Abstract
The bacterial heat shock response is characterized by the elevated expression of a number of chaperone complexes. Two-dimensional polyacrylamide gel electrophoresis revealed that GroEL expression in probiotic Lactobacillus paracasei NFBC 338 was increased under heat adaptation conditions (52°C for 15 min). Subsequently, the groESL operon of L. paracasei NFBC 338 was PCR amplified, and by using the nisin-inducible expression system, two plasmids, pGRO1 and pGRO2, were constructed on the basis of vectors pNZ8048 and pMSP3535, respectively. These vectors were transferred into Lactococcus lactis(pGRO1) and L. paracasei(pGRO2), and after induction with nisin, overexpressed GroEL represented 15 and 20% of the total cellular protein in each strain, respectively. Following heat shock treatment of lactococci (at 54°C) and lactobacilli (at 60°C), the heat-adapted cultures maintained the highest level of viability (5-log-unit increase, approximately) in each case, while it was found that the GroESL-overproducing strains performed only moderately better (1-log-unit increase) than the controls. On the other hand, the salt tolerance of both GroESL-overproducing strains (in 5 M NaCl) was similar to that of the parent cultures. Interestingly, both strains overproducing GroESL exhibited increased solvent tolerance, most notably, the ability to grow in the presence of butanol (0.5% [vol/vol]) for 5 h, while the viability of the parent strain declined. These results confirm the integral role of GroESL in solvent tolerance, and to a lesser extent, thermotolerance of lactic acid bacteria. Furthermore, this study demonstrates that technologically sensitive cultures, including certain probiotic lactobacilli, can potentially be manipulated to become more robust for survival under harsh conditions, such as food product development and gastrointestinal transit.
Probiotic strains must have demonstrable benefits to host health and have GRAS (generally recognized as safe) status. Furthermore, from a food processing perspective, such strains should be suitable for large-scale industrial production by possessing the ability to survive both food processing conditions and storage (36). Problems that are frequently associated with the incorporation of probiotic strains into food products include the poor temperature, salt, and oxygen tolerance of some species. Approaches taken to address these problems include the use of oxygen-impermeable containers, microencapsulation (6), incorporation of nutrients, and selection of stress-resistant strains (34). A well-characterized phenomenon among lactic acid bacteria (LAB) and many other microbial systems is sublethal stress, which can lead to an elevated state of resistance (7, 18, 29). In a previous study, we observed that when prestressed by either heat (52°C for 15 min) or salt (0.3 M for 30 min), the probiotic strain Lactobacillus paracasei NFBC 338 survived up to 300-fold better than unadapted control cells during heat stress and 18-fold better during spray drying (7).
Advances in genomics and proteomics have led to the identification of genes involved in Lactobacillus stress responses, such as the molecular chaperone groESL and dnaK (heat stress) (29, 33, 41), methionine sulfoxide reductase (oxygen stress) (42), and F1F0-ATPase (acid stress) (21) genes. In particular, the chaperone proteins are considered essential components of the heat shock response in that they guide proteins along the proper pathways for folding. Many are termed heat shock proteins, because they are produced in large amounts when cells are exposed to heat, which makes misfolding more common. The two major groups of chaperone proteins are the 70-kDa DnaK family and the 60-kDa GroE family (14). The latter GroEL-GroES complex forms an enclosed environment for the correct folding of proteins under normal growth conditions and under conditions of cellular stress (14). Indeed, levels of cellular stress can be measured by changes in GroESL levels of expression in some cases (30). Biochemical and structural studies have revealed that the GroEL chaperone acts with its partner, cochaperone GroES, as a two-stroke ATP-regulated folding machine. Two internal cavities, which can bind partially unfolded proteins, are created by the GroEL tetradecamer, which forms two back-to-back rings, each with seven subunits. When the substrate protein is bound, ATP binds to the nucleotide-binding site of the seven subunits, causing the hinge to open. In response to ATP binding, the GroES “lid” closes on the cavity, the protein is allowed to resume folding and is ejected by the binding of protein or ATP in the opposite cavity (43). It has been shown that the Escherichia coli chaperone GroEL can interact with up to 50% of soluble proteins when they are in a nonnative state, and indeed, in GroEL-deficient cells about 30% of proteins remain misfolded (43).
The L. paracasei NFBC 338 strain fulfills a number of the criteria outlined for the selection of probiotic strains, including being of human intestinal origin, exhibiting bile and acid tolerance, and being technologically compatible with cheese manufacture (12) and spray drying (13). Furthermore, data indicate that ingestion of L. paracasei NFBC 338 is associated with positive effects on gastrointestinal flora (9). In this study, we identified some of the proteins that were naturally overexpressed in this strain as a result of exposure to sublethal heat stress. Subsequently, the chaperone proteins GroES/EL were overexpressed in lactococci and probiotic lactobacilli in order to investigate their contribution to the bacterial survival mechanism when exposed to a variety of environmental conditions, such as heat, salt, and solvent stresses.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and media.
The probiotic strain Lactobacillus paracasei subsp. paracasei NFBC 338 (subsequently referred to as L. paracasei NFBC 338), was previously isolated from the human gastrointestinal tract (GIT) and obtained from University College, Cork, Ireland, under a restricted-material transfer agreement. L. paracasei NFBC 338 was routinely cultured overnight (∼17 h) in 5 ml of MRS broth (Oxoid Ltd., Hampshire, United Kingdom) from a 1% (vol/vol) stock inoculum and incubated at 37°C under anaerobic conditions using anaerobic jars containing Anaerocult A gas packs (Merck, Darmstadt, Germany). Lactococcus lactis NZ9800 (subsequently referred to as L. lactis) was propagated at 30°C in M17 (Difco Laboratories, Detroit, Mich.) broth and/or agar containing glucose (0.5% [vol/vol]). The harvested cells of both strains were stored at −80°C as stock solutions in 40% (vol/vol) aqueous glycerol.
Protein extraction, two-dimensional (2D) gel electrophoresis, and mass spectrometry.
Exponential-phase cells of L. paracasei NFBC 338 were grown for 3 to 3.5 h (optical density at 600 nm [OD600] of 0.3) in MRS broth (400 ml) and heat adapted at 52°C for 15 min as described previously (6). Cultures were then harvested (400 ml) at 6,000 × g for 5 min at 4°C and washed three times with 20 mM Tris-Cl (pH 7.1). Cell lysis was performed in 20 mM Tris-EDTA (pH 7.1) by bead beating in a Hybaid Ribolyser (Hybaid Ltd., Middlesex, United Kingdom) three times at maximum speed for 1 min each time, with 1-min intervals on ice. The cytosolic fraction was obtained by removal of cell debris at 39,000 × g for 1 h at 4°C.
Protein concentration in each of the samples was estimated using the Bio-Rad protein assay (Bio-Rad Laboratories Ltd., Hemel Hempstead, Hertfordshire, United Kingdom). To improve sample quality and remove soluble organic contaminants, the protein extracts were acetone precipitated. Protein samples of 200 μg were diluted in 3 to 4 volumes of ice-cold acetone, and proteins were allowed to precipitate for 2.5 h at −20°C. The resultant pellets were harvested and then resuspended in 20 mM Tris-Cl (pH 7.1).
The two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) procedure used was essentially the method of O'Farrell (26) and Klose (20), with minor modifications using the Multiphor II system (Amersham Pharmacia Biotech, Uppsala, Sweden). For the first-dimensional isoelectric focusing, protein samples were loaded onto precast Immobiline Drystrips (Amersham Pharmacia Biotech) with a linear pH gradient (pH 4 to 7), while the second dimension involved precast ExelGel sodium dodecyl sulfate (SDS)-polyacrylamide gels (Amersham Pharmacia Biotech) with a 12 to 14% (wt/vol) polyacrylamide gradient. Gels were stained with Coomassie brilliant blue 1× solution (Sigma Chemical Company, Poole, Dorset, United Kingdom), and protein expression was quantified by densitometry, using the software package Investigator HT Analyzer version 2.1 (Genomic Solutions Ltd., Huntingdon, Cambridgeshire, United Kingdom).
After 2D-PAGE, selected proteins were excised from gels, digested with trypsin, and analyzed by mass spectrometry (The Proteomics Facility, University of Aberdeen, Aberdeen, Scotland) by the method of Shevchenko et al. (35). A nonredundant protein sequence database such as Protein Prospector or Matrix Science Mascot was used for database searches using the peptide search software package. The search parameters used were as follows: cysteine as the S-carbamidomethyl derivative, maximum allowed peptide mass error between 50 and 150 ppm, more than five peptide mass hits required for a protein match, and protein scores greater than 72 were deemed significant (P < 0.5). No restriction was placed on either the isoelectric point or species of origin of the protein. A protein mass range between 0 and 150 kDa was allowed.
DNA manipulations, transformation, and plasmid construction.
Total DNA was extracted from an overnight-grown culture of L. paracasei NFBC 338 by the method of Hoffman and Winston (16). The QIAGEN Plasmid Mini kit (QIAGEN, West Sussex, United Kingdom) was used to isolate plasmid DNA from E. coli and L. lactis, with one minor modification for L. lactis, i.e., 0.02 mg of lysozyme per ml was added to P1 buffer and incubated for 20 min at 37°C. PCR products were purified using a Qiaquick PCR purification kit (QIAGEN). Restriction enzymes (New England Biolabs, Beverly, Mass.) were used per the manufacturer's instructions. T4 DNA ligase (New England Biolabs) was used for ligation reactions at 15°C overnight.
PCR was performed using the Expand High-Fidelity PCR system (Roche Diagnostics, East Sussex, United Kingdom). The groES and groEL genes were amplified from L. paracasei NFBC 338 by designing primers against a conserved region of groEL on the basis of multiple sequence alignments of the groESL operon from Lactobacillus johnsonii (GenBank accession no. AF214488), Lactobacillus helveticus (GenBank accession no. AF031929), and Lactobacillus zeae (GenBank accession no. AF010281) using the groF primer (GAA GGT ATG AAG AAC GTT AC) and groR primer [TT(A/G) GTT GGG TC(A/G) ATA ATA CC]. Sequencing this fragment from the L. paracasei NFBC 338 operon revealed that it was 99% similar to that of L. zeae (GenBank accession no. AF010281); therefore, primers for the full-length L. paracasei NFBC 338 operon (groESLF primer [ACATGCATGC GGA GGG ATT GCC TTG TT] and groESLR primer [GCTCTAGA TTA CAT CAT ACC GCC]) were designed on the basis of the known L. zeae sequence, and they also contain the restriction sites for the SphI and XbaI enzymes (underlined).
PCR was performed using a Hybaid PCR express unit (Hybaid Ltd., Hampshire, United Kingdom) in 50-μl volumes containing 2 μg of DNA, 17.5 mM MgCl2, 50 pmol of each primer, and 2.5 U of Expand High-Fidelity enzyme. Negative PCR controls without template DNA were also included. The authenticity of the groESL insert was verified in four individual pTOPO (Invitrogen Ltd., Paisley, United Kingdom) clones by sequencing with an automated DNA sequencer (MWG-BIOTECH Custom DNA Sequencing Service, Ebersberg, Germany) using standard M13 forward and reverse primers. Sequence analysis of the insert was performed using DNAStar software (DNAStar, Madison, Wis.). Only one of the four clones had the consensus sequence and so was deemed to have no PCR-generated errors. The resulting 1,965-bp product from this clone was then introduced into the SphI and XbaI sites in the multiple cloning site of the vectors pNZ8048 (4) and pMSP3535 (2), resulting in plasmids pGRO1 and pGRO2, respectively. These steps were performed using L. lactis as a host. L. lactis (pGRO1) was grown on GM17 agar containing chloramphenicol (5 μg/ml), and L. paracasei NFBC 338 (pGRO2) was grown on MRS agar containing erythromycin (10 μg/ml).
Electrocompetent L. lactis cells were prepared and transformed by the method of de Ruyter et al. (4), while electrocompetent L. paracasei NFBC 338 cells were prepared using 3.5× SMEB by the method of Luchansky et al. (23).
SDS-polyacrylamide gel analysis of GroESL overexpression.
GroESL from probiotic L. paracasei NFBC 338 was expressed in lactococci and lactobacilli using the nisin-inducible expression (NICE) system (4), which allows maximum protein expression at sublethal concentrations of nisin and is tightly controlled so that there is negligible expression in the absence of nisin (5). pGRO1 was introduced into L. lactis NZ9800, an MG1363 derivative containing the nisRK signal transduction genes integrated on the chromosome, while pGRO2, which was introduced into L. paracasei NFBC 338, is a plasmid that contains nisRK in addition to the promoter, PnisA.
To examine the levels of GroESL overexpression that could be achieved upon induction after introduction of the plasmids into L. lactis and L. paracasei NFBC 338, cells were induced with nisin as follows. Cultures at an OD600 of 0.5 were induced with nisin (10 ng/ml for L. lactis and 50 ng/ml for L. paracasei NFBC 338) for 1, 3, and 5 h. Aliquots were taken at intervals after nisin induction, and whole-cell protein extractions performed. The control in each case was the nisin-induced Lactococcus or Lactobacillus strain with pNZ8048 or pMSP3535, respectively. Cells (1 ml) were harvested and washed twice in 20 mM Tris-Cl (pH 7.5). The cells were sonicated three times for 30-s pulses at maximum amplitude, with 1-min intervals on ice. The cell lysate was then centrifuged for 5 min at 8,000 × g to remove cell debris. The protein extracts were analyzed by SDS-PAGE using a Mini Protean II cell unit (Bio-Rad) by the method of Laemmli (22) with a 17% acrylamide (appropriate for ∼60-kDa proteins, such as GroEL) or 20% acrylamide (appropriate for ∼10-kDa proteins, such as GroES) resolving gel. A prestained standard (range, 6.5 to 205 kDa; Sigma) was used as a molecular mass marker. Densitometric analysis of the protein spots was performed using the software package Investigator HT Analyzer version 2.1 (Genomic Solutions Ltd.).
Investigation of the stress tolerance of GroESL-overproducing L. lactis and L. paracasei NFBC 338.
Four cultures (40 ml each) were each grown to an OD600 of 0.5 and were then induced with nisin (10 ng/ml for 1 h for L. lactis and 50 ng/ml for 2 h for L. paracasei NFBC 338). Thermotolerance of L. lactis was investigated by monitoring survival in GM17 broth at 54°C for 30 min with constant agitation. Aliquots were removed at 0, 5, 10, 15, and 30 min, and the first seven serial dilutions were plated on GM17 agar and then incubated at 30°C for 48 h. The L. lactis cultures tested were as follows: (i) L. lactis harboring pNZ8048 [L. lactis(pNZ8048)], nisin induced; (ii) L. lactis(pNZ8048), nisin induced (and heat adapted at 40°C for 30 min); (iii) L. lactis pGRO1, uninduced; (iv) L. lactis(pGRO1), nisin induced; and (v) L. lactis(pGRO1), nisin induced (and heat adapted at 40°C for 30 min).
The thermotolerance of L. paracasei NFBC 338 was investigated at 60°C in MRS broth for 30 min with constant agitation. Aliquots were removed at 0, 5, 10, 15, and 30 min, and the first seven serial dilutions were plated on MRS agar and then incubated at 37°C for 72 h. The Lactobacillus cultures tested were as follows: (i) L. paracasei NFBC 338(pMSP3535), nisin induced; (ii) L. paracasei NFBC 338(pMSP3535), nisin induced (and heat adapted at 52°C for 15 min); (iii) L. paracasei NFBC 338(pGRO2), uninduced; (iv) L. paracasei NFBC 338(pGRO2), nisin induced; and (v) L. paracasei NFBC 338(pGRO2), nisin-induced (and heat adapted at 52°C for 15 min). Salt tolerance was determined upon the addition of 5 M NaCl, and culture viability was monitored for 1 h with aliquots taken at 10-min intervals. Solvent tolerance was determined upon the addition of butanol (0.5% [vol/vol]), and survival was monitored for 6 h at 37°C.
Nucleotide sequence accession number.
The nucleotide sequence data represented in this study has been submitted to GenBank and assigned accession number AY631040.
RESULTS
GroEL is upregulated during heat shock.
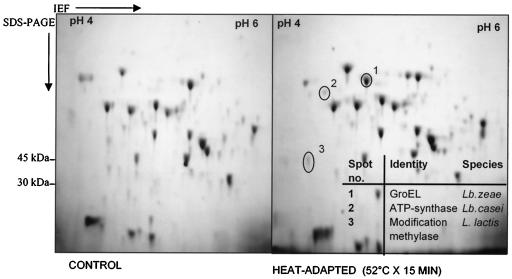
In an effort to identify intracellular proteins that contribute to the heat shock response, 2D electrophoretograms of heat-adapted (52°C for 15 min) and control probiotic L. paracasei NFBC 338 cultures in early log phase (OD600 of 0.3) were compared. Expression of at least 12 proteins had changed as a result of heat exposure (Fig. 1). The most dramatic change observed was the level of expression of a 60-kDa protein in the heat-adapted culture compared with that of the control culture. This protein was identified after trypsin digestion and mass spectrometry as GroEL, which received a protein score of 161 compared with GroEL of L. zeae (GenBank accession no. AF010281). GroEL from L. paracasei NFBC 338 displayed a molecular mass of 57.4 kDa and an isoelectric point of 4.89, similar to those reported for the GroE systems of Bacillus subtilis (32), Propionibacterium freudenreicheii (17), and Lactobacillus rhamnosus (29). Two of the remaining 11 proteins affected by heat exposure were identified as ATP synthase beta chain (EC 3.6.1.34) (protein score of 95) and modification methylase SCRF1-A (EC 2.1.1.73) (protein score of 73).
FIG. 1.
2D-PAGE used to investigate the mechanisms involved in the development of thermotolerance for probiotic L. paracasei NFBC 338. The electrophoretograms shown are representative of two independent trials. IEF, isoelectric focusing.
Sequencing of the groESL operon from L. paracasei NFBC 338.
groEL DNA was amplified from the chromosome of L. paracasei NFBC 338 using primers designed against a conserved region in three Lactobacillus groEL sequences on the basis of multiple sequence alignments. Sequencing of the resulting fragment (1,307 bp) revealed a high level of similarity (99%) to groESL of L. zeae (GenBank accession no. AF010281). Consequently, primers were designed on the basis of the L. zeae groESL sequence to PCR amplify the 1,953-bp groESL operon from the chromosome of L. paracasei NFBC 338. The sequence of the resulting fragment was found to be 99% similar to that of L. zeae with six nucleotide differences, which gave rise to two amino acid changes.
Overexpression of Lactobacillus GroESL in Lactococcus and Lactobacillus.
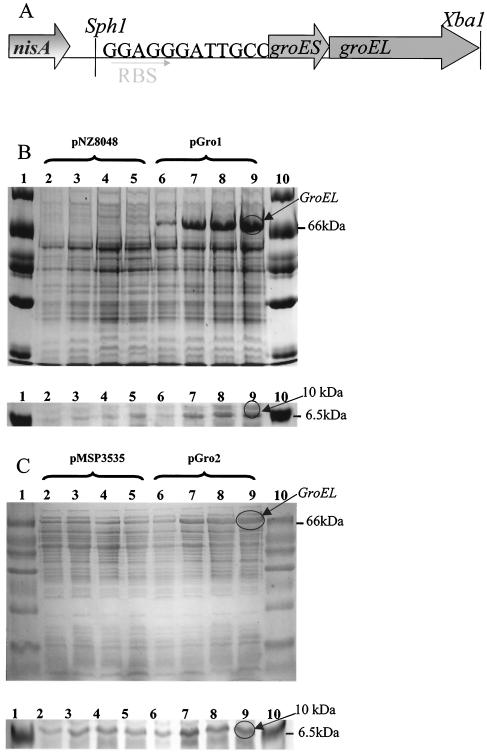
Given that GroEL appeared to be the main protein upregulated during the heat shock response of L. paracasei NFBC 338, a genetically modified LAB which overexpressed this protein was constructed. groESL from L. paracasei NFBC 338 was heterologously expressed in L. lactis NZ9800 and homologously expressed in the parent strain using the NICE system (4) (Fig. 2A). In the case of lactococci, this was achieved using the vector pNZ8048, resulting in plasmid pGRO1, where the host strain L. lactis NZ9800 constitutively expressed nisRK. To investigate the level of GroESL expression that could be achieved upon nisin induction, SDS-PAGE analysis was performed (Fig. 2B and C). Mid-log-phase cells of control L. lactis(pNZ8048) and L. lactis(pGRO1) were induced with 10 ng of nisin per ml, and samples were removed at the time of induction (0 h) and 1, 3, and 5 h after nisin addition. Following nisin induction, L. lactis(pGRO1) overexpressed a protein of approximately 60 kDa which was absent in the nisin-treated control (Fig. 2B). This protein band was excised and subjected to tryptic digestion and mass spectrometry, from which it was positively identified as GroEL, with a protein score of 107 compared to GroEL from L. zeae. The effect of nisin induction on the smaller GroES protein (10 kDa) was difficult to determine, as resolution of proteins below 14 kDa was poor, even on gels containing 20% (wt/vol) polyacrylamide. However, a band at approximately 10 kDa was evident in all L. lactis (pGRO1) samples treated with nisin (Fig. 2B).
FIG. 2.
(A) Schematic representation of the groESL operon under control of the NICE system. RBS, ribosome binding site. (B and C) SDS-PAGE illustrating the controlled expression of GroEL and GroES using the NICE system in L. lactis (B) using vector pNZ8048 and in L. paracasei NFBC 338 (C) using vector pMSP3535. Lanes for both gels: 1 and 10, protein standard marker; 2, plasmid control at time zero; 3, plasmid control plus nisin after 1 h; 4, plasmid control plus nisin after 3 h; 5, plasmid control plus nisin after 5 h; 6, GroESL-overproducing strain at time zero; 7, GroESL-overproducing strain plus nisin after 1 h; 8, GroESL overproducing strain plus nisin after 3 h; 9, GroESL-overproducing strain plus nisin after 5 h.
In the homologous host, the overexpression of GroESL was achieved using the vector pMSP3535, which itself encodes nisRK, resulting in the strain L. paracasei NFBC 338(pGRO2). Mid-log-phase cells of control L. paracasei NFBC 338(pMSP3535) and L. paracasei NFBC 338(pGRO2) were induced with 50 ng of nisin per ml, and samples were removed at the time of induction (0 h) and 1, 3, and 5 h after nisin addition. Following nisin induction, L. paracasei NFBC 338(pGRO2) cells overexpressed a protein of approximately 60 kDa compared with the nisin-treated vector control (Fig. 2C). As before, this protein was positively identified as GroEL, with a protein score of 121 compared with GroEL from L. zeae. A protein of ∼10 kDa was present in all nisin-treated samples (Fig. 2C).
Stress tolerance of GroESL-overproducing lactococci.
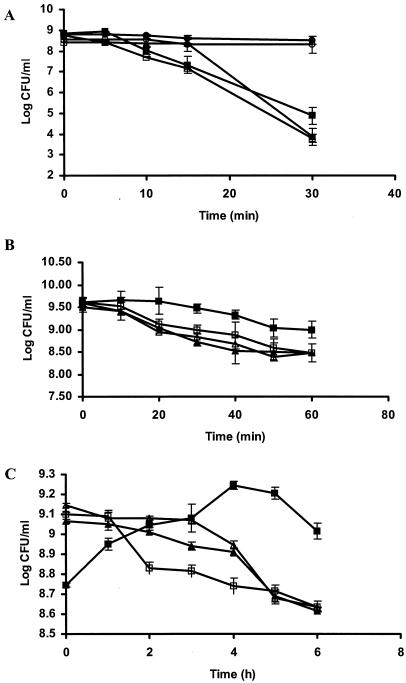
To determine whether GroESL overexpression resulted in improved stress tolerance in LAB, the heterologous host L. lactis was used as a model system. GroESL-overproducing cultures (along with appropriate controls) of Lactococcus were exposed to heat (54°C for 30 min), salt (5 M NaCl for 1 h), or solvent (0.5% [vol/vol] butanol) stress. The performance of both heat-adapted (40°C for 30 min) and unadapted parent cultures were included for comparative purposes in the thermotolerance study. The viability of both L. lactis(pNZ8048) and uninduced L. lactis(pGRO1) cultures was reduced by 4.0 log CFU/ml after heat stress at 54°C for 30 min (Fig. 3A). In contrast, the viability of the heat-adapted parent declined very little (≤0.3 log CFU/ml) under identical conditions. The induced L. lactis(pGRO1) culture, however, exhibited intermediate viability, declining by ≤3.3 log CFU/ml during heat stress.
FIG. 3.
Stress tolerance in GroESL-overproducing strains of L. lactis during (A) heat stress (54°C for 30 min), (B) salt stress (5 M NaCl for 1 h), and (C) butanol stress (0.5% [vol/vol] butanol for 6 h). Symbols: ▴, nisin-induced L. lactis(pNZ8048); ▵, L. lactis(pNZ8048); •, nisin-induced, heat-adapted L. lactis(pNZ8048); □, L. lactis(pGRO1); ▪, nisin-induced L. lactis(pGRO1); ○, nisin-induced, heat-adapted L. lactis(pGRO1). The values shown are the means ± standard errors (error bars) for three challenge trials.
It has been demonstrated that the synthesis of GroESL chaperone proteins is induced not only during mild heat stress but also after exposure to salt stress (1). For this reason, we examined the salt tolerance of L. lactis(pGRO1) when exposed to 5 M NaCl for 1 h. The viability of both L. lactis(pNZ8048) and uninduced L. lactis(pGRO1) was reduced by 1 log CFU/ml after salt stress (Fig. 3B), while the viability of induced L. lactis(pGRO1) declined only ≤0.6 log CFU/ml under identical conditions.
We also tested the hypothesis that overexpression of GroESL may increase solvent tolerance (0.5% [vol/vol] butanol) of LAB. Following butanol challenge of L. lactis(pNZ8048) and uninduced L. lactis(pGRO1), the viability of both controls declined after 1 h (≤0.2 log CFU/ml), and this trend continued over the 6-h period of solvent challenge (Fig. 3C). In contrast, induced L. lactis(pGRO1) exhibited the ability to grow (increasing by 0.5 log CFU/ml) during 5-h exposure to butanol stress.
Stress tolerance of GroESL-overproducing lactobacilli.
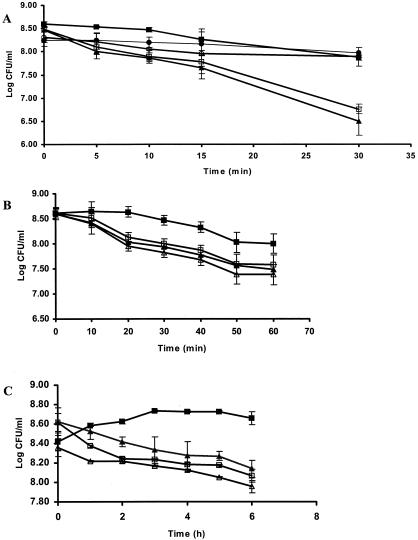
To determine whether GroESL overexpression resulted in improved stress tolerance in the homologous host, L. paracasei NFBC 338(pGRO2) was subjected to heat, salt, and solvent stress. Heat adaptation of L. paracasei NFBC 338 was performed at 52°C for 15 min. Exposure of L. paracasei NFBC 338(pMSP3535) and uninduced L. paracasei NFBC 338(pGRO2) to heat stress (60°C for 30 min) resulted in a decline in viability of 2 log CFU/ml, while the heat-adapted culture proved to be considerably more thermotolerant, declining in viability by only ≤0.5 log CFU/ml (Fig. 4A). The induced GroESL-overproducing strain, L. paracasei NFBC 338(pGRO2), performed moderately better than the controls, exhibiting a decline in viability of 1 log CFU/ml after heat stress.
FIG. 4.
Stress tolerance in GroESL-overproducing strains of L. paracasei NFBC 338 during (A) heat stress (60°C for 30 min), (B) salt stress (5 M NaCl for 1 h), and (C) butanol stress (0.5% [vol/vol] butanol for 6 h). Symbols: ▴, nisin-induced L. paracasei NFBC 338(pMSP3535); ▵, L. paracasei NFBC 338(pMSP3535); •, nisin-induced, heat-adapted L. paracasei NFBC 338(pMSP3535); □, L. paracasei NFBC 338(pGRO2); ▪, nisin-induced L. paracasei NFBC 338(pGRO2); ○, nisin-induced, heat-adapted L. paracasei NFBC 338(pGRO2). The values shown are the means ± standard errors (error bars) for three challenge trials.
Similar to the results obtained with GroESL-overproducing lactococci, the viability of L. paracasei NFBC 338(pMSP3535) and uninduced L. paracasei NFBC 338(pGRO2) was reduced by 1 log CFU/ml after salt stress (5 M NaCl for 1 h) (Fig. 4B). In contrast, induced L. paracasei NFBC 338(pGRO2) declined by ≤0.8 log CFU/ml under identical conditions.
The growth of nisin-induced L. paracasei NFBC 338(pGRO2) increased by 0.5 log CFU/ml during solvent stress (0.5% [vol/vol] butanol) (Fig. 4C). This represents a dramatic difference in the growth kinetics during solvent stress of induced L. paracasei NFBC 338(pGRO2) compared with parent cultures. The viability of all lactococcal and Lactobacillus strains declined after 5-h solvent exposure (Fig. 3C and 4C), with nisin-induced L. lactis(pGRO1), L. paracasei NFBC 338(pGRO2), and the controls declining in viability at the same rate during 30-h exposure to butanol (data not shown).
DISCUSSION
In order for probiotic strains to survive food processing and gastric transit, they have to overcome a wide range of environmental stresses. To withstand such adverse environments, bacteria have evolved complex stress-sensing defenses (40). This study was designed to identify proteins involved in thermal stress in the probiotic L. paracasei NFBC 338 strain using proteomics and to use this knowledge to engineer the probiotic bacteria to overexpress selected proteins involved in the stress response.
2D-PAGE revealed that the chaperone protein GroEL was among the most strongly expressed proteins in the cell under heat adaptation conditions. Indeed, densitometric analyses indicated an approximately 49.1-fold increase in cells that were preadapted to heat. Kim et al. (18) demonstrated that the transcriptional activity of both chaperones is increased dramatically in response to heat shock and is increased to a lesser extent by ethanol stress. Other studies have shown that when log-phase Lactobacillus cultures were subjected to heat stress, a 15-fold increase in GroEL synthesis was observed compared with only 1.5-fold increase in protein synthesis in stationary-phase cultures (29).
In this study, the groESL operon of L. paracasei NFBC 338 was PCR amplified, sequenced, and found to be highly homologous to that of other Lactobacillus species, such as L. zeae (GenBank accession no. AF010281) and L. johnsonii (41). The groESL operon in L. johnsonii and in many other microbial systems is preceded by a typical Sigma A promoter (24, 32, 41). In addition, one or more highly conserved palindromic 9-bp inverted repeats are typically found near the promoter. This structure is termed the CIRCE element (controlling inverted repeat of chaperone expression) and plays a dual role as both a regulatory element and a promoter-proximal operator (44). For the purpose of controlling the expression of GroESL, we did not employ the native Lactobacillus promoter, but instead cloned the operon under the control of the nisin promoter PnisA. Nisin is an antimicrobial peptide that can be used to induce transcription of genes under the control of PnisA and PnisF promoters via a two-component regulatory system consisting of the histidine protein kinase NisK and the response regulator NisR (19). The NICE system was developed originally for use in Lactococcus and was subsequently used in other gram-positive bacteria (10, 19, 25) and has proven very versatile in a wide range of applications. The NICE system has previously been used in lactobacilli to successfully synthesize therapeutic molecules (19, 27) and for the nisin-dependent induction of the fusion of a reporter gene, pepI, for the nisA promoter (25).
In an attempt to assign a phenotype to the GroESL-overproducing strains, we exposed the recombinant cultures to heat, salt, or solvent stress and compared their performance with those of control cultures. Moderate improvements in thermotolerance and osmotic tolerance were observed for both Lactococcus and Lactobacillus GroESL-overproducing strains. For heat tolerance in particular, the level of protection conferred by overexpressing GroESL alone was many orders of magnitude less than that of the heat-adapted cultures. Not surprisingly, better performance was observed in the homologous Lactobacillus GroESL-overproducing strain. Overproduction of GroESL also gave modest improvements in the performance of the recombinant Lactobacillus strain during both spray drying (10-fold increase in survival) and freeze-drying (14% increase in survival) compared with that of the control (unpublished data). However, none of the recombinant strains generated in this study gave a response equal to that of the heat-adapted parent cultures. Again, this may be expected, since the heat shock response involves a number of proteins, including DnaK, DnaJ, and GrpE chaperone proteins, tagatose-6-phosphate-aldolase, glyceraldehyde-3-phosphate dehydrogenase, and triose-phosphate isomerase (29). Thus, overproduction of an entire battery of stress-related proteins, such as DnaK, DnaJ, GrpE, Clp, HtrA, and FtsH, might give better survival, since it would more closely mimic the heat adaptation response of lactobacilli. Indeed, Prasad et al. (29) proposed that for L. rhamnosus, the heat or osmotic adaptation response that leads to an elevated state of cell resistance may be due to the mechanisms associated with stress proteins, glycolysis-related machinery, and other stationary-phase-related proteins and regulatory factors. From previous studies (1, 7), heat adaptation of live microorganisms prior to heat stress has been shown to improve the thermotolerance of lactococci and lactobacilli by up to 300-fold compared with untreated parent strains.
We demonstrated that development of solvent tolerance in both LAB strains was associated with overexpression of GroESL. Previous reports have indicated that growth is the most sensitive cellular activity to solvents; however, the synthesis of heat shock proteins has also been shown to be induced by toxic solvent concentrations (37), which naturally indicates a link between GroESL expression and solvent stress. Tomas and coworkers (38) reported that GroESL overexpression in Clostridium acetobutylicum increased solvent titers and solvent tolerance, most probably due to the stabilization of the biosynthetic machinery of the cell under solvent stress. This was further supported by the fact that the overall metabolic activity in the GroESL-overproducing strain was reported to be higher than that of the controls. Indeed, the GroESL machinery may even stabilize the cell to help overcome the stress associated with the presence of a plasmid, which is known to be a metabolic burden and a cause of cellular stress (31). It has been reported that the response of cells to heat shock and alkanols show significant similarities, because both stresses alter membrane fluidity (3, 28). In addition, organic solvents have been found to permeabilize the cell membrane and disrupt the function of embedded proteins in gram-negative bacteria. In this respect, a fraction of the cellular GroE pool has been shown to bind and associate with membrane lipids, leading to membrane stabilization (39). The degree of binding is influenced by the composition and physical state of the phospholipid bilayer, demonstrating a link between stress proteins and lipid membrane unsaturation (39). Increased solvent tolerance as exhibited by the recombinant Lactococcus and Lactobacillus strains in this study may be a useful feature in the development of ethanol-producing LAB (8) or in the development of probiotic products containing alcohol, where growth under harsh solvent stress conditions would be an attribute.
In conclusion, these data suggest that overexpression of stress-induced proteins has the potential to improve the performance of probiotic LAB. In particular, the GroES/EL chaperone complex can be exploited to prepare LAB for industrial processes. Indeed, the innate probiotic characteristics of the strain, such as adherence to the host cell wall and acid tolerance during gastric transit, may also be improved by the overexpression of GroESL. In this respect, the fact that the GroEL protein is involved in the attachment of bacteria to host cells and to each other is well established (11, 15). Furthermore, expression of the protein is induced by acid stress or cell contact in Clostridium difficile (15); thus, generation of a recombinant probiotic strain overproducing GroESL may afford the strain a selective advantage in the host GIT as well as promote its attachment there. Identification of other stress-induced proteins coupled with a more in-depth understanding of their mode of action could lead to their exploitation toward the generation of more robust probiotic bacteria.
Acknowledgments
We thank Gary Dunny, University of Minnesota, and Michel Kleerebezem, Wageningen Centre for Food Sciences, for the kind gifts of the nisin expression vectors.
C. Desmond is a recipient of a Teagasc Walsh fellowship. This work was funded in part by the Irish Government under the National Development Plan 2000-2006, the European Research and Development Fund, EU project QLK1-CT-2000-30042, and Science Foundation Ireland funds.
REFERENCES
- 1.Boutibonnes, P., C. Tranchard, A. Hartke, B. Thammavongs, and Y. Auffray. 1992. Is thermotolerance correlated to heat-shock protein synthesis in Lactococcus lactis subsp. lactis? Int. J. Food Microbiol. 16:227-236. [DOI] [PubMed] [Google Scholar]
- 2.Bryan, E. M., T. Bae, M. Kleerebezem, and G. M. Dunny. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183-190. [DOI] [PubMed] [Google Scholar]
- 3.Curran, B. P., and S. A. Khalawan. 1994. Alcohols lower the threshold temperature for the maximum activation of a heat shock expression vector in the yeast Saccharomyces cerevisiae. Microbiology 140:2225-2228. [DOI] [PubMed] [Google Scholar]
- 4.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Ruyter, P. G., O. P. Kuipers, W. C. Meijer, and W. M. de Vos. 1997. Food-grade controlled lysis of Lactococcus lactis for accelerated cheese ripening. Nat. Biotechnol. 15:976-979. [DOI] [PubMed] [Google Scholar]
- 6.Desmond, C., R. P. Ross, E. O'Callaghan, G. Fitzgerald, and C. Stanton. 2002. Improved survival of Lactobacillus paracasei NFBC 338 in spray dried powders containing gum acacia. J. Appl. Microbiol. 93:1003-1011. [DOI] [PubMed] [Google Scholar]
- 7.Desmond, C., C. Stanton, G. F. Gitzgerald, K. Collins, and R. P. Ross. 2001. Environmental adaptation of probiotic lactobacilli towards improved performance during spray drying. Int. Dairy J. 11:801-808. [Google Scholar]
- 8.Dien, B. S., N. N. Nichols, and R. J. Bothast. 2002. Fermentation of sugar mixtures using Escherichia coli catabolite repression mutants engineered for production of L-lactic acid. J. Ind. Microbiol. Biotechnol. 29:221-227. [DOI] [PubMed] [Google Scholar]
- 9.Dunne, C., L. O'Mahony, L. Murphy, G. Thornton, D. Morrissey, S. O'Halloran, M. Feeney, S. Flynn, G. Fitzgerald, C. Daly, B. Kiely, G. C. O'Sullivan, F. Shanahan, and J. K. Collins. 2001. In vitro selection criteria for probiotic bacteria of human origin: correlation with in vivo findings. Am. J. Clin. Nutr. 73:386S-392S. [DOI] [PubMed] [Google Scholar]
- 10.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frisk, A., C. A. Ison, and T. Lagergard. 1998. GroEL heat shock protein of Haemophilus ducreyi: association with cell surface and capacity to bind to eukaryotic cells. Infect. Immun. 66:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardiner, G., R. P. Ross, J. K. Collins, G. Fitzgerald, and C. Stanton. 1998. Development of a probiotic Cheddar cheese containing human-derived Lactobacillus paracasei strains. Appl. Environ. Microbiol. 64:2192-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardiner, G. E., E. O'Sullivan, J. Kelly, M. A. Auty, G. F. Fitzgerald, J. K. Collins, R. P. Ross, and C. Stanton. 2000. Comparative survival rates of human-derived probiotic Lactobacillus paracasei and L. salivarius strains during heat treatment and spray drying. Appl. Environ. Microbiol. 66:2605-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georgopoulos, C., and W. J. Welch. 1993. Role of the major heat shock proteins as molecular chaperones. Annu. Rev. Cell Biol. 9:601-634. [DOI] [PubMed] [Google Scholar]
- 15.Hennequin, C., F. Porcheray, A. Waligora-Dupriet, A. Collignon, M. Barc, P. Bourlioux, and T. Karjalainen. 2001. GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology 147:87-96. [DOI] [PubMed] [Google Scholar]
- 16.Hoffman, C. S., and F. Winston. 1987. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57:267-272. [DOI] [PubMed] [Google Scholar]
- 17.Jan, G., P. Leverrier, V. Pichereau, and P. Boyaval. 2001. Changes in protein synthesis and morphology during acid adaptation of Propionibacterium freudenreichii. Appl. Environ. Microbiol. 67:2029-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, W. S., L. Perl, J. H. Park, J. E. Tandianus, and N. W. Dunn. 2001. Assessment of stress response of the probiotic Lactobacillus acidophilus. Curr. Microbiol. 43:346-350. [DOI] [PubMed] [Google Scholar]
- 19.Kleerebezem, M., M. M. Beerthuyzen, E. E. Vaughan, W. M. de Vos, and O. P. Kuipers. 1997. Controlled gene expression systems for lactic acid bacteria: transferable nisin-inducible expression cassettes for Lactococcus, Leuconostoc, and Lactobacillus spp. Appl. Environ. Microbiol. 63:4581-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klose, J. 1975. Protein mapping by combined isoelectric focusing and electrophoresis of mouse tissues. A novel approach to testing for induced point mutations in mammals. Humangenetik 26:231-243. [DOI] [PubMed] [Google Scholar]
- 21.Kullen, M. J., and T. R. Klaenhammer. 1999. Identification of the pH-inducible, proton-translocating F1F0-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol. Microbiol. 33:1152-1161. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Luchansky, J. B., P. M. Muriana, and T. R. Klaenhammer. 1988. Application of electroporation for transfer of plasmid DNA to Lactobacillus, Lactococcus, Leuconostoc, Listeria, Pediococcus, Bacillus, Staphylococcus, Enterococcus and Propionibacterium. Mol. Microbiol. 2:637-646. [DOI] [PubMed] [Google Scholar]
- 24.Narberhaus, F., and H. Bahl. 1992. Cloning, sequencing, and molecular analysis of the groESL operon of Clostridium acetobutylicum. J. Bacteriol. 174:3282-3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neu, T., and B. Henrich. 2003. New thermosensitive delivery vector and its use to enable nisin-controlled gene expression in Lactobacillus gasseri. Appl. Environ. Microbiol. 69:1377-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 27.Pavan, S., P. Hols, J. Delcour, M. C. Geoffroy, C. Grangette, M. Kleerebezem, and A. Mercenier. 2000. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl. Environ. Microbiol. 66:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piper, P. W. 1995. The heat shock and ethanol stress responses of yeast exhibit extensive similarity and functional overlap. FEMS Microbiol. Lett. 134:121-127. [DOI] [PubMed] [Google Scholar]
- 29.Prasad, J., P. McJarrow, and P. Gopal. 2003. Heat and osmotic stress responses of probiotic Lactobacillus rhamnosus HN001 (DR20) in relation to viability after drying. Appl. Environ. Microbiol. 69:917-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rechinger, K. B., H. Siegumfeldt, I. Svendsen, and M. Jakobsen. 2000. “Early” protein synthesis of Lactobacillus delbrueckii ssp. bulgaricus in milk revealed by [35S]methionine labeling and two-dimensional gel electrophoresis. Electrophoresis 21:2660-2669. [DOI] [PubMed] [Google Scholar]
- 31.Ricci, J. C. D., and M. E. Hernandez. 2000. Plasmid effects on Escherichia coli metabolism. Crit. Rev. Microbiol. 20:79-108. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt, A., M. Schiesswohl, U. Volker, M. Hecker, and W. Schumann. 1992. Cloning, sequencing, mapping, and transcriptional analysis of the groESL operon from Bacillus subtilis. J. Bacteriol. 174:3993-3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt, G., C. Hertel, and W. P. Hammes. 1999. Molecular characterisation of the dnaK operon of Lactobacillus sakei LTH681. Syst. Appl. Microbiol. 22:321-328. [DOI] [PubMed] [Google Scholar]
- 34.Shah, N. P. 2000. Probiotic bacteria: selective enumeration and survival in dairy foods. J. Dairy Sci. 83:894-907. [DOI] [PubMed] [Google Scholar]
- 35.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 36.Stanton, C., C. Desmond, M. Coakley, J. K. Collins, G. Fitzgerald, and R. P. Ross. 2003. Challenges facing development of probiotic-containing functional foods, p. 27-58. In E. Farnworth (ed.), Probiotics and health. CRC Press, Boca Raton, Fla.
- 37.Terracciano, J. S., and E. R. Kashket. 1986. Intracellular conditions required for initiation of solvent production by Clostridium acetobutylicum. Appl. Environ. Microbiol. 52:86-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tomas, C. A., N. E. Welker, and E. T. Papoutsakis. 2003. Overexpression of groESL in Clostridium acetobutylicum results in increased solvent production and tolerance, prolonged metabolism, and changes in the cell's transcriptional program. Appl. Environ. Microbiol. 69:4951-4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Torok, Z., I. Horvath, P. Goloubinoff, E. Kovacs, A. Glatz, G. Balogh, and L. Vigh. 1997. Evidence for a lipochaperonin: association of active protein-folding GroESL oligomers with lipids can stabilize membranes under heat shock conditions. Proc. Natl. Acad. Sci. USA 94:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van de Guchte, M., P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich, and E. Maguin. 2002. Stress responses in lactic acid bacteria. Antonie Leeuwenhoek 82:187-216. [PubMed] [Google Scholar]
- 41.Walker, D. C., H. S. Girgis, and T. R. Klaenhammer. 1999. The groESL chaperone operon of Lactobacillus johnsonii. Appl. Environ. Microbiol. 65:3033-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter, J., N. C. Heng, W. P. Hammes, D. M. Loach, G. W. Tannock, and C. Hertel. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 69:2044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, J., and L. Chen. 2003. Domain motions in GroEL upon binding of an oligopeptide. J. Mol. Biol. 334:489-499. [DOI] [PubMed] [Google Scholar]
- 44.Yuan, G., and S. L. Wong. 1995. Regulation of groE expression in Bacillus subtilis: the involvement of the sigma A-like promoter and the roles of the inverted repeat sequence (CIRCE). J. Bacteriol. 177:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]