Abstract
The distribution of Escherichia coli O157 in bovine feces was examined by testing multiple samples from fecal pats and determining the density of E. coli O157 in immunomagnetic separation (IMS)-positive fecal samples. The density of E. coli O157 in bovine feces was highly variable, differing by as much as 76,800 CFU g−1 between samples from the same fecal pat. The density in most positive samples was <100 CFU g−1, the limit of reliable detection by IMS. Testing only one 1-g sample of feces per pat with IMS may result in a sensitivity of detection as low as 20 to 50%. It is therefore probable that most surveys have greatly underestimated the prevalence of E. coli O157 shedding in cattle and the proportion of farms with shedding cattle. The sensitivity of the detection of E. coli O157 in bovine feces can be as much as doubled by testing two 1-g samples per pat rather than one 1-g sample.
Shiga-toxigenic Escherichia coli O157 is a major public health concern. It is associated with human illnesses ranging from uncomplicated watery diarrhea to hemorrhagic colitis and hemolytic-uremic syndrome, which may result in death (22, 24, 31). Cattle are an important source of E. coli O157 (15, 25, 39), and many surveys around the world have been conducted to estimate the prevalence of E. coli O157 shedding by cattle (20).
Much research has gone into improving the sensitivity of laboratory methods for the detection of E. coli O157 and the introduction of immunomagnetic separation (IMS) and selective isolation media has greatly improved the sensitivity of E. coli O157 isolation from bovine feces (4, 5, 35). In contrast, the distribution of E. coli O157 in bovine feces and its impact on the accuracy of prevalence estimates reported in bovine fecal E. coli O157 shedding surveys has largely been ignored. A variety of sampling techniques have been used to collect bovine feces in surveys, including rectal swabs (11, 19), rectal grab samples (15, 39), and grab or swab samples from fecal pats (10, 12). In 27 surveys we reviewed, only one sample was taken from each animal or fecal pat. In 9 of these surveys, a swab from each animal or fecal pat was tested (1, 2, 10-12, 19, 29, 30, 38); in 7 surveys, 1 g of feces from each animal or pat was tested (8, 16, 25, 26, 28, 32, 36); in 10 surveys, ≥10 g of feces from each animal or pat was tested (3, 7, 9, 13-15, 18, 23, 34, 39). In the remaining survey, the amount of feces tested was not reported (17). The analysis of results in these 27 studies implicitly assumed homogenous distribution of E. coli in the fecal samples tested.
This study investigated the distribution of E. coli O157 in bovine feces and assessed its impact on the sensitivity of shedding survey results.
MATERIALS AND METHODS
Sampling.
Fecal samples were drawn from two separate studies. In both studies, convenience samples of fecal pats were taken, with pats being sampled without replacement. In the first study, multiple 10- to 25-g samples were taken from fecal pats in three separate pens of cattle housed on straw bedding. On 23 October 2001, three samples were taken from each of 58 pats in a pen of 34 yearlings on farm 1 (group A); on 5 November 2001, five samples were taken from each of 29 pats in a pen of 24 cows and calves also on farm 1 (group B); and on 7, 15, and 21 September 2002, 10 samples were taken from each of 59 pats in a pen of 27 finishing animals on farm 2 (group C). In the second study, a cross-sectional survey of Scottish cattle farms to determine the prevalence of E. coli O157 shedding in store and finishing cattle, single 10- to 25-g samples from each of 4,662 fresh fecal pats were submitted from 172 farms sampled in the period from 7 May 2002 to 14 January 2003, inclusive. In both studies, samples were collected from fresh, undisturbed fecal pats by project veterinary staff. Samples were taken either from the surface and subsurface of the pat or from deep within the pat following removal of the top few millimeters of the pat surface. When multiple samples were taken from the same pat, they were all taken from different locations of the pat. Each sample was collected into a sterile 30-ml polystyrene universal container by using a sterile plastic spatula (Bibby Sterilin, Stone, Staffs, United Kingdom). The source animal for each pat was either unknown or not recorded.
All samples were transported from the sampling location to the laboratory at ambient temperature, refrigerated at 5°C within 2 h of collection, and always tested within 48 h of collection. Samples from farm 1 were tested by IMS only. Samples from farm 2 and farms participating in the cross-sectional survey were also tested by IMS, and the density of E. coli O157 in IMS-positive samples was determined.
Duplicate testing of samples.
As part of a quality control exercise for this study, and other studies not reported in this paper which required IMS testing of fresh bovine fecal samples for E. coli O157, one in every 20 fecal samples submitted was tested by IMS in duplicate. Where duplicate testing was required, two 1-g aliquots were drawn from each fecal sample and tested independently for the presence of E. coli O157.
IMS.
Preenrichment in buffered peptone water, IMS procedures, and plating onto sorbitol MacConkey agar supplemented with cefixime (2.5 mg l−1) and potassium tellurite (0.05 mg l−1) (CT-SMAC) were conducted as described previously (27). Fecal samples were not homogenized prior to testing, and the 1-g aliquots of feces used to inoculate buffered peptone water were always obtained from the surface of the original 10- to 25-g fecal samples submitted to the laboratory. From CT-SMAC plates, not more than 10 non-sorbitol-fermenting colonies with morphologies typical of E. coli O157 were plated onto Chromocult coliform agar (Merck, Poole, Dorset, United Kingdom) and incubated overnight at 37°C. Distinctive red-pink colonies were tested by using anti-E. coli O157 antibody-coated latex reagent (Oxoid, Basingstoke, Hants, United Kingdom). No further confirmatory tests were conducted on E. coli O157 isolates from the first study, which examined multiple samples from fecal pats, or on E. coli O157 identified during duplicate testing of fecal samples. However, all E. coli O157 isolates recovered by IMS in the cross-sectional study were referred to the Scottish E. coli O157 Reference Laboratory for species and serogroup confirmation.
Estimation of E. coli O157 density.
The density of E. coli O157 was only estimated for samples that returned a positive IMS result. One gram of feces from each IMS-positive sample was suspended in 9 ml of maximum-recovery diluent (MRD) (Oxoid Ltd.), and 0.1 ml of suspension was spread onto each of two 9-cm-diameter CT-SMAC plates. Plates were incubated at 42°C for 24 h. Non-sorbitol-fermenting colonies with morphologies typical of E. coli O157 were counted manually, and a maximum of 10 such colonies were tested with anti-E. coli O157-coated latex reagent (Oxoid Ltd.). No further confirmatory tests were conducted on E. coli O157 isolates. If the number of colonies on each of the two plates was too large to count accurately, i.e., greater than around 100 colonies, a further 1 g of feces from the original sample was suspended in 9 ml of MRD and serially diluted up to six times by sequentially adding 1 ml of suspension to a further 9 ml of MRD. The series of suspensions were then plated, incubated, and counted in the same way as the original 1:9 suspension. The number of serial dilutions required for accurate counting of colonies was determined by growth on plates on which the original 1:9 suspension was spread. The density of E. coli O157 in the fecal sample was estimated by averaging the E. coli O157 colony count of the two plates and correcting for fecal dilution. If E. coli O157 could not be detected by this method but IMS testing of the same sample yielded a positive result, the density of E. coli O157 in the sample was deemed to be <100 CFU g−1.
Statistical analysis and Monte Carlo simulation.
Statistical analysis and Monte Carlo simulation were performed by using SAS, version 8.2 (SAS Institute Inc, Cary, N.C.). Associations between duplicate test results on fecal samples were assessed by using the exact test for paired data and the κ statistic (33). The values Pneg and Ppos (6) were calculated manually from the results of duplicate testing of fecal samples.
Using the results from groups A, B, and C, a simulation trial was conducted to determine the likelihood of taking at least one positive sample from a positive pat if only one or two samples per pat had been taken. For each group, 10,000 sampling iterations were conducted. During each iteration, one result from each fecal pat known to be positive for E. coli O157 was selected at random and the proportion of pats correctly determined to be positive, i.e., sensitivity, was calculated.
RESULTS
Multiple fecal pat samples.
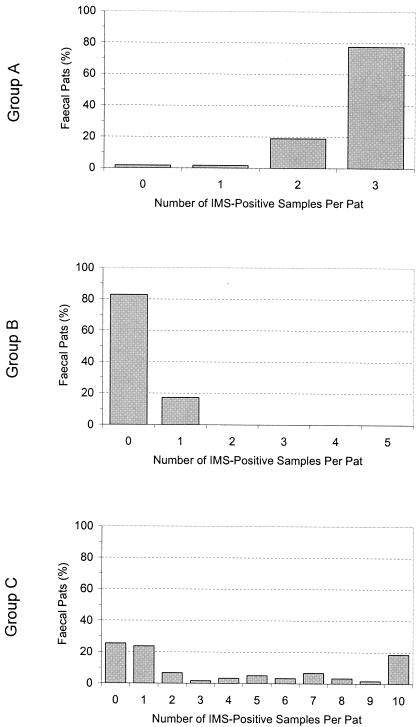
The results of IMS testing of multiple 1-g fecal samples from pats collected on farms 1 and 2, shown in Fig. 1, demonstrate that results can be highly variable within pats. In group A, E. coli O157 was detected in 57 of 58 (98.3%) fecal pats. For 1 of 58 (1.7%) pats, only one sample in three was positive; for 11 of 58 (19.0%) pats, two samples in three were positive; and for 45 of 58 (77.6%) pats, all three samples were positive. In group B, E. coli O157 was detected in only 5 of 29 (17.2%) pats; in each positive pat, E. coli O157 was detected in only one sample in five. In group C, E. coli O157 was detected in 44 of 59 (74.6%) pats. For 14 of 59 (23.7%) pats, only 1 sample in 10 was positive compared with 11 of 59 (18.6%) pats for which all 10 samples were positive. In 19 of 59 (32.2%) pats, 2 to 9 of the 10 samples from each pat were positive.
FIG. 1.
Proportion of samples per fecal pat in which E. coli O157 was detected by IMS. Group A, 3 samples taken per pat; group B, 5 samples taken per pat; group C, 10 samples taken per pat.
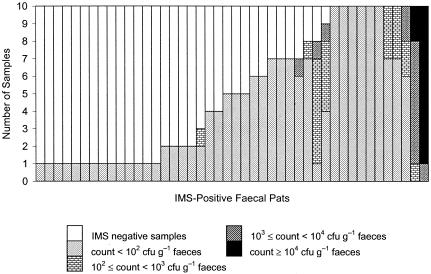
The densities of E. coli O157 in samples from the 44 pats with one or more IMS-positive samples originating from group C were generally low (Fig. 2). In 34 of 44 (77.3%) pats, the densities of E. coli O157 were <100 CFU g−1 in all IMS-positive samples; in a further 8 of 44 (18.2%) pats, the lowest densities of E. coli O157 among IMS-positive samples from each pat were <100 CFU g−1 while the highest densities among IMS-positive samples from each pat ranged from 150 to 7,300 CFU g−1. In the remaining 2 of 44 (4.6%) pats with one or more IMS-positive samples, the densities of E. coli O157 in IMS-positive samples ranged from 800 to 77,600 CFU g−1 for one pat and 9,200 to 26,200 CFU g−1 for the other pat.
FIG. 2.
E. coli O157 density in samples taken from IMS-positive fecal pats in group C. Each column shows results for 10 samples taken from a single fecal pat.
Scottish cattle farm cross-sectional survey.
E. coli O157 was detected by IMS in one or more fecal samples from each of 37 of 172 (21.5%) farms screened in the cross-sectional farm survey. Of the farms with detectable E. coli O157, 13 of 37 (35.1%) yielded only one IMS-positive fecal pat and 5 of 37 (13.5%) yielded only two IMS-positive fecal pats. The densities of E. coli O157 in samples from all pats collected on the 37 farms with IMS-detectable E. coli O157 are shown in Table 1. For 26 of 37 (70.3%) farms with IMS-detectable E. coli O157, the density of E. coli O157 was <100 CFU g−1 in all IMS-positive samples; on a further 5 (13.5%) farms with IMS-detectable E. coli O157, >65% of IMS-positive samples had an E. coli O157 density of <100 CFU g−1.
TABLE 1.
E. coli O157 density in fecal samples from cross-sectional survey farms with one or more IMS-positive fecal patsa
| Farm | Sampling date (day.mo.yr) | Locationb | No. of samples | % of samples:
|
||||
|---|---|---|---|---|---|---|---|---|
| IMS negative | IMS positive with E. coli O157 count (CFU g−1 feces) of:
|
|||||||
| n < 102 | 102 ≤ n < 103 | 103 ≤ n < 104 | n ≥ 104 | |||||
| E037 | 07.05.2002 | H | 52 | 98.1 | 1.9 | 0.0 | 0.0 | 0.0 |
| E018 | 12.06.2002 | P | 48 | 97.9 | 2.1 | 0.0 | 0.0 | 0.0 |
| E036 | 07.05.2002 | P | 80 | 97.5 | 2.5 | 0.0 | 0.0 | 0.0 |
| E345 | 17.09.2002 | P | 36 | 97.2 | 2.8 | 0.0 | 0.0 | 0.0 |
| E074 | 19.08.2002 | P | 27 | 96.3 | 3.7 | 0.0 | 0.0 | 0.0 |
| E303 | 27.05.2002 | P | 54 | 96.3 | 3.7 | 0.0 | 0.0 | 0.0 |
| E335 | 19.08.2002 | P | 48 | 95.8 | 0.0 | 4.2 | 0.0 | 0.0 |
| E053 | 25.06.2002 | P | 46 | 95.7 | 4.3 | 0.0 | 0.0 | 0.0 |
| E064 | 09.07.2002 | P | 23 | 95.7 | 4.3 | 0.0 | 0.0 | 0.0 |
| E061 | 03.07.2002 | P | 23 | 95.7 | 0.0 | 0.0 | 0.0 | 4.3 |
| E314 | 12.06.2002 | H | 20 | 95.0 | 5.0 | 0.0 | 0.0 | 0.0 |
| E373 | 06.01.2003 | P | 19 | 94.7 | 0.0 | 0.0 | 5.3 | 0.0 |
| E351 | 14.10.2002 | H | 15 | 93.3 | 6.7 | 0.0 | 0.0 | 0.0 |
| E090 | 01.10.2002 | P | 13 | 92.3 | 7.7 | 0.0 | 0.0 | 0.0 |
| E306 | 05.06.2002 | H + P | 37 | 91.9 | 5.4 | 2.7 | 0.0 | 0.0 |
| E341 | 09.09.2002 | H | 32 | 90.6 | 9.4 | 0.0 | 0.0 | 0.0 |
| E075 | 19.08.2002 | P | 27 | 88.9 | 11.1 | 0.0 | 0.0 | 0.0 |
| E097 | 08.10.2002 | H | 27 | 88.9 | 11.1 | 0.0 | 0.0 | 0.0 |
| E349 | 23.09.2002 | P | 9 | 88.9 | 11.1 | 0.0 | 0.0 | 0.0 |
| E091 | 01.10.2002 | P | 35 | 88.6 | 11.4 | 0.0 | 0.0 | 0.0 |
| E105 | 04.11.2002 | H | 33 | 87.9 | 12.1 | 0.0 | 0.0 | 0.0 |
| E081 | 09.09.2002 | H | 8 | 87.5 | 12.5 | 0.0 | 0.0 | 0.0 |
| E340 | 09.09.2002 | P | 31 | 87.1 | 12.9 | 0.0 | 0.0 | 0.0 |
| E059 | 03.07.2002 | P | 14 | 85.7 | 14.3 | 0.0 | 0.0 | 0.0 |
| E360 | 11.11.2002 | H | 7 | 85.7 | 0.0 | 0.0 | 14.3 | 0.0 |
| E060 | 03.07.2002 | H | 47 | 80.9 | 19.1 | 0.0 | 0.0 | 0.0 |
| E327 | 15.07.2002 | H | 15 | 80.0 | 20.0 | 0.0 | 0.0 | 0.0 |
| E355 | 21.10.2002 | H | 18 | 77.8 | 22.2 | 0.0 | 0.0 | 0.0 |
| E078 | 26.08.2002 | P | 64 | 71.9 | 26.6 | 1.6 | 0.0 | 0.0 |
| E006 | 10.06.2002 | H + P | 46 | 71.7 | 28.3 | 0.0 | 0.0 | 0.0 |
| E017 | 20.05.2002 | P | 54 | 68.5 | 31.5 | 0.0 | 0.0 | 0.0 |
| E365 | 18.11.2002 | H | 79 | 63.3 | 30.4 | 3.8 | 2.5 | 0.0 |
| E077 | 26.08.2002 | P | 24 | 62.5 | 4.2 | 16.7 | 4.2 | 12.5 |
| E361 | 11.11.2002 | H | 6 | 50.0 | 16.7 | 16.7 | 16.7 | 0.0 |
| E304 | 27.05.2002 | P | 19 | 42.1 | 57.9 | 0.0 | 0.0 | 0.0 |
| E353 | 14.10.2002 | P | 27 | 40.7 | 51.9 | 7.4 | 0.0 | 0.0 |
| E366 | 25.11.2002 | H | 32 | 40.6 | 43.8 | 9.4 | 0.0 | 6.3 |
One sample was taken per fecal pat.
H, cattle housed; P, cattle at pasture.
Duplicate testing of samples.
Five hundred forty-two samples were tested twice for E. coli O157 by the IMS procedure. The results of duplicate IMS testing are shown in Table 2. Twenty-seven samples gave a positive result on the first test, 23 gave a positive result on the second test, and 19 samples were positive on both tests. There was no statistically significant difference between the proportion of samples testing positive in the primary and secondary tests (P = 0.39, exact test for paired data). The level of agreement between tests on each sample was good, but the results were not always consistent (κ = 0.75; 95% confidence interval, 0.61 to 0.89).
TABLE 2.
Results of duplicate testing of fecal samples from cross-sectional farm survey for cattle shedding E. coli O157a
| Test 1 result | No. of samples with test 2 result of:
|
Total | |
|---|---|---|---|
| + | − | ||
| + | 19 | 8 | 27 |
| − | 4 | 511 | 515 |
| Total | 23 | 519 | 542 |
Paired exact test, P = 0.39; kappa statistic, 0.75; Pneg, 0.99; Ppos, 0.76.
Monte Carlo simulation.
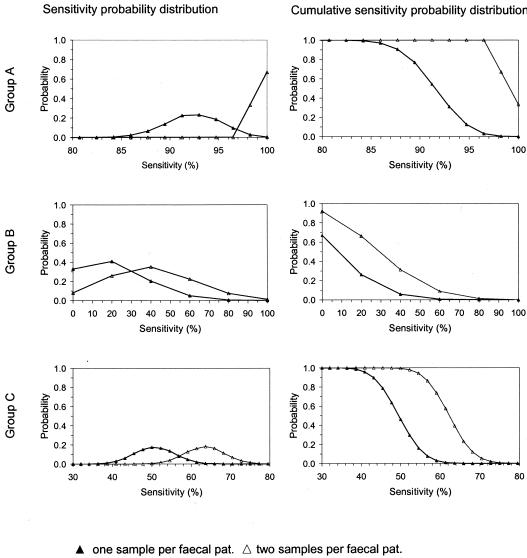
For groups A, B, and C, the results from Monte Carlo simulation predicted a higher sensitivity of E. coli O157 detection when separately testing 1 g of feces from each of two samples taken from different locations in a pat compared with testing 1 g of feces from only one sample per pat. The lower the sensitivity estimated by testing one sample per pat was, the higher the absolute and relative improvement in sensitivity predicted by testing two samples per pat was (Fig. 3). Group A had the highest predicted sensitivity, 93.0% (range, 80.7 to 100%), when only one sample was tested. When two samples were tested, the predicted sensitivity rose to 100% (range, 98.2 to 100%), representing an absolute increase in sensitivity of 7.0% and a relative increase of 7.5%. Group C had a predicted sensitivity of 50.0% (range, 34.1 to 70.5%) when only one sample was tested, midway between groups A and B. When two samples were tested, the predicted sensitivity rose to 63.6% (range, 45.5 to 84.1%), representing an absolute increase in sensitivity of 13.6% and a relative increase of 27.2%. Group B had the lowest predicted sensitivity, 20.0% (range, 0.0 to 100%), when only one sample was tested. When two samples were tested, the predicted sensitivity rose to 40% (range, 0.0 to 100%), an absolute increase in sensitivity of 20.0% and a relative increase of 100%.
FIG. 3.
Predicted sensitivity of detection of E. coli O157 in bovine fecal pats from groups A, B, and C when taking one or two samples per pat.
DISCUSSION
Our results show clearly that E. coli O157 is not evenly distributed in bovine feces and that the density of these bacteria can vary substantially between different sites in a fecal pat. Recent work demonstrating the principal location of E. coli O157 in the bovine gut (21) provides a likely explanation for these findings. With E. coli O157 located on the mucosal surface of the terminal rectum, there is limited opportunity for the organisms to mix homogenously through the feces. We are not aware of any studies on fecal dynamics, but we believe that as a fecal bolus is voided from an infected animal, the surface would be coated in mucus containing E. coli O157 and partial mixing would occur as the bolus deforms at the time of impact with the ground or some other surface. We do not know the extent to which motile strains migrate from pockets of contaminated mucus subsequent to fecal voiding.
Duplicate IMS testing of samples showed that negative IMS results are highly repeatable, presumably because when a negative result is returned, E. coli O157 is either absent or generally well below detectable levels. In contrast, positive IMS results are less repeatable. One possible explanation for the lower repeatability of positive IMS results is that the density of E. coli O157 can vary from a level undetectable by IMS to a level detectable by IMS even between two aliquots of feces taken from the same sample. A second possible explanation follows from E. coli O157 densities found in fecal samples from group C and the cross-sectional study. In samples from these studies in which E. coli O157 was present, the density was typically <100 CFU g−1. The manufacturer's documentation for Dynal IMS beads and work by Omisakin et al. (23) indicate that 100 CFU g−1 is the limit of reliable detection by IMS. It is possible, therefore, that lower repeatability of positive IMS results arises because the densities of E. coli O157 in IMS-positive samples are lower than 100 CFU g−1, the limit of reliable detection. Unfortunately, it is not possible to test this second possibility by using direct plating techniques, as the limit of enumeration with this method is 100 CFU g−1 feces (39). Dilution of feces with MRD at a ratio lower than 1:9 would hinder even dispersion of bacteria in the suspension. Plating a volume of suspension of >0.1 ml would leave agar plates too wet and risk smudging of colonies and overgrowth with organisms other than E. coli O157. A most-probable number technique could be used, but these techniques are very labor intensive.
With the exception of isolates from the cross-sectional survey, confirmation of the identity of E. coli O157 isolates in this study was limited to slide agglutination tests with anti-E. coli O157 antibody-coated latex reagent. During the course of our studies on E. coli O157 over the past 4.5 years, our laboratory staff have used this test on slightly over 1,900 putative E. coli O157 colonies, of which 1,012 tested positive. Colonies yielding a positive reaction were subcultured, and isolates were referred to the Scottish E. coli O157 Reference Laboratory for confirmation of species and serogroup. Only 7 submissions proved not to be E. coli O157. The specificity of this test in the hands of our laboratory staff therefore appears to be very high.
Storage and transport of samples are important issues in almost all field prevalence surveys of bovine fecal E. coli O157 shedding. Evidence from studies on the persistence of E. coli O157 in bovine fecal samples show that at 4 to 5°C, populations of E. coli O157 in bovine feces decline only slightly over 7 days (37). Our samples were rapidly refrigerated and processed within 48 h of collection.
The distribution of E. coli O157 in fecal samples is of practical significance. When we tested multiple samples from fecal pats, E. coli O157 was usually detectable in only a fraction of samples from positive pats. This indicates that some parts of positive pats contained either no E. coli O157 or undetectable numbers. Monte Carlo simulation showed that testing only a single 1-g sample of feces by IMS can result in a sensitivity of detection of as low as 20%. This low level of sensitivity was related to samples from cattle in group B, which comprised cows and young calves, the age group of cattle in which the prevalence of E. coli O157 is usually lowest (14, 26, 39). In fecal samples from group C and the cross-sectional study, we found that when E. coli O157 was present, the density was typically <100 CFU g−1. Because of the uneven distribution and typically low density of E. coli O157 in fecal pats and the consequent likelihood of some fecal samples with E. coli O157 yielding negative results, if a single 1-g sample is analyzed per fecal pat, bovine shedding of E. coli O157 is less likely to be detected on farms with few shedding cattle than on farms with many shedding cattle. For the 37 farms in our cross-sectional survey found to have pats containing E. coli O157, it was usual for less than 20% of pats to be positive and half of the farms were declared positive based on one or two positive pats only. A corollary of these findings is that most surveys to date are likely to have greatly underestimated the prevalence of E. coli O157 shedding in cattle and the proportion of farms with shedding cattle.
In six of the 27 E. coli O157 prevalence surveys we reviewed, samples were taken from fecal pats (8, 11, 12, 18, 28, 32). We assessed these studies to see whether we could apply our results to adjust estimates of prevalence. Regrettably, the extraordinary variety in the type and size of samples taken and the differences in test procedures would have made retrospective adjustment of survey results by using our findings inappropriate. Standardization of sampling and test procedures for E. coli O157 surveys is therefore highly desirable to permit comparison of results and to allow the application of correction factors.
Our results show that substantial improvement to the detection of E. coli O157 in bovine fecal pats can be achieved by increasing the number of samples tested per pat. Increasingly, surveys are testing single fecal samples containing more than 1 g of feces; 10 of the recent surveys we reviewed tested single 10- to 25-g fecal samples rather than swabs or single 1-g samples (3, 7, 9, 13-15, 18, 23, 34, 39). Overall, this probably does increase the survey sensitivity, but paradoxically, it may also reduce the chance of detection in fecal samples if the additional feces is free of E. coli O157 and merely dilutes a previously detectable density of E. coli O157 to an undetectable density. Research is required to determine what effect testing larger single samples has on the sensitivity of detection. An alternative is to test multiple small fecal samples, since we have shown that testing more than one sample can lead to a marked improvement in the sensitivity of detection. We recommend that, within practicable limits, surveys of E. coli O157 prevalence in cattle should maximize the number of samples tested per fecal sampling unit to increase the accuracy of prevalence estimates, but we recognize the not inconsiderable penalty in laboratory time and increased consumable costs incurred by doing so.
Acknowledgments
We thank Dominic Mellor for assistance in the collection of fecal samples and Roger Humphry for advice and assistance in the preparation of this paper.
This study is a part of the International Partnership Research Award in Veterinary Epidemiology (IPRAVE), “Epidemiology and Evolution of Enterobacteriaceae Infections in Humans and Domestic Animals,” funded by the Wellcome Trust. The Scottish Agricultural College receives financial support from the Scottish Executive Environment and Rural Affairs Department (SEERAD).
REFERENCES
- 1.Besser, T. E., D. D. Hancock, L. C. Pritchett, E. M. McRae, D. H. Rice, and P. I. Tarr. 1997. Duration of detection of fecal excretion of Escherichia coli O157 in cattle. J. Infect. Dis. 175:726-729. [DOI] [PubMed] [Google Scholar]
- 2.Blanco, M., J. E. Blanco, J. Blanco, E. A. Gonzalez, A. Mora, C. Prado, L. Fernandez, M. Rio, J. Ramos, and M. P. Alonso. 1996. Prevalence and characteristics of Escherichia coli serotype O157:H7 and other verotoxin-producing E. coli in healthy cattle. Epidemiol. Infect. 117:251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonardi, S., E. Maggi, A. Bottarelli, M. L. Pacciarini, A. Ansuini, G. Vellini, S. Morabito, and A. Caprioli. 1999. Isolation of verocytotoxin-producing Escherichia coli O157:H7 from cattle at slaughter in Italy. Vet. Microbiol. 67:203-211. [DOI] [PubMed] [Google Scholar]
- 4.Chapman, P. A., C. A. Siddons, P. M. Zadik, and L. Jewes. 1991. An improved selective medium for the isolation of Escherichia coli O157. J. Med. Microbiol. 35:107-110. [DOI] [PubMed] [Google Scholar]
- 5.Chapman, P. A., D. J. Wright, and C. A. Siddons. 1994. A comparison of immunomagnetic separation and direct culture for the isolation of verocytotoxin-producing Escherichia coli O157 from bovine faeces. J. Med. Microbiol. 40:424-427. [DOI] [PubMed] [Google Scholar]
- 6.Cicchetti, D. V., and A. R. Feinstein. 1990. High agreement but low kappa. II. Resolving the paradoxes. J. Clin. Epidemiol. 43:551-558. [DOI] [PubMed] [Google Scholar]
- 7.Faith, N. G., J. A. Shere, R. Brosch, K. W. Arnold, S. E. Ansay, M.-S. Lee, J. B. Luchansky, and C. W. Kaspar. 1996. Prevalence and clonal nature of Escherichia coli O157:H7 on dairy farms in Wisconsin. Appl. Environ. Microbiol. 62:1519-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galland, J. C., D. R. Hyatt, S. S. Crupper, and D. W. Acheson. 2001. Prevalence, antibiotic susceptibility, and diversity of Escherichia coli O157:H7 isolates from a longitudinal study of beef cattle feedlots. Appl. Environ. Microbiol. 67:1619-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gannon, V. P. J., T. A. Graham, R. King, P. Michel, S. Read, K. Ziebell, and R. P. Johnson. 2002. Escherichia coli O157:H7 infection in cows and calves in a beef cattle herd in Alberta, Canada. Epidemiol. Infect. 129:163-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock, D. D., T. E. Besser, M. L. Kinsel, P. I. Tarr, D. H. Rice, and M. G. Paros. 1994. The prevalence of Escherichia coli O157:H7 in dairy and beef cattle in Washington state. Epidemiol. Infect. 113:199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hancock, D. D., T. E. Besser, D. H. Rice, D. E. Herriott, and P. I. Tarr. 1997. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiol. Infect. 118:193-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hancock, D. D., D. H. Rice, L. A. Thomas, D. A. Dargatz, and T. E. Besser. 1997. Epidemiology of Escherichia coli O157:H7 in feedlot cattle. J. Food Prot. 60:462-465. [DOI] [PubMed] [Google Scholar]
- 13.Heuvelink, A. E., F. L. A. M. Van den Biggelaar, E. De Boer, R. G. Herbes, W. J. G. Melchers, J. H. J. Huis In 'T Veld, and L. A. H. Monnens. 1998. Isolation and characterization of verocytotoxin-producing Escherichia coli O157 strains from Dutch cattle and sheep. J. Clin. Microbiol. 36:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heuvelink, A. E., F. L. A. M. Van den Biggelaar, J. T. M. Zwartkruis-Nahuis, R. G. Herbes, R. Huyben, N. Nagelkerke, W. J. G. Melchers, L. A. H. Monnens, and E. De Boer. 1998. Occurrence of verocytotoxin-producing Escherichia coli O157 on Dutch dairy farms. J. Clin. Microbiol. 36:3480-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laegreid, W. W., R. O. Elder, and J. E. Keen. 1999. Prevalence of Escherichia coli O157:H7 in range beef calves at weaning. Epidemiol. Infect. 123:291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lahti, E., M. Keskimäki, L. Rantala, P. Hyvönen, A. Siitonen, and T. Honkanen-Buzalski. 2001. Occurrence of Escherichia coli O157 in Finnish cattle. Vet. Microbiol. 79:239-251. [DOI] [PubMed] [Google Scholar]
- 17.Lahti, E., O. Ruoho, L. Rantala, M.-L. Hänninen, and T. Honkanen-Buzalski. 2003. Longitudinal study of Escherichia coli O157 in a cattle finishing unit. Appl. Environ. Microbiol. 69:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LeJeune, J. T., T. E. Besser, D. H. Rice, J. L. Berg, R. P. Stilborn, and D. D. Hancock. 2004. Longitudinal study of fecal shedding of Escherichia coli O157:H7 in feedlot cattle: predominance and persistence of specific clonal types despite massive cattle population turnover. Appl. Environ. Microbiol. 70:377-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mechie, S. C., P. A. Chapman, and C. A. Siddons. 1997. A fifteen month study of Escherichia coli O157:H7 in a dairy herd. Epidemiol. Infect. 118:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyer-Broseta, S., S. N. Bastian, P. D. Arné, O. Cerf, and M. Sanaa. 2001. Review of epidemiological surveys on the prevalence of contamination of healthy cattle with Escherichia coli serogroup O157:H7. Int. J. Hyg. Environ. Health 203:347-361. [DOI] [PubMed] [Google Scholar]
- 21.Naylor, S. W., J. C. Low, T. E. Besser, A. Mahajan, G. J. Gunn, M. C. Pearce, I. J. McKendrick, D. G. E. Smith, and D. L. Gally. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect. Immun. 71:1505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neill, M. A., P. I. Tarr, C. R. Clausen, D. L. Christie, and R. O. Hickman. 1987. Escherichia coli O157:H7 as the predominant pathogen associated with the hemolytic uremic syndrome—a prospective study in the Pacific northwest. Pediatrics 80:37-40. [PubMed] [Google Scholar]
- 23.Omisakin, F., M. MacRae, I. D. Ogden, and N. J. C. Strachan. 2003. Concentration and prevalence of Escherichia coli O157 in cattle feces at slaughter. Appl. Environ. Microbiol. 69:2444-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pai, C. H., N. Ahmed, H. Lior, W. M. Johnson, H. V. Sims, and D. E. Woods. 1988. Epidemiology of sporadic diarrhea due to verocytotoxin-producing Escherichia coli: a two-year prospective study. J. Infect. Dis. 157:1054-1057. [DOI] [PubMed] [Google Scholar]
- 25.Paiba, G. A., J. C. Gibbens, S. J. S. Pascoe, J. W. Wilesmith, S. A. Kidd, C. Byrne, J. B. M. Ryan, R. P. Smith, I. M. McLaren, R. J. Futter, A. C. S. Kay, Y. E. Jones, S. A. Chappell, G. A. Willshaw, and T. Cheasty. 2002. Faecal carriage of verocytotoxin-producing Escherichia coli O157 in cattle and sheep at slaughter in Great Britain. Vet. Rec. 150:593-598. [DOI] [PubMed] [Google Scholar]
- 26.Paiba, G. A., J. W. Wilesmith, S. J. Evans, S. J. S. Pascoe, R. P. Smith, S. A. Kidd, J. B. M. Ryan, I. M. McLaren, S. A. Chappell, G. A. Willshaw, T. Cheasty, N. P. French, T. W. H. Jones, H. F. Buchanan, D. J. Challoner, A. D. Colloff, M. P. Cranwell, R. G. Daniel, I. H. Davies, J. P. Duff, R. A. T. Hogg, F. D. Kirby, M. F. Millar, R. J. Monies, M. J. Nicholls, and J. H. Payne. 2003. Prevalence of faecal excretion of verocytotoxigenic Escherichia coli O157 in cattle in England and Wales. Vet. Rec. 153:347-353. [DOI] [PubMed] [Google Scholar]
- 27.Pearce, M. C., C. Jenkins, L. Vali, A. W. Smith, H. I. C. T. Knight, H. R. Smith, G. J. Gunn, M. E. J. Woolhouse, S. G. B. Amyes, and G. Frankel. 2004. Temporal shedding patterns and virulence factors of Escherichia coli serogroups O26, O103, O111, O145, and O157 in a cohort of beef calves and their dams. Appl. Environ. Microbiol. 70:1708-1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renter, D. G., J. M. Sargeant, R. D. Oberst, and M. Samadpour. 2003. Diversity, frequency, and persistence of Escherichia coli O157 strains from range cattle environments. Appl. Environ. Microbiol. 69:542-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice, D. H., E. D. Ebel, D. D. Hancock, T. E. Besser, D. E. Herriott, and L. V. Carpenter. 1997. Escherichia coli O157 in cull dairy cows at farm and at slaughter. J. Food Prot. 60:1386-1387. [DOI] [PubMed] [Google Scholar]
- 30.Richards, M. S., J. D. Corkish, A. R. Sayers, I. M. McLaren, S. J. Evans, and C. Wray. 1998. Studies of the presence of verocytotoxic Escherichia coli O157 in bovine faeces submitted for diagnostic purposes in England and Wales and on beef carcasses in abattoirs in the United Kingdom. Epidemiol. Infect. 120:187-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riley, L. W., R. S. Remis, S. D. Helgerson, H. B. McGee, J. G. Wells, B. R. Davis, R. J. Herbert, E. S. Olcott, L. M. Johnson, N. T. Hargrett, P. A. Blake, and M. L. Cohen. 1983. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 308:681-685. [DOI] [PubMed] [Google Scholar]
- 32.Sargeant, J. M., M. W. Sanderson, R. A. Smith, and D. D. Griffin. 2003. Escherichia coli O157 in feedlot cattle feces and water in four major feeder-cattle states in the USA. Prev. Vet. Med. 61:127-135. [DOI] [PubMed] [Google Scholar]
- 33.Stokes, M. E., C. S. Davis, and G. G. Koch. 2000. Categorical data analysis using the SAS system. SAS Institute, Cary, N.C.
- 34.Tutenel, A. V., D. Pierard, J. Uradzinski, E. Jozwik, M. Pastuszczak, J. Van Hende, M. Uyttendaele, J. Debevere, T. Cheasty, J. Van Hoof, and L. De Zutter. 2002. Isolation and characterization of enterohaemorrhagic Escherichia coli O157:H7 from cattle in Belgium and Poland. Epidemiol. Infect. 129:41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tutenel, A. V., D. Pierard, D. Vandekerchove, J. Van Hoof, and L. De Zutter. 2003. Sensitivity of methods for the isolation of Escherichia coli O157 from naturally infected bovine faeces. Vet. Microbiol. 94:341-346. [DOI] [PubMed] [Google Scholar]
- 36.Vold, L., B. Klungseth Johansen, H. Kruse, E. Skjerve, and Y. Wasteson. 1998. Occurrence of shigatoxinogenic Escherichia coli O157 in Norwegian cattle herds. Epidemiol. Infect. 120:21-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, G. D., G. T. Zhao, and M. P. Doyle. 1996. Fate of enterohemorrhagic Escherichia coli O157:H7 in bovine feces. Appl. Environ. Microbiol. 62:2567-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yilmaz, A., H. Gun, and H. Yilmaz. 2002. Frequency of Escherichia coli O157:H7 in Turkish cattle. J. Food Prot. 65:1637-1640. [DOI] [PubMed] [Google Scholar]
- 39.Zhao, T., M. P. Doyle, J. Shere, and L. Garber. 1995. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 61:1290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]