Abstract
The New Zealand native legume flora are represented by four genera, Sophora, Carmichaelia, Clianthus, and Montigena. The adventive flora of New Zealand contains several legume species introduced in the 19th century and now established as serious invasive weeds. Until now, nothing has been reported on the identification of the associated rhizobia of native or introduced legumes in New Zealand. The success of the introduced species may be due, at least in part, to the nature of their rhizobial symbioses. This study set out to address this issue by identifying rhizobial strains isolated from species of the four native legume genera and from the introduced weeds: Acacia spp. (wattles), Cytisus scoparius (broom), and Ulex europaeus (gorse). The identities of the isolates and their relationship to known rhizobia were established by comparative analysis of 16S ribosomal DNA, atpD, glnII, and recA gene sequences. Maximum-likelihood analysis of the resultant data partitioned the bacteria into three genera. Most isolates from native legumes aligned with the genus Mesorhizobium, either as members of named species or as putative novel species. The widespread distribution of strains from individual native legume genera across Mesorhizobium spp. contrasts with previous reports implying that bacterial species are specific to limited numbers of legume genera. In addition, four isolates were identified as Rhizobium leguminosarum. In contrast, all sequences from isolates from introduced weeds aligned with Bradyrhizobium species but formed clusters distinct from existing named species. These results show that native legume genera and these introduced legume genera do not have the same rhizobial populations.
Rhizobia are soil-inhabiting bacteria that form symbiotic relationships with plant legume species in root nodules. The bacteria fix nitrogen from the atmosphere to form ammonia, which is assimilated by the plant. This relationship has been exploited by agriculture to enhance legume crop growth without the addition of nitrogen-containing fertilizers. For this reason, the majority of research in this field has focused on the herbaceous crop legumes of agricultural significance. In contrast, few studies have been made of rhizobial associations among noncrop legumes, despite the fact that they may be important in ecological interactions.
Worldwide, there are an estimated 17,000 to 19,000 legume species (19). However, symbiotic bacterial species have only been identified for a small proportion of these. To date, 45 symbiotic nodulating bacterial species have been identified in 10 genera: Azorhizobium, Blastobacter, Bradyrhizobium, Burkholderia, Devosia, Mesorhizobium, Methylobacterium, Ralstonia, Rhizobium, and Sinorhizobium (28). Most of the species are in the genera Rhizobium, Azorhizobium, Bradyrhizobium, Mesorhizobium, and Sinorhizobium and are related to one another in the order Rhizobiales, with Rhizobium and Sinorhizobium in the family Rhizobiaceae, and Mesorhizobium in the Phyllobacteriaceae (28).
The symbiotic relationships of nitrogen-fixing members of the Rhizobiaceae have been intensively studied (for reviews, see references 5, 6, 15, and 16). Some of these rhizobial species are reported to form host specific associations with particular legumes. For example, Rhizobium leguminosarum biovar trifolii is considered specific to clover (Trifolium spp.) (12), and Rhizobium galegae is reported only on goat's rue (Galega spp.) (16). Many other Rhizobium spp. and Mesorhizobium spp. appear to be more or less “promiscuous,” nodulating more than one plant genus, and most nodulate two or more plant genera (5, 16). Sinorhizobium fredii strain NGR234 forms rhizobial associations with 232 species in 112 genera (12, 26, 27). Generally, rhizobia of herbaceous host species are reported to be more promiscuous than those of woody legumes (24). Despite more than a century of research, the host ranges for rhizobial species are known for fewer than 200 plant species, most being crop, forage, or grain legumes (16, 17).
New Zealand has a number of native (naturally occurring) woody legumes comprising 34 species in the genera Carmichaelia, Clianthus, Montigena, and Sophora (9, 10). There are also over 100 naturalized woody legume species that were introduced into New Zealand since colonization by Europeans in the 19th century. These include the woody legumes Cytisus spp. (brooms), gorse (Ulex europaeus) from Europe, and various Acacia and Albizia spp. (wattles) from Australia (21), introduced as ornamental or hedge plants. In their native habitats, these shrubs are in equilibrium with their natural flora, but in New Zealand, they have become serious invasive noxious weeds. The New Zealand native legumes, together with broom and gorse, are members of the subfamily Faboideae (Papilionoideae), distinct from Acacia and Albizia in the Mimosoideae.
The ability of legume plants to become established in soils of low fertility and to compete successfully with other plants can be attributed in part to the symbiotic associations that give them the capacity to fix atmospheric nitrogen. The plant-rhizobial association usually forms immediately following germination if the nodulating rhizobia are present naturally in the soil. Because New Zealand became geographically isolated about 80 million years ago (29), it is postulated that the native legume genera coevolved with nitrogen-fixing bacterial symbionts in isolation from the regions of major legume evolution. However, the source of rhizobial symbionts of introduced legumes is unknown. Either these rhizobia were introduced at the same time as the plants, or the plants were able to use indigenous rhizobia associated with native legumes. Alternatively, they made use of a diazotrophic bacterial population preexisting in New Zealand soils.
The primary objective of this research was to identify the rhizobial symbionts of both native and introduced New Zealand legumes and to determine whether these are putatively indigenous or cosmopolitan strains. To this end, rhizobial isolates have been obtained from native and introduced legumes and sets of gene sequences from them were compared in order to establish the identity and relationships of the bacteria to their legume hosts.
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains (Table 1) were either obtained from the International Collection of Microorganisms from Plants (ICMP, Landcare Research, Auckland, New Zealand; http://www.landcareresearch.co.nz/research/biodiversity/fungiprog/icmp.asp) or directly isolated from wild plants (usually as young seedlings). Nodules of native legumes were obtained from pristine sites on conservation lands, distant from agricultural plantings, throughout the country. Introduced legume samples were obtained from arable lands or from conservation lands.
TABLE 1.
Genomic grouping of rhizobia isolated from native and introduced plants in New Zealand
| Legumes | ICMP no. | Host legume | Genomic group | GenBank accession no.a
|
|||
|---|---|---|---|---|---|---|---|
| 16S rDNA | atpD | glnII | recA | ||||
| Native | 11727 | Carmichaelia australis | E | AY491060 | ND | ND | ND |
| 12687 | Carmichaelia australis | E | AY491061 | ND | ND | ND | |
| 13190 | Carmichaelia australis | D | AY491071 | AY493456 | AY494808* | AY494822 | |
| 15054 | Carmichaelia australis | A | AY491068 | ND | ND | ND | |
| 11708 | Carmichaelia nana | D | AY491073 | AY493454 | AY494792 | AY494818 | |
| 11722 | Carmichaelia nana | D | AY491072 | AY493458 | AY494810 | AY494814 | |
| 14319 | Carmichaelia odorata | D | AY491074 | AY493459 | AY494812 | AY494817 | |
| 12635 | Carmichaelia petriei | D | AY491075 | AY493457 | AY494811 | AY494815 | |
| 12649 | Carmichaelia petriei | A | AY491064 | ND | ND | ND | |
| 11541 | Clianthus puniceus | D | AY491070 | AY493455 | AY494809 | AY494821 | |
| 11542 | Clianthus puniceus | E | AY491059 | ND | ND | ND | |
| 11721 | Clianthus puniceus | C | AY491077 | ND | ND | ND | |
| 11726 | Clianthus puniceus | C | AY491078 | ND | ND | ND | |
| 12685 | Montigena novae-zelandiae | B | AY491069 | AY493452 | AY494793 | AY494823 | |
| 12690 | Montigena novae-zelandiae | D | AY491076 | ND | ND | ND | |
| 14642 | Sophora chathamica | E | AY491062 | AY493451 | AY494795 | AY494813 | |
| 11736 | Sophora microphylla | A | AY491063 | ND | ND | ND | |
| 12637 | Sophora microphylla | A | AY491066 | ND | ND | ND | |
| 14330 | Sophora microphylla | A | AY491067 | AY493461 | AY494806 | AY494820 | |
| 11719 | Sophora tetraptera | A | AY491065 | X | AY494805 | AY494819 | |
| Introduced | 12835 | Acacia dealbata | G | AY491090 | AY493444 | AY494799 | AY494826 |
| 14754 | Acacia longifolia | G | AY491094 | AY493448 | AY494801 | AY494832 | |
| 14755 | Acacia longifolia | G | AY491089 | AY493449 | AY494802 | AY494828 | |
| 14752 | Albizia julibrissin | J | AY491081 | AY493443 | AY494798 | AY494830 | |
| 14753 | Albizia julibrissin | F | AY491082 | AY493447 | AY494797 | AY494831 | |
| 12624 | Cytisus scoparius | J | AY491079 | ND | ND | ND | |
| 14291 | Cytisus scoparius | J | AY491084 | AY493442 | AY494796 | AY494829 | |
| 14309 | Cytisus scoparius | J | AY491087 | ND | ND | ND | |
| 14310 | Cytisus scoparius | J | AY491088 | ND | ND | ND | |
| 14328 | Cytisus scoparius | J | AY491086 | ND | ND | ND | |
| 12674 | Ulex europaeus | I | AY491080 | ND | ND | ND | |
| 14292 | Ulex europaeus | J | AY491085 | ND | ND | ND | |
| 14304 | Ulex europaeus | H | AY491091 | ND | ND | ND | |
| 14306 | Ulex europaeus | J | AY491083 | ND | ND | ND | |
| 14320 | Ulex europaeus | I | AY491092 | ND | ND | ND | |
| 14533 | Ulex europaeus | G | AY491093 | AY493445 | AY494800 | AY494827 | |
It was not always possible to obtain both sequences for glnII. * indicates relative position of the missing section. ND, not determined. X, PCR amplification failed.
Bacterial isolation.
Root nodules were dissected from roots, rinsed thoroughly in water and Tween 80 (0.001%), surface sterilized by immersion in a 5% solution of commercial sodium hypochlorite (3% active hypochlorite) for 30 min, and rinsed in sterile water. Nodules were individually comminuted, and the suspension was streaked onto surface-dried yeast mannitol agar plates (YMA; 10 g of active dried baker's yeast, 10 g of mannitol [Sigma], 2.5 g of peptone [Difco], 15 g of agar, 1 liter of deionized water). Plates were incubated at 28°C for 3 to 10 days. Individual colonies were selected, restreaked onto YMA plates, and subcultured onto slopes of YMA supplemented with 3 g of calcium carbonate and 1.5 g of calcium gluconate per liter for short-term storage at 8°C. The strains used and reported in this study were deposited in the ICMP.
DNA extraction, PCR amplification, and sequencing.
DNA was isolated from bacterial cultures with a standard phenol-chloroform extraction method (2). Primers for PCR amplification, with their sources and sequences, are shown in Table 2. All amplification conditions were performed as specified by their authors. Either 16S-1F or 16S-PB36 was used as the forward primer for the 16S ribosomal DNA (rDNA) sequence. No recA PCR products for Bradyrhizobium spp. were obtained with the published recA primers (7).
TABLE 2.
Primers used in this study
| Primer | Sequence | Target gene | Reference |
|---|---|---|---|
| 16S-1F | AGCGGCGGACGGGTGAGTAATG | 11 | |
| 16S-485F | CAGCAGCCGCGGTAA | 11 | |
| 16S-1100R | GGGTTGCGCTCGTTG | 16S rDNA | 11 |
| 16S-1509R | AAGGAGGGGATCCAGCCGCA | 11 | |
| 16S-PB36 | AGRGTTTGATCMTGGCTCAG | 4 | |
| atpD-273F | SCTGGGSCGYATCMTGAACGT | 7 | |
| atpD-771R | GCCGACACTTCCGAACCNGCCTG | ATP synthase beta-subunit | 7 |
| atpD-294F | ATCGGCGAGCCGGTCGACGA | 7 | |
| GSII-1 | AACGCAGATCAAGGAATTCG | 34 | |
| GSII-2 | ATGCCCGAGCCGTTCCAGTC | 34 | |
| GSII-3 | AGRTYTTCGGCAAGGGYTC | Glutamine synthetase II | 34 |
| GSII-4 | GCGAACGATCTGGTAGGGGT | 34 | |
| recA-6F | CGKCTSGTAGAGGAYAAATCGGTGGA | 7 | |
| recA-555R | CGRATCTGGTTGATGAAGATCACCAT | 7 | |
| recA-63F | ATCGAGCGGTCGTTCGGCAAGGG | DNA recombinase A | 7 |
| recA-504R | TTGCGCAGCGCCTGGCTCAT | 7 | |
| recA-BF | CGTACTGTCGAAGGTTCTTCCATGGA | This study |
After aligning the sequences, it was found that the forward primer was placed in a variable region of the gene. A new forward primer, recA-BF, was designed based on the recA sequence from the related bacterium Rhodopseudomonas palustris (GenBank accession no. D84467). With this primer, recA PCR products were obtained for all New Zealand Bradyrhizobium strains and the type strain of Bradyrhizobium liaoningense. However, no PCR product was obtained from Bradyrhizobium elkanii or Bradyrhizobium japonicum. PCR amplifications were performed with an Applied Biosystems 9700 thermal cycler. PCR products were column purified with a Roche High Pure PCR product purification kit. Purified products were cycle sequenced with the appropriate primers with BigDye terminator ready reaction mix (ABI) (version 3.0 or 3.1), and sequences were obtained in both directions with an ABI 310 Prism genetic analyzer. Sequences were assembled and edited with Sequencher 3.11 (Gene Codes Corp.).
Phylogenetic analysis.
Nucleotide alignments were constructed with ClustalX 1.83 (19) and edited manually with GeneDoc 2.6.02 (33). The four primers for glnII amplify two overlapping sections. However, when one of the two sequences for some glnII genes was not amplified, the standard method of replacing missing data with the symbol “?” was used. The alignments were checked for chimeras with the Bellerophon server (http://foo.maths.uq.edu.au/∼huber/bellerophon.pl) (13). None were found. GenBank sequences from the type strains of representative species from Mesorhizobium, Rhizobium, and Sinorhizobium were also included for comparison (Table 3). The outgroup for each alignment of 16S rDNA, atpD, and recA was the appropriate sequence from Caulobacter crescentus strain CB15, obtained from the complete genome (GenBank accession no. NC_002696) in which homologues for sequences were found. There is no outgroup for glnII because there is too little homology between the glutamine synthase II gene of the rhizobia and other taxa that could act as an appropriate outgroup.
TABLE 3.
Type strains of Mesorhizobium spp., Rhizobium spp., Sinorhizobium spp., and Caulobacter crescentus, showing the GenBank records of sequences used in analyses
Type strain indicated by T; —, strain not present in the ICMP.
Italicized sequences were obtained from GenBank. It was not always possible to obtain both sequences for glnII. *, relative position of the missing section; X, PCR amplification failed; —, sequence data not available.
Preliminary analyses were performed with the ClustalX and PAUP* neighbor-joining methods, with 1,000 bootstrap replicates. Maximum likelihood was compared with neighbor joining and was chosen as the preferred method of analysis because, although more computationally demanding, the assumptions of this model are more rigorous (4). The appropriate maximum-likelihood parameters were selected with the application MODELTEST, version 3.5 (25). This computer program tests nucleotide alignments against 56 models of DNA evolution with maximum likelihood. The resultant negative log likelihood (−lnL) scores and associated parameters were subjected to a hierarchical likelihood ratio test to determine which model best fitted the sequence data. In the 16S rDNA analysis, the TrN+I+Γ model of DNA evolution was selected. In the atpD, glnII, and recA analyses, the GTR+I+Γ model of DNA evolution was selected. The model parameters (base frequencies, proportion of invariable sites, gamma distribution shape parameter, and substitution rate matrix) were then specified in PAUP* 4.0b10 (32) to build phylograms with tree-bisection-reconnection heuristics.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences reported in this study are AY491059 to AY491094 (16S rDNA), AY493442 to AY493461 (atpD), AY494791 to AY494812 (glnII), and AY494813 to AY494833 (recA).
RESULTS
The 16S rDNA sequences were obtained from all isolates. Sequences were obtained for the atpD, glnII, and recA genes from at least one isolate from each of the 14 legume species considered. Sequences of glnII could only be partially amplified for four strains: Mesorhizobium plurifarium and strain ICMP 14753 (48% sequence coverage obtained) and Mesorhizobium amorphae and strain ICMP 13190 (67% sequence coverage obtained). Gaps were treated as missing data as described in Materials and Methods.
Maximum-likelihood and neighbor-joining trees for each gene had similar overall tree topologies (neighbor joining trees, with bootstrap values, are available as supplementary data at http://www.rhizobia.co.nz/papers.html). Groups were selected on the basis of the minimum standard changes between named species in the 16S rDNA phylogram (Fig. 1), and all groups were well supported in both maximum-likelihood and neighbor-joining analyses except for the monotypic groups H and F, which had less than 50% bootstrap support in the neighbor-joining tree. The sequences from rhizobia isolated from New Zealand legumes are distributed in 10 genomic groups (A to J). Sequences from native legumes, Carmichaelia, Clianthus, Montigena, and Sophora spp., were distributed in groups A to D, together with the reference sequences representing Mesorhizobium spp. Other sequences from native legumes also formed a clade (group E) with Rhizobium leguminosarum. All rhizobia isolated from introduced legumes, Acacia, Albizia, Cytisus, and Ulex spp., were in the Bradyrhizobium clade (groups F to J).
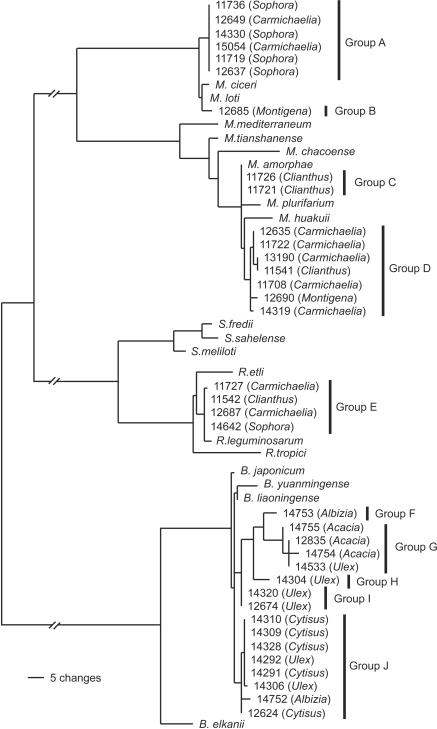
FIG. 1.
Maximum-likelihood tree based on 16S rDNA gene sequence data (1,321 bp), showing the relationships of rhizobial isolates from New Zealand legume flora to type strains of rhizobia. The isolate names and the accession number in the ICMP culture collection are shown. The genus of the legume from which the bacteria were isolated is shown in parentheses. Genomic grouping is shown by the vertical bars. The value of −lnL for this tree is 4,565.84.
Group A comprised strains 14330 (Sophora sp.), 11719 (Sophora sp.), 11736 (Sophora sp.), 12637 (Sophora sp.), 12649 (Carmichaelia sp.), and 15054 (Carmichaelia sp.). These strains were most closely grouped to Mesorhizobium ciceri and Mesorhizobium loti.
Group B comprised a single strain 12685 (Montigena sp.) as an outgroup to Mesorhizobium ciceri and Mesorhizobium loti.
Group C comprised 11726 and 11721, both from Clianthus. These strains were most closely grouped to Mesorhizobium amorphae.
Group D comprised strains 11708 (Carmichaelia sp.), 14319 (Carmichaelia sp.), 12690 (Montigena sp.), 12635 (Carmichaelia sp.), 13190 (Carmichaelia sp.), 11722 (Carmichaelia sp.), and 11541 (Clianthus sp.). These strains were most closely grouped to Mesorhizobium huakuii.
Group E comprised 14642 (Sophora sp.), 12687 (Carmichaelia sp.), 11542 (Clianthus sp.), and 11727 (Carmichaelia sp.). These strains were members of the Rhizobium leguminosarum clade.
Group F comprised the single isolate 14753 (Albizia sp.).
Group G comprised 14754 (Acacia sp.), 14755 (Acacia sp.), 12835 (Acacia sp.) and 14533 (Ulex sp.), an outlier group to Bradyrhizobium liaoningense and Bradyrhizobium yuanmingense.
Group H comprised the single isolate 14304 (Ulex sp.).
Group I comprised isolates 14320 (Ulex sp.) and 12674 (Ulex sp.).
Group J comprised isolates 14309 (Cytisus sp.), 14310 (Cytisus sp.), 14291 (Cytisus sp.), 14328 (Cytisus sp.), 14292 (Ulex sp.), 14306 (Ulex sp.), 14752 (Albizia sp.), and 12624 (Cytisus sp.).
The trees with the other partial gene sequences (atpD, glnII, and recA) all concurred with the 16S gene tree with respect to placement of the strains into the three genera, Rhizobium, Mesorhizobium, and Bradyrhizobium (Fig. 2, 3, and 4). However, the branching order of the individual sequences in groups A to J represented by 16S rDNA differed from that represented by the 16S rDNA tree.
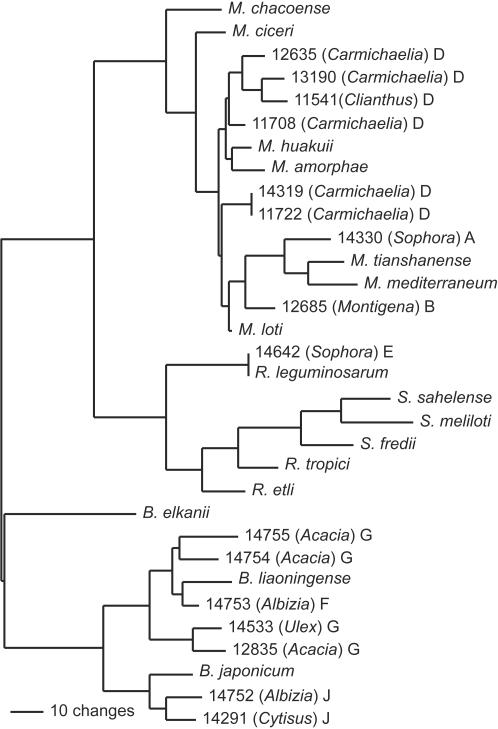
FIG. 2.
Maximum-likelihood tree based on atpD gene sequence data (464 bp), showing the relationships of rhizobial isolates from New Zealand legume flora to type strains of rhizobia. The isolate names and the accession number in the ICMP culture collection are shown. The genus of the legume from which the bacteria were isolated is shown in parentheses. The letter following the parentheses indicates the genomic grouping as defined by the 16S rDNA data. The value of −lnL for this tree is 3,633.07.
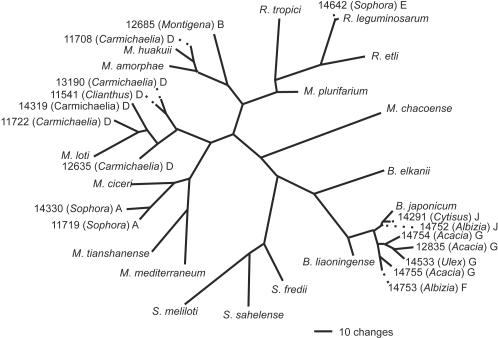
FIG. 3.
Maximum-likelihood tree based on glnII gene sequence data (828 bp), showing the relationships of rhizobial isolates from New Zealand legume flora to type strains of rhizobia. The isolate names and the accession number in the ICMP culture collection are shown. The genus of the legume from which the bacteria were isolated is shown in parentheses. The letter following the parentheses indicates the genomic grouping as defined by the 16S rDNA data. The value of −lnL for this tree is 6,404.27.
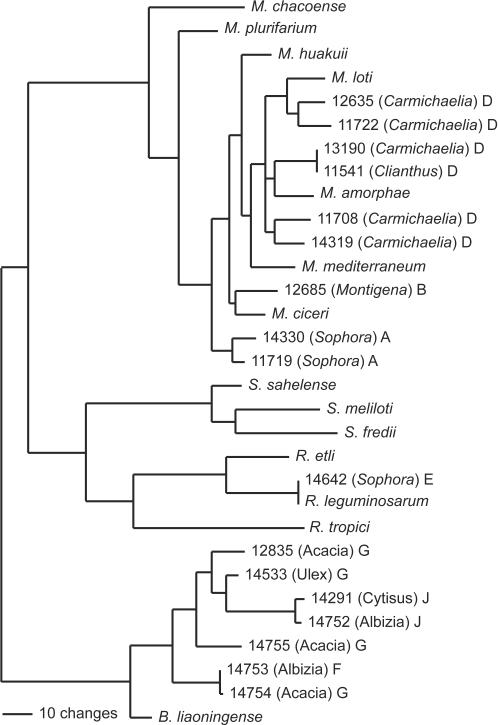
FIG. 4.
Maximum-likelihood tree based on recA gene sequence data (828 bp), showing the relationships of rhizobial isolates from New Zealand legume flora to type strains of rhizobia. The isolate names and the accession number in the ICMP culture collection are shown. The genus of the legume from which the bacteria were isolated is shown in parentheses. The letter following the parentheses indicates the genomic grouping as defined by the 16S rDNA data. The value of −lnL for this tree is 4,114.36.
A comparison of individual base variations in 16S rDNAs between sequences within the different groups showed that there were no base differences in group A (100% similarity), 4 bases (99.7% similarity) for group D in the Mesorhizobium spp., 4 bases (99.7% similarity) within group E (R. leguminosarum), and 27 bases (98.0% similarity) for all strains within the Bradyrhizobium clade.
DISCUSSION
Rhizobial isolates of the three most common and geographically widespread species of Carmichaelia and Sophora and from the monotypic genera Clianthus and Montigena (Table 1) were used to infer the phylogenetic relationships of the rhizobia of the native legume genera in New Zealand. These were compared with the rhizobia of important introduced legumes, Acacia, Cytisus, and Ulex spp., which are noxious weeds in New Zealand. Reference sequences from Bradyrhizobium, Mesorhizobium, Rhizobium, and Sinorhizobium type strains were included. Phylogenetic inference, as an approach to clarifying bacterial relationships, is usually based on the comparative analysis of 16S rDNA sequences and has been used in past investigations of rhizobia (5, 6, 15, 16, 17, 37). In this study, three additional genes were used to derive more reliable phylogenetic inferences (1, 7, 11).
Tree topologies for Mesorhizobium, Rhizobium, and Bradyrhizobium.
Partial sequences of the three housekeeping genes (atpD, glnII, and recA) were also used to generate phylograms, which were then compared. The topologies of all four trees are congruent in indicating that New Zealand's native legumes are nodulated by members of the genera Mesorhizobium and Rhizobium. Based on the analysis of 16S rDNA, individual rhizobial strains were assigned to 10 groups (A to J). Sequences representing rhizobial strains from a single plant genus are distributed between groups. With the exception of group C, which includes two strains from Clianthus spp., and group D, which is dominated by strains from Carmichaelia spp., the groups generally do not represent bacterial strains from particular host legumes. Homogeneous groups such as group C are probably a reflection of the small sample size of this legume genus. The presence of an outlying Clianthus strain in group D suggests that larger representations of strains will result in groups that are more heterogeneous. All other groups are heterogeneous with respect to the host sources of strains. For instance, group A comprises four strains from Sophora and two from Carmichaelia plants.
Groups A to D are in a clade represented entirely by known Mesorhizobium spp. The clade formed by strains in group E, from Sophora, Carmichaelia, and Clianthus plants, includes the sequence representing Rhizobium leguminosarum. Group G in Bradyrhizobium contains all sequences from Acacia spp.. The topologies of the different sequences place the strains isolated from New Zealand native legumes in the genus Mesorhizobium and in Rhizobium leguminosarum and those from all the introduced legumes in the genus Bradyrhizobium. However, consideration of the individual gene trees shows that they are not mutually congruent at the species level.
In some cases, sequences are as similar to one another as to the neighboring known species and therefore they may be members of these species. For instance, the sequences in group C may represent strains of Mesorhizobium amorphae. The placement of many strains into clusters that are distinct from existing named species indicates possible novel species. However, the absence of criteria relating sequences directly to taxonomic differences means that further data must be obtained by other methods before these strains can be properly classified (35).
These data confirm a preliminary study which showed that isolates from Carmichaelia plants were members of the genus Mesorhizobium (N. McCallum and C. W. Ronson, personal communication).
Establishment of symbioses.
Studies of Lotus corniculata have shown that this legume species was not nodulated in pristine soils because no effective bacteria were present (8). Nodulation and fixation were initiated when Lotus plants were inoculated with an effective rhizobial strain (22). Since then, it has been shown that the effective rhizobial symbiont of Lotus corniculata, Mesorhizobium loti, carries nodulating and nitrogen-fixing genes on a large transmissible element, a symbiosis island, of 500 kb (31). This symbiosis island can be transmitted to and incorporated by a range of Mesorhizobium strains already present in the soil, converting them into effective strains (30). Symbiosis islands may therefore also be involved with transfer and fixation in the native New Zealand Mesorhizobium spp. If so, then the observation that sequences representative of isolates from Carmichaelia and Sophora plants are distributed across the Mesorhizobium clade indicates either that a single symbiosis island with a broad host range is responsible for nodulation and fixation of several native legume genera or that symbiosis islands specific for each native legume genus are distributed across the genus. By their nature, symbiosis islands are incorporated into the bacterial genome of recipient strains. It seems clear that these genes may be transferred to many, if not all, Mesorhizobium species. The distribution of sequences in the genus Mesorhizobium, apparently representing several species, raises a fundamental question concerning the specificity of the association of the effective nodulating strains. Further studies will establish the extent and genetic basis of the host specificity of these strains.
It appears that many, if not all, known Mesorhizobium spp. reported in other countries (5) are present in New Zealand and have the capacity to nodulate the native legumes. Presumably these species were present prior to the separation of New Zealand from the regions of major legume evolution 80 million years ago (29).
Rhizobium leguminosarum.
An exception to the general association of native legume strains with Mesorhizobium spp. was four isolates from Sophora chathamica, Carmichaelia australis, and Clianthus puniceus that had very high sequence identity to Rhizobium leguminosarum. This demonstrates the ability of this species to nodulate native legumes. These strains of R. leguminosarum may have acquired Sym plasmids that enable wider nodulating host ranges than are currently recognized, including woody legumes. Alternatively, they may have acquired specific nodulating plasmids, one or more of which enable the nodulation of Carmichaelia, Clianthus, and Sophora plants. As yet it is not known if the strains from the native legumes represent one or more of the known R. leguminosarum biovars or if they are novel and specific. These Rhizobium strains may be endemic in New Zealand, or they may be strains introduced as commercial inoculum to enhance crop legume development and have acquired either the necessary symbiosis genes alone or the entire symbiosis island from a Mesorhizobium sp. The recorded host range of R. leguminosarum includes Lathyrus spp., Lens spp., Phaseolus spp., Pisum spp., Trifolium spp., and Vicia spp., allocated to three biovars, named according to the host plants with which they are associated (16). These isolations represent extensions of the known host range of R. leguminosarum.
A primary question of this research was to determine if the legume weeds (broom, gorse, and wattle) introduced into New Zealand were being nodulated by rhizobia that were cosmopolitan and already present in New Zealand, were introduced with them during colonization, or were able to take advantage of a native New Zealand rhizobial flora. This study indicates that most rhizobia isolated from New Zealand native legumes are members of the genus Mesorhizobium, and all isolates obtained from the introduced legumes studied are members of the genus Bradyrhizobium. Therefore it is clear that the two groups of legume plants from different origins are nodulated by unrelated rhizobial populations. The nodulating bradyrhizobia may have been transmitted in the course of dispersal of the plants (36). For instance, Bradyrhizobium spp. could be introduced either with adventive legumes, in soil imported with other plants, or with seed (23). Alternatively, these bacteria may occur naturally in New Zealand soils without being involved in symbiotic associations but have been available to nodulate the introduced legumes. The heterogeneity of the Bradyrhizobium sequences, which is substantially greater than the recorded difference between B. liaoningense and B. yuanmingense, may be an indication of a long presence and evolution in New Zealand rather than of a small recent founder population. Similar heterogeneity has been recorded for Bradyrhizobium spp. elsewhere (14, 18).
Acknowledgments
This study was supported by a grant from the Marsden Fund of the Royal Society of New Zealand under contract 97-LAN-LFS-002 and by a grant from the Non-Specific Output Fund of Landcare Research.
P. J. Bellingham, P. B. Heenan, and the staff of the Department of Conservation assisted in identifying native legume sampling sites. T. Armstrong, H. M. Harman, and R. Howitt offered constructive comments on the manuscript.
REFERENCES
- 1.Anzai, Y., H. Kim, J.-Y. Park, H. Wakabayashi, and H. Oyaizu. 2000. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 50:1563-1589. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, A. Moore, J. G. Seidman, J. A. Smith, and K. Struh. 1987. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, Sunderland, Mass.
- 3.Baxevanis, A. D., and B. F. Oulette. 2001. Bioinformatics: a practical guide to the analysis of genes and proteins, 2nd ed. John Wiley and Sons, Inc., New York, N.Y.
- 4.Bell, P. J. L., A. Sunna, M., D Gibbs, N. C. Curach, H. Nevalainen, and P. L. Bergquist. 2002. Prospecting for novel lipase genes using PCR. Microbiology 148:2283-2291. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W. X., E. T. Wang, and L. D. Kuykendall. Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, in press. Springer-Verlag, New York, N.Y.
- 6.De Lajudie, P., A. Willems, B. Pot, et al. 1994. Polyphasic taxonomy of rhizobia: emendation of the genus Sinorhizobium and description of Sinorhizobium meliloti comb. nov., Sinorhizobium saheli sp. nov. and Sinorhizobium teranga sp. nov. Int. J. Syst. Bacteriol. 44:715-733. [Google Scholar]
- 7.Gaunt, M. W., S. L. Turner, L. Rigottier-Gois, S. A. Lloyd-Macgilp, and J. P. Young. 2001. Phylogenies of atpD and recA support the small subunit rRNA-based classification of rhizobia. Int. J. Syst. Evol. Microbiol. 51:2037-2048. [DOI] [PubMed] [Google Scholar]
- 8.Greenwood, R. M., and C. E. Pankhurst. 1977. The Rhizobium component of the nitrogen-fixing symbiosis. Proc. N. Z. Grassland Assoc. 38:147-150. [Google Scholar]
- 9.Heenan, P. B. 1998. Phylogenetic analysis of the Carmichaelia complex, Clianthus, and Swainsona (Fabaceae), from Australia and New Zealand. N. Z. J. Bot. 36:21-40. [Google Scholar]
- 10.Heenan, P. B., P. J., de Lange, and A. D. Wilton. 2001. Sophora (Fabaceae) in New Zealand: taxonomy, distribution, and biogeography N. Z. J. Bot. 39:17-34. [Google Scholar]
- 11.Hilario, E., T. R. Buckley, and J. M. Young. 2004. Improved resolution of the phylogenetic relationships among Pseudomonas by the combined analysis of atpD, carA, recA and 16S rDNA. Antonie van Leeuwenhoek 86:51-64. [DOI] [PubMed] [Google Scholar]
- 12.Hirsch, A. M., M. R. Lum, and J. Downie. 2001. What makes the rhizobia-legume symbiosis so special. Plant Physiol. 127:1484-1492. [PMC free article] [PubMed] [Google Scholar]
- 13.Hugenholtz, P., and T. Huber. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int. J. Syst. Evol. Microbiol. 53:289-293. [DOI] [PubMed] [Google Scholar]
- 14.Jarabo-Lorenzo, A., R. Perez-Galdona, J. Donate-Correa, et al. 2003. Genetic diversity of bradyrhizobial population from diverse geographic origins that nodulate Lupinus spp. and Ornithopus spp. Syst. Appl. Microbiol. 26:611-623 [DOI] [PubMed] [Google Scholar]
- 15.Jarvis, B. D. W., P. van Berkum, W. X. Chen, Nour, S. M., M. Fernandez, J.-C. Cleyet-Marel, and M. Gillis. 1997. Transfer of Rhizobium loti, Rhizobium huakuii, Rhizobium ciceri, Rhizobium mediterraneum, and Rhizobium tianshansense to Mesorhizobium gen. nov. Int. J. Syst. Bacteriol. 47:895-898. [Google Scholar]
- 16.Kuykendall, L. D., J. M. Young, E. Martínez-Romero, A. Kerr, and H. Sawada. Bergey's manual of systematic bacteriology, 2nd ed., vol. 2. Springer-Verlag, New York, N. Y., in press.
- 17.Kuykendall, L. D., F. M. Hashem, and E. T. Wang. Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, in press. Springer-Verlag, New York, N.Y.
- 18.Lafay, B., and J. J. Burdon. 1998. Molecular diversity of rhizobia occurring on native shrubby legumes in southeastern Australia. Appl. Environ. Microbiol. 64:3989-3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martínez-Romero, E., and J. Caballero-Mellado. 1996. Rhizobium phylogenies and bacterial genetic diversity. Crit. Revs. Plant Sci. 15:113-140. [Google Scholar]
- 20.Nicholas, K. B., H. B. Nicholas, Jr., and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 21.Parsons, M. J., P. Douglas, and J. McMillain. 1998. Current names for wild plants in New Zealand. Manaaki Whenua Press, Lincoln, New Zealand.
- 22.Patrick, H. N., and W. L. Lowther. 1992. Response of Lotus corniculatus to inoculation and pelleting on a range of Otago tussock grassland environments. Proc. N. Z. Grassland Assoc. 54:105-109. [Google Scholar]
- 23.Pérez-Romírez, N. O., M. A. Rogel, E. Wang, J. Z. Castellanos, and E. Martinez-Romero. 1998. Seeds of Phaseolus vulgaris bean carry Rhizobium etli. FEMS Microbiol. Ecol. 26:289-296. [Google Scholar]
- 24.Perret, X., C. Staehelin, and W. J. Broughton. 2000. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 64:180-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 26.Pueppke, S. G., and W. J. Broughton. 1999. Rhizobium sp. NGR234 and R. fredii USDA257 share exceptionally broad, nested host-ranges. Mol. Plant-Microbe Interact. 12:293-318. [DOI] [PubMed] [Google Scholar]
- 27.Saldana, G., V. Martinez-Alcantara, J. M. Vinardell, R. Bellogin, J. E. Ruiz-Sainz, and P. A. Balatti. 2003. Genetic diversity of fast-growing rhizobia that nodulate soybean ( Glycine max L. Merr). Arch. Microbiol. 180:45-52. [DOI] [PubMed] [Google Scholar]
- 28.Sawada, H., L. D. Kuykendall, and J. M. Young. 2003. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J. Gen. Appl. Microbiol. 49:155-179. [DOI] [PubMed] [Google Scholar]
- 29.Stevens, G., M. McGlone, and B. McCulloch. 1988. Prehistoric New Zealand. Heinemann Reed, Auckland, New Zealand.
- 30.Sullivan, J. T., H. N. Patrick, W. L. Lowther, D. B. Scott, and C. W. Ronson. 1995. Nodulating strains of Rhizobium loti arose through chromosomal symbiotic gene transfer in the environment. Proc. Natl. Acad. Sci. USA 92:8985-8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan, J. T., and C. W. Ronson. 1998. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA 95:5145-5149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swofford, D. L. 2003. PAUP* phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 33.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turner, S. L., and J. P. Young. 2000. The glutamine synthetases of rhizobia: phylogenetics and evolutionary implications. Mol. Biol. Evol. 17:309-319. [DOI] [PubMed] [Google Scholar]
- 35.Vandamme, P., B. Pot, M. Gillis, P. De Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, E. T., F. L. Kan, Z. Y. Tan, I. Toledo, W. X. Chen, and E. Martínez-Romero. 2003. Diverse Mesorhizobium plurifarium populations native to Mexican soils. Arch. Microbiol. 180:444-454. [DOI] [PubMed] [Google Scholar]
- 37.Young, J. M., L. D. Kuykendall, E. Martínez-Romero, A. Kerr, and H. Sawada. 2001. A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. rubi, R. undicola, and R. vitis. Int. J. Syst. Evol. Microbiol. 51:89-103. [DOI] [PubMed] [Google Scholar]