fatty acid analysis and frequency of biological control activity. Plant Soil. 1992;139:74–84. [Google Scholar]
- 24.McInnis T., Kluepfel D., Zehr E. Suppression of Criconemella xenoplax on peach by rhizosphere bacteria. Second International Nematology Congress, Abstracts of Papers 106; Veldhoven, The Netherlands; 1990. [Google Scholar]
- 25.Kluepfel D.A., McInnis T.M., Zehr E.I. Involvement of root-colonizing bacteria in peach orchard soils suppressive of the nematode Criconemella xenoplax. Phytopathology. 1993;83(11):1240–1245. [Google Scholar]
- 26.Westcott S.W., Kluepfel D. Inhibition of Criconemella xenoplax egg hatch by Pseudomonas aureofaciens. Phytopathology. 1993;83:1245–1249. [Google Scholar]
- 27.Zavaleta-Mejia E., Van Gundy S.D. Effects of rhizobacteria on Meloidogyne infection. J Nematol. 1982;14(4):475–476. [Google Scholar]
- 28.Duffy B., Schouten A., Raaijamakers J.M. Pathogen self-defense: mechanisms to counteract microbial antagonism. Annu Rev Phytopathol. 2003;41:501–538. doi: 10.1146/annurev.phyto.41.052002.095606. [DOI] [PubMed] [Google Scholar]
- 29.Aalten P.M., Vitour D., Blanvillain S.R., Gowen S.R., Sutra L. Effect of rhizosphere fluorescent Pseudomonas strains on plant parasitic nematodes Radopholus similes and Meloidogyne spp. Lett Appl Microbiol. 1998;27:357–361. [Google Scholar]
- 30.Dunne C., Moenne-Loccoz Y., de Bruijn F.J., ÓGara F. Overproduction of an inducible extracellular serine protease improves biological control of Pythium ultimum by Stenotrophomonas maltophilia strain W81. Microbiology. 2000;146:2069–2078. doi: 10.1099/00221287-146-8-2069. [DOI] [PubMed] [Google Scholar]
- 31.Feifilova E.P. The fungal cell wall: modern concepts of its composition and biological function. Microbiology. 2010;79(6):711–750. [PubMed] [Google Scholar]
- 32.Hallmann J., Rodríguez-Kábana R., Kloepper J.W. Chitin-mediated changes in bacterial communities of the soil, rhizosphere and within roots of cotton in relation to nematode control. Soil Biol Biochem. 1999;31:551–560. [Google Scholar]
- 33.Slimene I.B., Tabbene O., Gharbi D. Isolation of a chitinolytic Bacillus licheniformis S213 strain exerting a biological control against Phoma medicaginis infection. Appl Biochem Biotechnol. 2015;175(7):3494–3506. doi: 10.1007/s12010-015-1520-7. [DOI] [PubMed] [Google Scholar]
- 34.Brzezinska S.M., Jankiewicz U., Burkowska A., Walczak M. Chitinolytic microorganisms and their possible application in environmental protection. Curr Microbiol. 2014;68:71–81. doi: 10.1007/s00284-013-0440-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu S.C., Lockwood J.L. Powdered chitin agar as a selective medium to enumeration of actinomycetes in water and soil. Appl Microbiol. 1975;29:422–426. doi: 10.1128/am.29.3.422-426.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siddiqui I.A., Haas D., Heeb S. Extracellular protease of Pseudomonas fluorescens CHA0, a biocontrol factor with activity against the root-knot nematode Meloidogyne incognita. Appl Environ Microbiol. 2005;71(9):5646–5649. doi: 10.1128/AEM.71.9.5646-5649.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fravel D.R. Role of antibiosis in the biocontrol of plant diseases. Annu Rev phytopathol. 1988;26:75–91. [Google Scholar]
- 38.McSorley R. Alternative practices for managing plant-parasitic nematodes. Am J Altern Agric. 1998;13:98–104. [Google Scholar]
- 39.Ritchie D.F., Clayton C.N. Peach tree short life: a complex of interacting factors. Plant Dis. 1981;65(6):462–469. [Google Scholar]
- 40.Araújo F.F., Silva J.F.V., Araújo A.S.F. Influência de Bacillus subtilis na eclosão, orientação e infecção de Heterodera glycines em soja. Ciência Rural. 2002;32(2):197–202. [Google Scholar]
- 41.Dawar S.M., Tariq M., Zaki M.J. Application of Bacillus species in control of Meloidogyne javanica (Treub) Chitwood on cowpea and mash bean. Pakistan J Bot. 2008;40(1):439–444. [Google Scholar]
- 42.Jacobsen B.J., Zidack N.K., Larson B.J. The role of Bacillus-based biological control agents in integrated pest management systems: plant diseases. Phytopathology. 2004;94:1272–1275. doi: 10.1094/PHYTO.2004.94.11.1272. [DOI] [PubMed] [Google Scholar]
- 43.Velmurugan N., Choi M.S., Han S.S., Lee Y.S. Evaluation of antagonistic activities of Bacillus subtilis and Bacillus licheniformis against wood-staining fungi: in vitro and in vivo experiments. J Microbiol. 2009;47:385–392. doi: 10.1007/s12275-009-0018-9. [DOI] [PubMed] [Google Scholar]
- 44.Chun J., Bae K.S. Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Anton Van Leeuw J Microbiol. 2000;78:123–127. doi: 10.1023/a:1026555830014. [DOI] [PubMed] [Google Scholar]