Abstract
Microbial infections of medical implants occur in more than 2 million surgical cases each year in the United States alone. These increase patient morbidity and mortality, as well as patient cost and recovery time. Many treatments are available, but none are guaranteed to remove the infection. In many cases, the device infections are caused by the adhesion of microbes to the implant, ensuing growth, pathogenesis, and dissemination. The purpose of this work is to examine the initial events in microbial adhesion by simulating the approach and contact between a planktonic cell, immobilized on an atomic force microscope (AFM) cantilever, and a biomaterial or biofilm substrate. The two model microbes used in this study, Candida parapsilosis (ATCC 90018) and Pseudomonas aeruginosa (ATCC 10145), were chosen for both their clinical relevance and their ease of acquisition and handling in the laboratory setting. Attractive interactions exist between C. parapsilosis and both unmodified silicone rubber and P. aeruginosa biofilms. Using C. parapsilosis cells immobilized on AFM cantilevers with a silicone substrate, we have measured attractive forces of 4.3 ± 0.25 nN in the approach portion of the force cycle. On P. aeruginosa biofilms, the magnitude of the attractive force decreases to 2.0 ± 0.40 nN and is preceded by a 2.0-nN repulsion at approximately 75 nm from the cell surface. These data suggest that C. parapsilosis may adhere to both silicone rubber and P. aeruginosa biofilms, possibly contributing to patient morbidity and mortality. Characterization of cell-biomaterial and cell-cell interactions allows for a quantitative link between the physicomechanical and physicochemical properties of implant materials and the nanoscale interactions leading to microbial colonization and infection.
The development of the microbial biofilm and its importance in medical implant infections have been thoroughly discussed (13, 40). Causing over 2 million infections annually (22), which generate over $11 billion in additional patient costs (36), the biofilm, in a biomedical context, is a system which demands attention.
The application of atomic force microscopy (AFM) to biological systems seems advantageous and has been examined by a number of groups, beginning with early work on DNA (26) that was later extended to whole-cell systems. AFM was used to examine the physicochemical properties of microbial surfaces (17) and to characterize lectin-carbohydrate interactions at the nanoscale (42). AFM probes, functionalized with either biomaterial spheres or confluent microbial lawns, were used to characterize bacterial-biomaterial interactions (34, 35).
Single microbes, immobilized on AFM probes, have also been used to study a variety of surfaces. Bowen et al. (6) bound metabolically active Saccharomyces cerevisiae to the probe using a chemical adhesive. Also, Benoit et al. (4) measured discrete intercellular interactions by using Dictyostelium discoideum attached to the probe via a lectin. We propose to extend these prior techniques by attaching viable, clinically relevant microbes to the AFM cantilever. These cell probes will then be used to measure local interaction forces between the immobilized cell and both biomaterial and biofilm surfaces.
Many infectious systems can be polymicrobial, each strain contributing to the whole in some significant way (22, 24). Therefore, our model microbial system is designed to study the interactions between Pseudomonas aeruginosa, a bacterium responsible for 99% of bacteremia cases, keratitis, and infections in cystic fibrosis patients (28), and Candida parapsilosis, an emerging nosocomial pathogen isolated in infections of central venous catheters and bronchoscopes (27, 31). In our experiments, Candida cells are chemically immobilized onto AFM probes and used to quantify interactions with bare biomaterials and with biomaterials coated with a bacterial biofilm.
MATERIALS AND METHODS
Microbial samples, storage, and growth.
Freeze-dried samples of C. parapsilosis (ATCC 90018) and P. aeruginosa (ATCC 10145) were acquired from the American Type Culture Collection (Manassas, Va.). C. parapsilosis was maintained on Sabouraud dextrose agar (Emmons' modification; Sigma) plates and grown in complete liquid medium (2% [wt/vol] peptone, 1% [wt/vol] yeast extract, 2% [wt/vol] glycerol [Sigma]). P. aeruginosa was maintained on tryptic soy agar (Sigma) plates and grown in tryptic soy broth (Sigma). Cells were grown until mid-exponential growth phase in liquid medium (optical density at 600 nm [OD600] ≅ 0.5).
Characterization of cell morphology, force interactions, and cantilever treatment.
Cell cultures were examined with a Dimension 3100 atomic force microscope with a Nanoscope IIIa controller (Digital Instruments, Santa Barbara, Calif.) by using silicon nitride cantilevers (DNPS; Digital Instruments; spring constants of 0.13 ± 0.02 N · m−1 [11] or 0.25 ± 0.01 N · m−1 [47]). Cantilevers were cleaned by exposure to UV light (5 min) to remove adsorbed water and/or hydrocarbons. The unit was configured for intermittent contact mode in liquid per the manufacturer's instructions.
To attach cells, each slide was coated with 1 ml of poly-l-lysine and dried in a laminar flow hood for 10 min. Cellular solutions were centrifuged (1,350 × g, 15 min), washed with 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) buffer (Aldrich; pH 7.1), vortexed, and poured over the dry glass slides in a petri dish. The covered petri dish was placed on a shaker table for 20 min to allow attachment.
AFM images were captured for each cell found, and five force cycles per cell were recorded with drive amplitude set to zero (approximating contact mode). All force curves were recorded at approximately the same point on the cell, defined as the highest point midway down the length of the cell (48).
For cell probe experiments, cantilevers were cleaned by placement under UV light (30 s). Cantilevers were further cleaned by sonication (1 min in double-distilled water and 1 min in reagent-grade ethanol [Aldrich]). To allow cell immobilization, 50 μl of a 1-hexadecanethiol (HDT) stock solution (10 mM in reagent-grade ethanol) was placed on a polytetrafluoroethylene block, after which the cantilevers were placed into the droplet, removed, and allowed to dry.
Cell probe preparation.
Single C. parapsilosis cells were attached to AFM cantilevers by using a custom-designed triaxial micromanipulator with extension arm (stages from Edmund Industrial Optics, Barrington, N.J.). A single silicon nitride chip, with two triangular cantilevers, was attached to the extension arm by using double-sided tape. Twenty-five microliters of Candida culture (∼1011 cells · ml−1) was pipetted to a channel of comparable volume etched into a polytetrafluoroethylene block. The cantilever was oriented over the cell culture droplet such that both the tips on the cantilever and the droplet were visible under a stereoscope (Eclipse SMZ-10A; Nikon) and moved vertically downwards to the droplet until the tips were submerged. Tips were left for 5 min to allow cell adhesion to the HDT monolayer. The withdrawn cantilever was dried for 5 min in a laminar flow hood and viewed with an optical microscope (Eclipse E400; Nikon) equipped with a UV (λ = 330 to 380 nm) filter cube to verify attachment. Further, adhesion, number of cells adhered, and cell location were verified using scanning electron microscopy (SEM). The short, fat cantilever gave the best reproducibility for cell immobilization.
SEM.
Six cellular probes were viewed via SEM (Amray 1610 Turbo; Bedford, Mass.) after dehydration and coating with gold. Each cantilever was examined at the tip, base, and legs (magnification, ×5,000 to ×15,000).
Approach interactions at the cell-biomaterial interface.
A 2-cm2 area of medical-grade silicone rubber tubing (VWR; 0.25-in. inside diameter) was cut and sliced axially and taped to the bottom half of a petri dish, to which MES buffer was added. Force cycles were recorded for 8 to 10 different areas with three data sets taken on each area. Silicone was examined with the Candida probe and with the unmodified probe and HDT-coated probe, the latter two serving as controls.
Approach interactions at the cell-biofilm interface.
The parallel plate flow cell (BioSurface Technologies Corporation, Bozeman, Mont.) was opened and cleaned, after which a coupon of silicone rubber was fixed into the channel with double-sided tape. The flow cell was assembled and sterilized according to the manufacturer's instructions and mounted on a support brace in an orbital shaker bath (Lab-Line) at 37°C. Sixty milliliters of P. aeruginosa (OD600 ≈ 0.5) was centrifuged and resuspended in 0.1 M MES buffer and was then continuously pumped through the flow cell at 380 cm · min−1 (Reynolds number, ∼200). After biofilms were allowed to form for 1 or 3 days in the flow cell, the chamber was opened and the silicone was examined with AFM by using the Candida probe, the unmodified cantilever, and the HDT-coated cantilever.
Electrophoretic mobility analysis.
The electrophoretic mobilities and surface potentials of C. parapsilosis and P. aeruginosa were measured using a zeta potential analyzer (Zeta PALS; Brookhaven, Holtsville, N.Y.). Mid-exponential-growth-phase cultures were centrifuged (1,350 × g) and resuspended in MES buffer (25°C, pH 7.1) at several ionic strengths (0.02, 0.04, 0.06, 0.08, and 0.1 M) and in deionized water (∼10−6 M). All measurements were recorded four times and averaged.
Zeta potentials reported by the software are automatically calculated according to the Smoluchowski equation (45). Since recent work has shown that this formulation overestimates surface potentials for soft materials, such as bacteria (23, 30), we also applied the soft-particle DLVO (Derjaguin-Landau-Vervey-Overbeek) theory of Ohshima et al. (23, 30). A thorough description of the application of these models to our system may be found elsewhere (19).
Interaction energy calculations.
Advancing contact angles were recorded for each microbe under water, formamide, and diiodomethane, by using a Rame-Hart NRL contact angle goniometer (model 100; Mountain Lakes, N.J.) with 80 readings taken on both sides of each liquid droplet and averaged. The Hamaker constants were obtained from the microbial apolar surface free energy, by using the van Oss-Chaudhury-Good equation (44-46). We calculate the interaction energy as a function of particle separation according to the two DLVO theories, which can demonstrate London-van der Waals and electrostatic interactions.
Modeling of steric interactions with the microbial polymer brush.
A steric model of the total force experienced by two interacting surfaces, one covered with grafted polymers and the other bare, was developed to describe the total force (Fst) due to steric interactions of an AFM probe with a polymer brush (3, 10, 14):
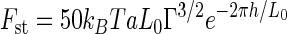 |
(1) |
where kB is the Boltzmann constant, L0 is the equilibrium polymer brush length, h is the distance between the probe and the sample, T is absolute temperature, a is tip radius of curvature, and Γ is grafted polymer density. While the probe has a nominal radius of curvature of 40 ± 20 nm, its behavior in terms of long-range interaction energies (van der Waals, electrostatic, etc.) is that of a much larger (130- to 380-nm) sphere (15, 37). For a similar microbial system, a larger radius (250 nm) was appropriate for the modeling of steric interactions (11), and so 250 nm was used in this study. By using nonlinear regression software, the model was fitted to the approach curves.
RESULTS
Characterization of cell morphologies and force interactions.
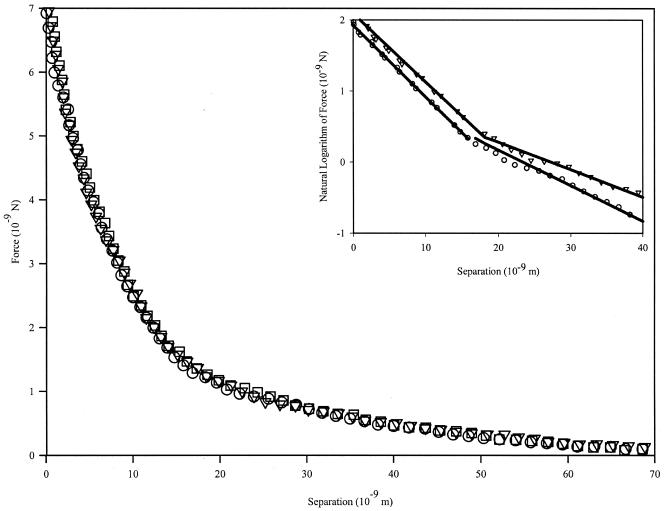
P. aeruginosa cells are rod shaped (2 to 3 μm by 800 nm; images not shown). Measurements of the forces between the unmodified silicon nitride probe and individual cells of P. aeruginosa during the approach of the probe to the cell show interactions beginning at 70 nm from the cell surface (Fig. 1). Only repulsive interactions exist for this cell during the approach portions of the force cycle, which reach a maximum value of 6 ± 1 nN at the point of zero separation.
FIG. 1.
P. aeruginosa approach curves with unmodified silicon nitride probe. Data shown represent three measurements on a single cell. Interactions begin at ∼70 nm from the cell surface and are purely repulsive in nature. Repulsion at zero separation distance reaches a value of 7.0 ± 1.4 nN. The inset figure demonstrates the two slopes seen when plotting the natural logarithm of the force data versus separation. This behavior suggests that the cantilever is detecting two separate polymer brushes grafted to the cell surface.
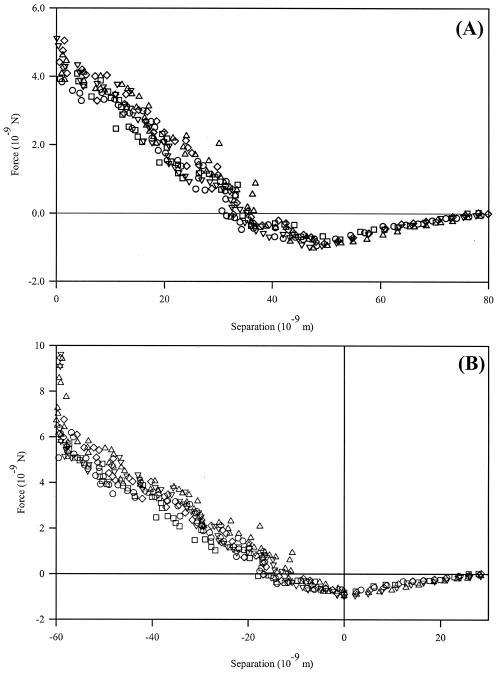
C. parapsilosis cells are spherical (diameters, 4 to 6 μm; images not shown). The interactions between an unmodified silicon nitride probe and individual cells of C. parapsilosis are attractive, reaching 0.9 ± 0.16 nN approximately 55 nm from the origin (Fig. 2A). This is in contrast with the findings of Bowen et al. (6), who showed no attractive behavior in the approach interactions of S. cerevisiae probes on mica. Differences in experimental conditions (e.g., substrate, pH, and microbe surface polymers) could explain the differences between these data sets.
FIG. 2.
C. parapsilosis approach curves with unmodified silicon nitride probe. Data shown represent three measurements on a single cell. Based on the method of Ducker and Senden (16), the curves appear as in panel A, with the constant compliance region aligned with the vertical axis. However, according to contact mechanics (9), we may also define the minimum of the attractive force, viz., where all force gradients acting on the cantilever sum to zero, as the cell surface (B). Whether either method accurately describes the polymer layer on the surface has yet to be determined.
High-force physical agitation of the cells (viz., the centrifugation and vortexing steps used to prepare cells) may be a concern, as this agitation could dislodge cell surface structures. Pembrey et al. (32) have shown, however, that cell surface properties (e.g., electrophoretic mobility, structural integrity, and viability) are not significantly altered by agitation at or below 5,000 × g. The centrifugation and vortexing protocols utilized in these experiments do not exert a force of >1,350 × g on the cells. Therefore, we do not expect cell surface properties to be altered by our protocols.
Interpretation of the distance of these interactions is a difficult and important issue, since several methods exist to define the cell surface. For example, the data in Fig. 2A were calibrated according to the method proposed by Ducker and Senden (16), in which the constant compliance region of the curve is aligned with the force axis. An alternate approach, following contact mechanical methods (8, 9), defines the sample surface as the minimum point of the attraction, where all forces acting on the cantilever sum to zero (Fig. 2B). Neither method, however, explicitly accounts for the extracellular polymer layer expected for a microbial sample. Such interpretation issues of attractive interactions in approach curves are discussed below.
Cell probe preparation and SEM analysis.
After preparation of the cellular probes, we verified adhesion and placement of the cells on the cantilevers by SEM.
Approach interactions at the cell-biomaterial interface.
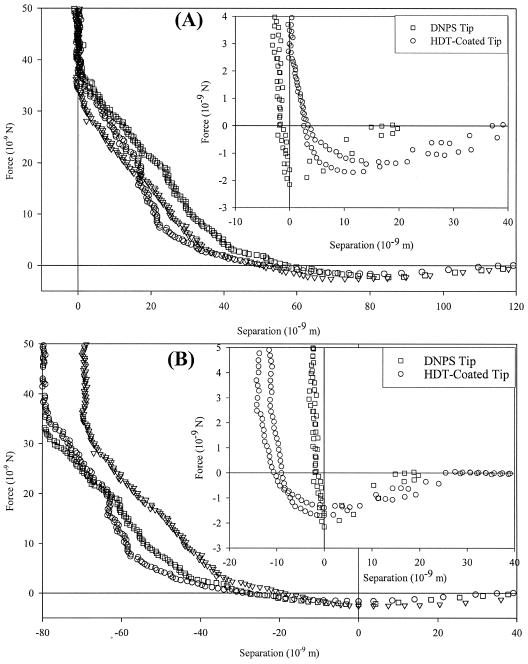
The interactions between silicone rubber and the cellular modified probe are attractive (Fig. 3), beginning at 120 nm from the cell surface with a magnitude of 2.3 ± 0.25 nN at a range of 78 nm. Both the magnitude and the range of these attractions are larger than those for bare and HDT-modified control cantilevers. If we directly apply the method of Ducker and Senden, then a strong steric repulsion occurs (Fig. 3A), beginning at ∼50 nm from the cell surface and reaching a magnitude of ∼35 nN at the silicone rubber surface. Contact mechanical analysis (Fig. 3B) eliminates this repulsive interaction, showing a total range of the attraction as ∼40 nm.
FIG. 3.
Approach curves for C. parapsilosis-modified cantilever approaching a bare silicone rubber surface. The inset figures show curves for a bare cantilever (squares) and an HDT-coated cantilever (circles) on the same material. Attractive events exist for each system. Panel A shows the data analyzed according to the Ducker and Senden method (16), while panel B was analyzed by contact mechanics (9). The modified probe has the strongest interactions with the cell-modified probes and the weakest interactions with the HDT-modified probes. Note that the different data analysis techniques do not change the interaction for a bare probe on silicone rubber.
Approach interactions at the cell-biofilm interface.
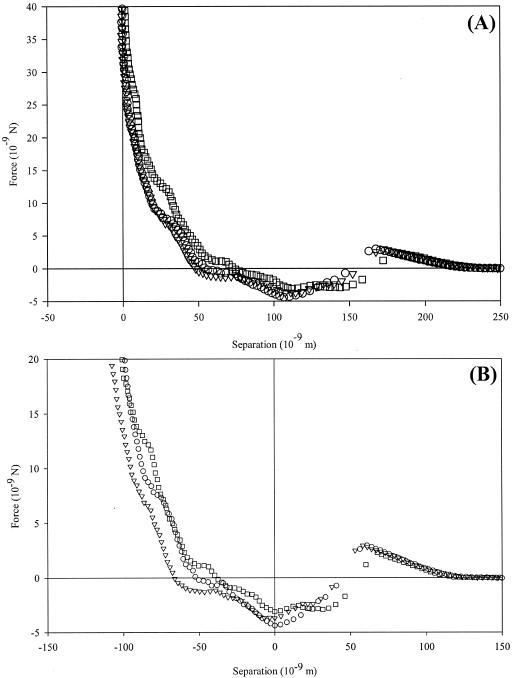
For a representative 3-day biofilm, AFM approach curves of a modified Candida probe examining the P. aeruginosa biofilm show both attractive and repulsive regions (Fig. 4). By the Ducker and Senden method of determining the origin of the force cycles (Fig. 4A), interactions begin at ∼250 nm from the biofilm surface, initially showing repulsion that reaches a maximum of 1.7 nN at 165 nm. At a range of ∼115 nm, there is an attractive interaction with a magnitude of 3.7 ± 0.64 nN. By use of the contact mechanical models, the surface repulsion is again ignored, and the total interaction range is truncated to 130 nm (Fig. 4B). Data were identical for biofilm growth experiments lasting 1 and 3 days in the parallel plate flow chamber, suggesting that adhesion and detachment of bacteria to the surface reach a steady state relatively quickly.
FIG. 4.
Approach curves with a C. parapsilosis-modified probe in examining a bacterial biofilm grown for 3 days. Panel A shows the data analyzed according to the Ducker and Senden method (16), while panel B was analyzed by contact mechanics (9). A repulsive interaction begins at ∼250 nm from the cell surface. Repulsions are followed by attractive events, which reach magnitudes of 3.7 ± 0.64 nN at ∼115 nm.
Electrophoretic mobility analysis.
Over the range of ionic strengths, the zeta potential for P. aeruginosa increases from −8.0 mV at 20 mM to −3.4 mV at 100 mM (Table 1). We see more variation as a function of ionic strength among the data for C. parapsilosis (−6.3 to −3.4 mV). As expected, for both microbes, lower values for zeta potentials are calculated by using soft-particle theory than by using the zeta potential as the surface potential via the Smoluchowski equation. The exception is at low ionic strengths, where soft-particle DLVO theory is least able to represent experimental results.
TABLE 1.
Zeta potential measurements for C. parapsilosis and P. aeruginosaa
| Organism and concn (mM) | pH | ζSmolb (103 V) | EMc (108 m2 · [V · s]−1) | ψ0d (103 V) |
|---|---|---|---|---|
| C. parapsilosis | ||||
| 20 | 6.81 | −5.1 | −0.39 ± 0.01 | −6.2 |
| 40 | 6.84 | −4.5 | −0.35 ± 0.18 | −3.1 |
| 60 | 6.99 | −3.4 | −0.26 ± 0.11 | −2.1 |
| 80 | 7.01 | −6.3 | −0.49 ± 0.10 | −1.6 |
| 100 | 7.03 | −4.7 | −0.37 ± 0.10 | −1.3 |
| P. aeruginosa | ||||
| 20 | 6.95 | −8.0 | −0.62 ± 0.14 | −10.3 |
| 40 | 6.98 | −6.7 | −0.52 ± 0.07 | −5.3 |
| 60 | 6.99 | −4.4 | −0.34 ± 0.08 | −3.6 |
| 80 | 7.01 | −5.4 | −0.42 ± 0.28 | −2.7 |
| 100 | 7.01 | −3.4 | −0.26 ± 0.10 | −2.2 |
All samples were grown at 37°C until the OD600 was approximately 0.5 and suspended in MES buffer with varying ionic strength to a cell concentration of approximately 108 cells/ml.
Zeta potential as calculated from the Smoluchowski equation.
Average electrophoretic mobility of microbial culture (n = 4) with SDs of repeated data sets.
Surface potential as calculated from soft-particle DLVO theory; this is equivalent to the zeta potential of the solution (36).
Interaction energy calculations.
By using two polar liquids (water and formamide) and one nonpolar liquid (diiodomethane), contact angles on microbial lawns may be translated into surface free energy components. Both microbes are relatively hydrophilic, but C. parapsilosis (θW = 15.17 ± 11.5° [standard deviation (SD)]) is more hydrophilic than P. aeruginosa (θW = 24.42 ± 1.5° [SD]) (Table 2). These values, and the corresponding surface free energy components, differ significantly from published values for P. aeruginosa Olin (21) and C. parapsilosis strains 294 and 289 (20), but such differences could be related to different culture or environmental conditions. Poortinga et al. (33) showed that DLVO theory in biological systems may be modified by modeling the cell surface as a soft, ion-penetrable particle. The models, however, would be specific to each microbial strain examined, since the interactions predicted by DLVO theory are more complex in a biological system.
TABLE 2.
Microbial contact angle and surface tension dataa
| Parameter | C. parapsilosis | P. aeruginosa |
|---|---|---|
| pH | 7.03 | 7.01 |
| θW (°)b | 15.17 ± 11.5 | 24.42 ± 1.5 |
| θF (°)c | 13.41 ± 1.4 | 32.28 ± 3.9 |
| θD (°)d | 46.63 ± 1.2 | 61.69 ± 0.3 |
| γMLW (mJ · m−2)e | 36.13 ± 0.65 | 27.56 ± 0.15 |
| γM+ (mJ · m−2)f | 1.96 ± 0.39 | 2.05 ± 0.57 |
| γM− (mJ · m−2)g | 51.94 ± 6.42 | 60.29 ± 3.55 |
| A (1020 J) | 6.71 ± 0.12 | 5.12 ± 0.03 |
Microbes were examined under three liquids (water, formamide, and diiodomethane). By using the van Oss-Chaudhury-Good equation for the three liquids (45), the three components of surface tension may be calculated. Hamaker constants (A) follow directly from the values of γMLW. SDs of repeated data sets are shown.
Contact angle of microbial lawn under double-distilled water.
Contact angle of microbial lawn under formamide.
Contact angle of microbial lawn under diiodomethane.
Apolar (Lifshitz-van der Waals) component of microbial surface free energy.
Electron-accepting parameter of the polar microbial surface free energy.
Electron-donating parameter of the polar microbial surface free energy.
Simultaneous solution of three instances of the van Oss-Chaudhury-Good equation (45) yields the three surface tension components (Table 2). The values of the apolar surface tension component are then used to calculate the Hamaker constants (45), which have values of (6.71 ± 0.12) × 10−20 J (SD) for C. parapsilosis and (5.12 ± 0.03) × 10−20 J (SD) for P. aeruginosa. Values for the two strains are in good agreement (<10−21-J variation) with reported values of similar strains (20, 21).
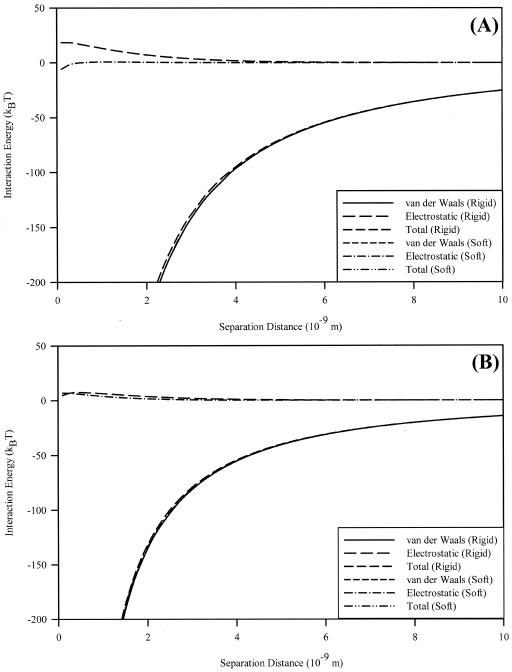
Interaction energy profiles for the interaction of each microbe with silicon nitride were calculated (Fig. 5). C. parapsilosis shows no energy barrier preventing adhesion by soft-particle DLVO theory but rather an attractive energy of 5.86 kBT. An electrostatic repulsion of 18.4 kBT, starting at 0.3 nm, exists in the calculation based on the rigid-particle calculation. P. aeruginosa shows a 4.83-kBT electrostatic repulsion at 0.1 nm by the rigid-particle model, while at the same distance this repulsion has a magnitude of 7.00 kBT by the soft-particle model. For both microbes, these small repulsions are greatly outweighed by van der Waals attractive interactions at very small separation distances. Neither classical nor soft-particle DLVO theory agrees with the behavior seen in AFM force curves (Fig. 1 and 2). Also, the closeness of the predictions based on the two models indicates that the ion penetrability of the sample is unimportant in the overall interaction. As such, the behavior shown in the force curves must be due to additional physicochemical or physicomechanical phenomena.
FIG. 5.
Interaction energies for C. parapsilosis (A) and P. aeruginosa (B) in 100 mM MES buffer. Shown are total energy and the individual contributions for van der Waals and electrostatic interactions, with the differences between soft-particle (points) and rigid-particle (lines) DLVO theories. The two show nearly identical results, save for small differences in electrostatic interactions at very short (<4-nm) separation distances.
Modeling of steric interactions with the cellular polymer brush.
The approach curves were fitted using the polymer scaling model, with the grafted polymer density and equilibrium polymer length as fitting parameters. Steric interactions are relevant only in systems that show repulsive interactions. When force curves are analyzed according to the method of Ducker and Senden, the steric model is applicable to all systems.
The steric model was in good agreement with experimental data (Table 3), except for the silicone rubber-DNPS tip system. Since no polymer brush is present on this surface, it was necessary to process the data according to contact mechanical models, viz., aligning the minimum of the attractive interaction with the point of zero separation.
TABLE 3.
Quantification of equilibrium polymer length and polymer grafting density for P. aeruginosa as obtained by the steric modela
| Parameter | Sample and probe
|
||||
|---|---|---|---|---|---|
| P. aeru- ginosa and DNPS | C. parap- silosis and DNPS | Silicone and HDT | Silicone and C. parap- silosis | Biofilm and C. parap- silosis | |
| Γ1 (10−16 m−2)b | 2.48 | 3.12 | 12.4 | 11.1 | 11.5 |
| Γ2 (10−16 m−2)b | 7.49 | ||||
| L1 (nm)c | 92.6 | 157 | 0.88 | 130 | 109 |
| L2 (nm)c | 24.0 | ||||
| R2 | 1.00 | 0.91 | 0.88 | 0.96 | 0.98 |
Five measurements were taken for each sample-probe combination and fitted with either the basic or modified steric model. R2 values are also presented. Γ2 and L2 were calculated only for P. aeruginosa.
Polymer grafting density per unit area.
Equilibrium polymer length of microbial surface features.
We noted that, for P. aeruginosa, although steric model fits agreed acceptably with the collected data (data not shown), the model failed to fit the data points at separation distances of ∼20 nm from the surface. When the data were plotted as the natural logarithm of the force versus the separation distance (Fig. 1, inset), an inflection point could be seen at the distance where the steric model fails to fit the data well. As such, we formulated an extension to the steric model which accounts for two polymer layers, each having a different grafting density and equilibrium polymer brush length. This is represented mathematically by changing the term L0Γ3/2 e−2πh/L0 in equation 1 to L1Γ13/2 e−2πh/L1+L2Γ23/2 e−2πh/L2. Fitting parameters for this extended model may be seen in Table 3. Modeling the cell as a cylinder with a 1-μm radius and two hemispherical end caps with 1-μm radii, the grafting densities for P. aeruginosa are 7.86 × 104 and 3.96 × 105 polymers · cell−1 for the long and short polymers, respectively.
DISCUSSION
Interpretation of AFM force curves.
AFM force curves may be interpreted in several ways, depending upon the reference points used to define the sample surface and baseline interaction force. For example, Ducker and Senden (16) align deflection data with the axes of a Cartesian plane, with the assumption that the cantilever is significantly more compliant than the sample surface, with the use of two regions of the curve as reference points. The linear constant compliance region is aligned with the vertical force axis, while the zero interaction region is aligned with the horizontal separation axis. Contact mechanical models, such as those presented in reference 9, define the reference points mathematically. These different models are discussed more thoroughly elsewhere (19). Analysis of the constant compliance regions recorded in this study (data not shown) shows that they agree within 5% with those recorded for glass by using cantilevers of comparable stiffness. This justifies the assumption that the cantilever is significantly more compliant than the substrate and validates our use of the Ducker method of force curve analysis.
Approach curves shown in Fig. 2 illustrate the differences between the Ducker and Senden method and a contact mechanical model defining the surface as the attractive minimum, respectively, for C. parapsilosis probed with an unmodified DNPS tip. The contact mechanical model employed requires that the surface be at this minimum and ignores the shape of the force curve. For many probe-sample combinations, this is a logical treatment, since the constant compliance region begins directly after the attractive minimum (Fig. 3, inset), and all cantilever deflection is due to scanner motion. However, for many systems, especially those involving polymer brushes, application of these models suggests significant indentation into the cell wall. This negates the primary assumption that the cantilever is more compliant than the sample surface.
It is therefore our argument that, since the constant compliance region is used as the reference point in all systems not involving polymer brushes (9, 16), the same reference should be used for those with polymer brushes, enabling comparison between disparate systems. Data in this paper have been reported according to this latter method, and data treated according to the contact mechanical model are shown for comparison.
Cell probe preparation and SEM analysis.
The present immobilization technique was developed to minimize the area of the cell that would be chemically treated, thus reducing artifacts due to chemical treatments. Previous work from our laboratory has shown that chemical treatments with fluorescent and visible-spectrum stains alter the magnitudes and locations of AFM force interactions for these microbes (data not shown). It is therefore likely that other chemical treatments (e.g., attachment protocols) would also introduce artifacts in the recorded data.
Examination of the Candida cellular probes with SEM showed cells bound to the cantilever with multiple cells present in some cases. In each case, however, only single cells were present near the cantilever tip and available to interact with the sample surface. Since the height of the tips is small (∼1 μm) relative to the height of the cells, the chemisorbed Candida cells are the major bodies interacting with the sample surfaces. It was also noted that, while the cells were not perfectly aligned at the apex of each cantilever, the tips themselves were, in all cases, off center. Any changes in force curves due to this misalignment of the tip are corrected for when defining the coordinate system. The same technique should be directly applicable to correcting for an off-center cell chemisorbed to the cantilever.
Approach interactions at the cell-biomaterial interface.
For a C. parapsilosis-modified probe interacting with silicone rubber, strong adhesive interactions exist. It is possible that irregularities in the silicone surface may account for the variation in attraction magnitude, causing microbial surface structures to interact with different regions of the material surface. There is little variation in the distance over which the attractions occur, however, which reinforces SEM results of single cells interacting with the substrate. The role of such structures in lectin-ligand binding to specific sites has been investigated by a number of groups (39, 42). While no specific structures have been identified for C. parapsilosis, various other yeasts (Kluyveromyces bulgaricus [18], S. cerevisiae [12], and Candida albicans [38]) produce them.
Retraction curves for the various systems were also analyzed (data not shown). For each system, adhesive events occurred and may be qualitatively correlated to the systems by their magnitude and distance from the surface. Also, we saw no variation with time or number of measurements in approach curves, suggesting that none of the cell surface molecules involved in the interaction are lost by adhesion to the substrate. Further information may be found in reference 19.
Approach interactions at the cell-biofilm interface.
Interactions between a modified probe and a nascent P. aeruginosa biofilm also exhibit attraction on approach. Immediately before this attraction occurs, long-range (∼250-nm) repulsion occurs, reaching a maximum at ∼175 nm from the biofilm surface. This can be attributed to a combination of electrostatic double-layer effects associated with the polymer brushes on each microbe, as well as steric effects. Indeed, fitting this data with a single-polymer steric model yields an equilibrium polymer brush length of ∼175 nm and a grafting density of 9.7 × 1017 polymers · m−2. There is little variation in the distance over which the attractions occur, suggesting that the probe experiences an “average” interaction with a substrate expected to be topographically heterogeneous.
As the Candida-modified probe moves closer to the surface, strong attractive interactions begin to dominate. This suggests that an initial energy barrier must be overcome to reach an energetic minimum favoring adhesion. Therefore, the planktonic microbe must have sufficient force, associated with bulk flow, gravitational settling, or inherent mobility, to initially bind to a biofilm. The attractions are also of much higher (as much as 3× greater) magnitude than those seen for a modified probe-bare biomaterial system, indicating that planktonic cells have a higher affinity for binding to surfaces on which a biofilm is already growing.
Microbial interaction energy analyses.
Microbial surface potentials were calculated using classical and soft-particle DLVO theories, with DLVO calculations based on the rigid-particle model showing higher values for surface potential than potentials calculated from the soft-particle equations. For C. parapsilosis (Fig. 5A), there was poor agreement with soft-particle DLVO theory, in terms of calculating the surface potential for the cell. Plotting calculated versus experimental electrophoretic mobilities (data not shown), followed by a linear regression of these data, gave a value of R2 = 0.22 and a spatial charge density (ZN) of −0.0196 mol · liter−1. Fungal cell walls often contain cellulose, chitin, or both (29), which greatly increase cell wall rigidity. So, while the potentials of various bacterial strains, such as those of Escherichia coli (2), Pseudomonas putida (1), P. aeruginosa (reference 23 and this study) (Fig. 5B), and Streptococcus salivarius (5), can be better described with soft-particle DLVO theory, C. parapsilosis would presumably interact as a more rigid particle.
Vadillo-Rodriguez et al. (43) have begun a series of relevant studies, utilizing AFM to determine relationships between macroscopic cell properties (fluid contact angles, electrophoretic mobility, etc.) and microscopic-nanoscopic cell properties (double-layer interactions, hydrogen bonding, etc.). Initial results from this work seem promising, and future work by this and other groups will hopefully lead to quantitative relations between macroscopic and microscopic-nanoscopic cell properties.
Modeling of steric interactions with the microbial polymer brush.
In most of the biological systems that we examined, for either microbe individually or for the cell-cell interactions, steric repulsions of varying magnitude were observed in approach curves. Applying the single-brush steric model to these repulsions, we see the role of the microbial exopolymers in the force-distance interactions. The model predicts grafting densities 1 order of magnitude higher for the interactions between the C. parapsilosis probe and the P. aeruginosa biofilm than for the interactions between C. parapsilosis and glass. Values of the equilibrium polymer brush length, however, remain comparable.
The two-brush steric model fits the repulsive regions of the approach curves for P. aeruginosa very well (R2 > 0.99), indicating that steric forces, attributable to two separate types of polymer brushes, play a significant role as the probe approaches the cells (Fig. 1). We believe that a two-brush model is valid based on the presence of an inflection point at ∼20 nm from the sample surface (Fig. 1, inset). The two separate slopes suggest that the probe is interacting with polymer samples having disparate mechanical properties (viz., rigidity and elasticity).
P. aeruginosa is known to produce a variety of polymeric materials in both its planktonic and sessile states, including type IV pili (41), alginate (7), lipopolysaccharides (25), and possibly a variety of others. Given this assortment of secreted materials, it is likely that their presence is detectable by the AFM cantilever, especially if the cantilever is very compliant. In actuality, we likely have more than two types of polymers present. However, they can be grouped into two populations with various mechanical properties, and this describes the data better than assuming that all polymers should be of the same length and density.
There were no attractive interactions seen during the approach of the silicon nitride tip with P. aeruginosa. Qualitatively similar results have been observed from AFM measurements on E. coli JM109 (2), Burkholderia cepacia G4, and P. putida KT2442 (11). The hydrophobicity and surface potentials of P. putida and P. aeruginosa are similar, and it is not surprising that strong repulsive interactions are observed between the polymer brush and the AFM probe for both bacterial strains.
Conclusions.
We examined two medically important microbes in order to characterize their affinity for biomaterial and biofilm surfaces. Although steric interactions play significant roles in the approach of silicon nitride probes to P. aeruginosa, they cannot explain the adhesive interactions seen in C. parapsilosis systems. Further, such adhesions are not explainable via soft- or rigid-particle DLVO analyses. C. parapsilosis shows attractive interactions with bare and P. aeruginosa biofilm-coated silicone rubber. The biofilm systems show interactions beginning at distances more than twice that of bare silicone rubber, with similar attractive magnitudes. Further, biofilms demonstrate an initial repulsive force that is a combination of electrostatic and steric interactions between the polymer brushes of cell probe and the film.
We have also demonstrated that established methods of force curve analysis show inconsistencies for systems involving a polymer brush on either the substrate or the probe. Additional experimentation is necessary to accurately quantify the role of the polymer brush in AFM approach curves involving attraction. In this case, the point of zero separation was defined according to the method of Ducker and Senden, in which the constant compliance region is aligned with the vertical axis. This does not truly represent contact with the cell wall, as the polymer brush must take up some space, but instead represents a vertical offset including the maximum compressible thickness of the brush at the point of constant compliance.
By using our model system and techniques, cell-biomaterial and cell-cell interactions are characterized at the scales of force and distance at which they occur.
Acknowledgments
The following persons have contributed to this research through discussions or experimental assistance: from Worcester Polytechnic Institute, Nancy Burnham, George Pins, Robert Thompson, Bahriye Akis, Bhupinder S. Arora, Nehal I. Abu-Lail, Giacomo Ferraro, and Douglas White, and, from Smith College, Domenico Grasso and Jayne Morrow.
This work has been funded in part by the National Science Foundation through grants DGE-9355019 and a CAREER Award to T.A.C. (BES-0238627). We also acknowledge the donors of the Petroleum Research Fund of the American Chemical Society for partial support of this work (PRF grant 38988-G2).
REFERENCES
- 1.Abu-Lail, N. I., and T. A. Camesano. 2003. Role of ionic strength on the relationship of biopolymer conformation, DLVO contributions and steric interactions to bioadhesion of Pseudomonas putida KT2442. Biomacromolecules 4:1000-1012. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Lail, N. I., and T. A. Camesano. 2003. Role of lipopolysaccharides in the adhesion, retention and transport of Escherichia coli JM109. Environ. Sci. Technol. 37:2173-2183. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, S. 1977. Adsorption of chain molecules with a polar head: a scaling description. J. Phys. (Paris) 38:983-987. [Google Scholar]
- 4.Benoit, M., D. Gabriel, G. Gerisch, and H. E. Gaub. 2000. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nat. Cell Biol. 2:313-317. [DOI] [PubMed] [Google Scholar]
- 5.Bos, R., H. C. van der Mei, and H. J. Busscher. 1998. ′Soft-particle' analysis of the electrophoretic mobility of a fibrillated and non-fibrillated oral streptococcal strain: Streptococcus salivarius. Biophys. Chem. 74:251-255. [DOI] [PubMed] [Google Scholar]
- 6.Bowen, W. R., R. W. Lovitt, and C. J. Wright. 2001. Atomic force microscopy study of the adhesion of Saccharomyces cerevisiae. J. Colloid Interface Sci. 237:54-61. [DOI] [PubMed] [Google Scholar]
- 7.Boyd, A., and A. M. Chakrabarty. 1995. Pseudomonas aeruginosa biofilms: role of the alginate exopolysaccharide. J. Ind. Microbiol. 15:162-168. [DOI] [PubMed] [Google Scholar]
- 8.Burnham, N. A., R. J. Colton, and H. M. Pollock. 1993. Interpretation of force curves in atomic force microscopy. Nanotechnology 4:64-80. [Google Scholar]
- 9.Burnham, N. A., and A. J. Kulik. 1999. Surface forces and adhesion, p. 247-271. In B. Bhushan (ed.), Handbook of micro/nanotribology, 2nd ed. CRC Press, Boca Raton, Fla.
- 10.Butt, H.-J., M. Kappl, H. Mueller, R. Paiteri, W. Meyer, and J. Ruhe. 1999. Steric forces measured with the atomic force microscope at various temperatures. Langmuir 15:2559-2565. [Google Scholar]
- 11.Camesano, T. A., and B. E. Logan. 2000. Probing bacterial electrosteric interactions using atomic force microscopy. Environ. Sci. Technol. 34:3354-3362. [Google Scholar]
- 12.Cappellaro, C., C. Baldermann, R. Rachel, and W. Tanner. 1994. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: cell wall attachment and active sites of a- and α-agglutinin. EMBO J. 13:4737-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 14.de Gennes, P. G. 1987. Polymers at an interface: a simplified view. Adv. Colloid Interface Sci. 27:189-209. [Google Scholar]
- 15.Drummond, C. J., and T. J. Senden. 1994. Examination of the geometry of long-range tip-sample interactions in atomic force microscopy. Colloids Surf. A Physicochem. Eng. Aspects 87:217-234. [Google Scholar]
- 16.Ducker, W. A., and T. J. Senden. 1992. Measurement of forces in liquids using a force microscope. Langmuir 8:1831-1836. [Google Scholar]
- 17.Dufrêne, Y. F., C. J. P. Boonaert, H. C. van der Mei, H. J. Busscher, and P. G. Rouxhet. 2001. Probing molecular interactions and mechanical properties of microbial cell surfaces by atomic force microscopy. Ultramicroscopy 86:113-120. [DOI] [PubMed] [Google Scholar]
- 18.Dumont-Hornebeck, B., J.-P. Joly, J. Coulon, and Y. Chapleur. 1999. Synthesis of ethoxy-linked pseudo-disaccharides incorporating a crown ether macrocycle and lectin recognition. Carbohydr. Res. 321:214-227. [Google Scholar]
- 19.Emerson, R. J. 2004. Microbial adhesion to medical implant materials: an atomic force microscopy study. M.S. thesis. Worcester Polytechnic Institute, Worcester, Mass.
- 20.Gallardo-Moreno, A. M., E. Garduno, M. L. Gonzalez-Martin, C. Perez-Giraldo, J. M. Bruque, and A. C. Gomez-Garcia. 2003. Analysis of the hydrophobic behaviour of different strains of Candida parapsilosis under two growth temperatures. Colloids Surf. B Biointerfaces 28:119-126. [Google Scholar]
- 21.Grasso, D., B. F. Smets, K. A. Strevett, B. D. Machinist, C. J. van Oss, R. F. Giese, and W. Wu. 1996. Impact of physiological state on surface thermodynamics and adhesion of Pseudomonas aeruginosa. Environ. Sci. Technol. 30:3604-3608. [Google Scholar]
- 22.Gristina, A. G. 1987. Biomaterial-centered infection: microbial adhesion versus tissue integration. Science 237:1588-1595. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi, H., S. Tsuneda, A. Hirata, and H. Sasaki. 2001. Soft particle analysis of bacterial cells and its interpretation of cell adhesion behaviors in terms of DLVO theory. Colloids Surf. B Biointerfaces 22:149-157. [DOI] [PubMed] [Google Scholar]
- 24.Khardori, N., and M. Yassien. 1995. Biofilms in device-related infection. J. Ind. Microbiol. 15:141-147. [DOI] [PubMed] [Google Scholar]
- 25.Knirel, Y. A., O. V. Bystrova, A. S. Shashkov, B. Lindner, N. A. Kocharova, S. N. Senchenkova, H. Moll, U. Zähringer, K. Hatano, and G. B. Pier. 2001. Structural analysis of the lipopolysaccharide core of a rough, cystic fibrosis isolate of Pseudomonas aeruginosa. Eur. J. Biochem. 268:4708-4719. [DOI] [PubMed] [Google Scholar]
- 26.Lee, G. U., L. A. Chrisey, and R. J. Colton. 1994. Direct measurement of the forces between complementary strands of DNA. Science 266:771-773. [DOI] [PubMed] [Google Scholar]
- 27.Levin, A. S., S. F. Costa, N. S. Mussi, M. Basso, S. I. Sinto, C. Machado, D. C. Geiger, M. C. B. Villares, A. Z. Schreiber, A. A. Barone, and M. L. M. Branchini. 1998. Candida parapsilosis fungemia associated with implantable and semi-implantable central venous catheters and the hands of healthcare workers. Diagn. Microbiol. Infect. Dis. 30:243-249. [DOI] [PubMed] [Google Scholar]
- 28.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2000. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes Infect. 2:1051-1060. [DOI] [PubMed] [Google Scholar]
- 29.Madigan, M. T., J. M. Martinko, and J. Parker. 2000. Biology of microorganisms, 9th ed. Prentice Hall, Upper Saddle River, N.J.
- 30.Ohshima, H., and T. Kondo. 1989. Approximate analytic expression for the electrophoretic mobility of colloidal particles with surface-charge layers. J. Colloid Interface Sci. 130:281-282. [DOI] [PubMed] [Google Scholar]
- 31.Palabiyikoglu, I., M. Oral, and M. Tulunay. 2001. Candida colonization in mechanically ventilated patients. J. Hosp. Infect. 47:239-242. [DOI] [PubMed] [Google Scholar]
- 32.Pembrey, R. S., K. C. Marshall, and R. P. Schneider. 1999. Cell surface analysis techniques: what do cell preparation protocols do to cell surface properties? Appl. Environ. Microbiol. 65:2877-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poortinga, A. T., R. Bos, W. Norde, and H. J. Busscher. 2002. Electric double layer interactions in bacterial adhesion to surfaces. Surf. Sci. Rep. 47:1-32. [Google Scholar]
- 34.Razatos, A., Y.-L. Ong, F. Boulay, D. L. Elbert, J. A. Hubbell, M. M. Sharma, and G. Georgiou. 2000. Force measurements between bacteria and poly(ethylene glycol)-coated surfaces. Langmuir 16:9155-9158. [Google Scholar]
- 35.Razatos, A., Y.-L. Ong, M. M. Sharma, and G. Georgiou. 1998. Evaluating the interaction of bacteria with biomaterials using atomic force microscopy. J. Biomater. Sci. Polym. Ed. 9:1361-1373. [DOI] [PubMed] [Google Scholar]
- 36.Schierholz, J. M., and J. Beuth. 2001. Implant infections: a haven for opportunistic bacteria. J. Hosp. Infect. 49:87-93. [DOI] [PubMed] [Google Scholar]
- 37.Senden, T. J., and C. J. Drummond. 1995. Surface chemistry and tip-sample interactions in atomic force microscopy. Colloids Surf. A Physicochem. Eng. Aspects 94:29-51. [Google Scholar]
- 38.Shibata, N., K. Ikuta, T. Imai, Y. Satoh, R. Satoh, A. Suzuki, C. Kojima, H. Kobayashi, K. Hisamichi, and S. Suzuki. 1995. Existence of branched side chains in the cell wall mannan of pathogenic yeast, Candida albicans: structure-antigenicity relationship between the cell wall mannans of Candida albicans and Candida parapsilosis. J. Biol. Chem. 270:1113-1122. [DOI] [PubMed] [Google Scholar]
- 39.Singh, R. S., A. K. Tiwary, and J. F. Kennedy. 1999. Lectins: sources, activities and applications. Crit. Rev. Biotechnol. 19:145-178. [Google Scholar]
- 40.Stewart, P. S., and J. W. Costerton. 2001. Antibiotic resistance of bacteria in biofilms. Lancet. 358:135-138. [DOI] [PubMed] [Google Scholar]
- 41.Sundin, C., M. C. Wolfgang, S. Lory, Å. Forsberg, and E. Frithz-Lindsen. 2002. Type IV pili are not specifically required for contact dependent translocation of exoenzymes by Pseudomonas aeruginosa. Microb. Pathog. 33:265-277. [DOI] [PubMed] [Google Scholar]
- 42.Touhami, A., B. Hoffmann, A. Vasella, F. A. Denis, and Y. F. Dufrêne. 2003. Probing specific lectin-carbohydrate interactions using atomic force microscopy imaging and force measurements. Langmuir 19:1745-1751. [Google Scholar]
- 43.Vadillo-Rodriguez, V., H. J. Busscher, W. Norde, J. de Vries, and H. C. van der Mei. 2003. On relations between microscopic and macroscopic physicochemical properties of bacterial cell surfaces: an AFM study on Streptococcus mitis strains. Langmuir 19:2372-2377. [Google Scholar]
- 44.van Oss, C. J. 1993. Acid-base interfacial interactions in aqueous media. Colloids Surf. A Physicochem. Eng. Aspects 78:1-49. [Google Scholar]
- 45.van Oss, C. J. 1994. Interfacial forces in aqueous media. Marcel Dekker, Inc., New York, N.Y.
- 46.van Oss, C. J., M. K. Chaudhury, and R. J. Good. 1988. Additive and non-additive surface tension components and the interpretation of contact angles. Langmuir 4:884-891. [Google Scholar]
- 47.Velegol, S. B., and B. E. Logan. 2002. Contributions of bacterial surface polymers, electrostatics and cell elasticity to the shape of AFM force curves. Langmuir 18:5256-5262. [Google Scholar]
- 48.Velegol, S. B., S. Pardi, X. Li, D. Velegol, and B. E. Logan. 2003. AFM imaging artifacts due to bacterial cell height and AFM tip geometry. Langmuir 16:851-857. [Google Scholar]