Abstract
1-Aminocyclopropane-1-carboxylate (ACC) deaminase has been found in various plant growth-promoting rhizobacteria, including rhizobia. This enzyme degrades ACC, the immediate precursor of ethylene, and thus decreases the biosynthesis of ethylene in higher plants. The ACC deaminase of Rhizobium leguminosarum bv. viciae 128C53K was previously reported to be able to enhance nodulation of peas. The ACC deaminase structural gene (acdS) and its upstream regulatory gene, a leucine-responsive regulatory protein (LRP)-like gene (lrpL), from R. leguminosarum bv. viciae 128C53K were introduced into Sinorhizobium meliloti, which does not produce this enzyme, in two different ways: through a plasmid vector and by in situ transposon replacement. The resulting ACC deaminase-producing S. meliloti strains showed 35 to 40% greater efficiency in nodulating Medicago sativa (alfalfa), likely by reducing ethylene production in the host plants. Furthermore, the ACC deaminase-producing S. meliloti strain was more competitive in nodulation than the wild-type strain. We postulate that the increased competitiveness might be related to utilization of ACC as a nutrient within the infection threads.
For more than a decade now, the phytohormone ethylene has been known as an inhibitor of nodulation in various legumes, including both those that produce indeterminate nodules and those that produce determinate nodules (37). Application of exogenous ethylene or 1-aminocyclopropane-1-carboxylic acid (ACC), the immediate precursor of ethylene in higher plants, inhibits nodulation in Pisum sativum (29), Medicago truncatula (38), and Medicago sativa, Macroptilium atropurpureum, and Lotus japonicus (37). Also, nodulation can be promoted by treating these plants with ethylene biosynthesis or perception inhibitors (6, 29, 37, 41, 52).
Either direct or indirect involvement of ethylene has been implicated in the regulation of several processes in different stages of nodulation (21). For example, Oldroyd et al. (38) showed that ethylene inhibited early plant responses to Nod factor in Medicago truncatula, suggesting that ethylene could modulate the activation of the Nod factor pathway by acting directly on the calcium spiking or by inhibiting a component in the signal transduction pathway upstream of calcium spiking. Ethylene is also involved in the development of infection threads. It was proposed that a low level of ethylene is required for successful entry of infection threads into the outermost layer of cortical cells by allowing proper disposition of cytoskeleton and the formation of cytoplasmic bridges (21, 48); furthermore, ethylene could have an indirect effect on cellulose microfibril deposition (1, 21).
On the other hand, higher levels of ethylene might induce the abortion of the infection threads. In alfalfa, about 95 to 99% of infection thread progressions abort in the cortex (49). Ethylene is known to be able to stimulate the synthesis of the enzyme peroxidase (1). Diamine oxidase in peas could serve as a source of H2O2, which induces the cross-linking of the matrix glycoproteins in the infection threads and the congealing of the infection threads to result in their abortion (21, 51). A brz mutant of P. sativum, which has a potential ethylene-oversensitive phenotype, has a larger number of aborted infection threads than wild-type plants (22), whereas in an ethylene-insensitive and hypernodulating mutant of M. truncatula, sickle, almost all of the infection threads eventually form nodules (39). Ethylene has also been reported to be involved in determining the positioning of nodule primodia (24).
Some rhizobial strains enhance nodulation by lowering the ethylene levels in their host legumes. An ethylene biosynthesis inhibitor, rhizobitoxine, produced by Bradyrhizobium elkanii decreases ethylene production in plant roots and enhances nodulation on M. atropurpureum Urb. cv. Siratro (52). 1-Aminocyclopropane-1-carboxylate (ACC) deaminase, which has recently been found in several rhizobial strains, such as Rhizobium leguminosarum bv. viciae strains, Rhizobium hedysari (31), and Mesorhizobium loti (28, 47) could be another strategy that is employed by some rhizobial strains to regulate nodulation by modulating ethylene levels in plants. ACC deaminase has been found in various bacteria, yeasts, and fungi, and it can convert ACC into α-ketobutyrate and ammonia (19). Plant growth-promoting bacteria containing ACC deaminase and attached to the surface of plant roots or seeds can take up and degrade some of the ACC exuded from the plant by acting as a sink for ACC and therefore decrease ethylene synthesis in plants (18, 40). The lowering of ethylene levels by ACC deaminase is considered one of the major mechanisms employed by plant growth-promoting bacteria to facilitate plant growth (19).
The role of ACC deaminase in nodulation in rhizobia has been studied in the symbiotic interaction of P. sativum L. cv. Sparkle and R. leguminosarum bv. viciae 128C53K, from which the ACC deaminase structural gene (acdS) and its regulatory region were cloned (32). A leucine-responsive regulatory protein (LRP)-like gene (lrpL), which is located immediately upstream of acdS, is required for the expression of ACC deaminase in 128C53K. Neither the acdS nor the lrpL gene knockout mutant of 128C53K produces ACC deaminase, and both of the mutants showed decreased ability (approximately 30% lower than wild type) to nodulate Sparkle (32). This result suggests that ACC deaminase in R. leguminosarum bv. viciae 128C53K enhances nodulation on pea plants, likely by decreasing the ethylene levels in its host legumes.
Here, we report use of the Sinorhizobium meliloti-alfalfa system to further elaborate the effect of ACC deaminase on nodulation. The wild-type S. meliloti strain Rm1021 nodulates alfalfa (Medicago sativa), and this process is promoted by the application of ethylene inhibitors such as l-α-(2-aminoethyoxyvinyl) glycine (41) and silver thiosulfate (6). Genomic sequencing data indicated that Rm1021 did not have a potential ACC deaminase gene (2, 8, 13, 17). In addition, our previous experimental evidence confirmed that neither the ACC deaminase protein nor the enzyme activity was detected in strain Rm1021 after induction by ACC (31).
In this study, the ACC deaminase structural gene from R. leguminosarum bv. viciae 128C53K along with its upstream regulatory gene was introduced into S. meliloti through a plasmid vector as well as by in situ transposon replacement. Nodulation assay results showed that the engineered ACC deaminase-producing strains of S. meliloti nodulated alfalfa to a 40% greater extent than the wild-type strain. Furthermore, the mutant was much more competitive than the wild-type strain in nodulating alfalfa due to the action of ACC deaminase.
MATERIALS AND METHODS
Bacterial strains, antibiotics, and plant growth conditions.
The bacterial strains used in this study are listed in Table 1. R. leguminosarum bv. viciae 128C53K and S. meliloti strains were grown at 25°C in TY medium (3) or M9 glucose minimal medium (36) supplemented with 0.3 μg of biotin ml−1. Escherichia coli strains DH5α and S17-1 and transformants with different plasmids were grown at 37°C in Luria broth (LB) medium (Difco Laboratories, Detroit, Mich.).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Sinorhizobium meliloti | ||
| Rm1021 | SU47/Smr Acd− | 35 |
| Rm5356 | Rm1021 ω5033::Tn5-233, Smr Gmr Spr Acd− | 9 |
| Rm1021(pSP329) | Acd− Smr Tcr | This work |
| Rm1021(pWM2) | Acd+ Smr Tcr | This work |
| Rm11466 | Acd+ Smr Nmr | This work |
| Escherichia coli | ||
| DH5α | SupE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 23 |
| S17-1 | Mobilizing strain with chromosomally integrated RP4 derivative | 45 |
| MT616 | MT607(pRK600), mobilizing strain | 12 |
| Plasmids | ||
| pRK2013 | ColE1 replicon containing RK2 transfer region, Kmr | 11 |
| pRK600 | pRK2013 npt::Tn9, Cmr | 12 |
| pSP329 | RP4 replicon, broad-host-range vector, Tcr | 7 |
| pGS220 | Tn5 carried by pBR322 derivative, Nmr Apr | 10 |
| pWM1 | pUC18 containing the acdS and IrpL genes of R. leguminosarum bv. viciae 128C53K on a 4-kb DNA BamHI fragment | 32 |
| pWM2 | pSP329 containing the acdS and lrpL genes of R. leguminosarum bv. viciae 128C53K, Tcr | 32 |
| pWM3 | pGS220 containing the acdS and lrpL genes of R. leguminosarum bv. viciae 128C53K on a 4-kb DNA fragment | This work |
Sm, streptomycin; Gm, gentamicin; Sp, spectinomycin; Tc, tetracyeline; Nm, neomycin; Km, kanamycin; Cm, chloramphenicol; Ap, ampicillin.
Antibiotics were used at the following concentrations for both E. coli and Rhizobium strains: neomycin, 100 μg/ml; gentamicin, 30 μg/ml; streptomycin, 200 μg/ml; and tetracycline, 15 μg/ml.
Plant growth conditions.
Seeds of alfalfa (Medicago sativa var. Iroquois) were surface sterilized with 95% ethanol for 2 min, followed by 2.5% sodium hypochlorite for 15 min, and then washed thoroughly with sterile distilled water. After seed germination on 1% water agar plates in the dark for 2 days, the seedlings were aseptically transferred to Leonard's jars (three seedlings in each jar) filled with vermiculite wetted with 25 ml of nitrogen-free Jensen's medium (27). The plants were grown in a growth chamber under pink and white fluorescent lights (25 μmol m−2 s−1) at 25 to 20°C and a 16-h light/8-h dark regimen; sterile distilled deionized water was used to water the plants when necessary (approximately 80 ml every 4 days).
Construction and characterization of the ACC deaminase-producing mutants of S. meliloti.
Since the lrpL gene is required for the expression of acdS (32), a 4-kb DNA fragment containing both of the genes from R. leguminosarum bv. viciae 128C53K was inserted into plasmid pSP329 (7) to construct pWM2. To introduce pWM2 into S. meliloti Rm1021, triparental conjugation was conducted by using E. coli DH5α(pWM2) as the donor, S. meliloti Rm1021 as the recipient, and E. coli HB101(pRK2013) as a helper (11). TY plates supplemented with streptomycin and tetracycline were used to select the transconjugants. The vector pSP329 was also introduced into S. meliloti Rm1021 as a negative control.
ACC deaminase-encoding genes were also introduced into the genomic DNA of S. meliloti strain Rm5356, which has a transposon Tn5-233 inserted in its megaplasmid, pRmeSU47b (pSymb) (9). In order to determine the exact insertion site of the transposon in pRmeSU47b, genomic DNA from Rm5356 was isolated and digested with EcoRI, which cuts once in the spectinomycin-streptomycin resistance gene in Tn5-233 (10). The EcoRI-digested DNA fragments were ligated into pUC19 and introduced into competent cells of E. coli DH5α to construct a genomic DNA library. One recombinant plasmid with an approximately 4-kb DNA insertion in pUC19 from the library gave a positive signal when probed with digoxigenin-labeled Tn5 by Southern hybridization. The 4-kb fragment contains about 200 bp of flanking region from pRmeSU47b of Rm5356, which is linked to transposon Tn5-233, in addition to a 3.8-kb partial Tn5-233 fragment. The DNA sequence of the 200 bp fragment was determined by a primer designed according to the IS50 sequence of Tn5-233 (5′-CGGGAAAGGTTCCGTTCAGG-3′). The DNA sequencing data showed that transposon Tn5-233 is inserted into the 714th nucleotide of an open reading frame designated SMb21144, which is located from nucleotides 888553 to 889473 in pRmeSU47b of S. meliloti Rm1021 (12).
SMb21144 encodes a putative choline uptake ATP-binding cassette (ABC) transporter periplasmic solute-binding protein precursor (12). Solute-binding protein-dependent ABC transporters are found exclusively in prokaryotes and are required for the uptake of a variety of small molecules, including amino acids, metal ions, and sugars (26). In S. meliloti, choline can be up taken by three kinetically distinct transport systems (42) and is then oxidized to glycine betaine, a common solute used by bacteria to serve as an osmoprotectant and energy source at inhibitory osmolarities (46); however, choline itself has no direct osmoprotectant activity (43). Since the involvement of SMb21144 in nodulation is not known, the ACC deaminase-producing strain was only compared with the parent strain, Rm5356, for nodulation efficiency, which should eliminate any possible interference (if any) from the disruption of SMb21144 with the nodulation ability of the S. meliloti strains.
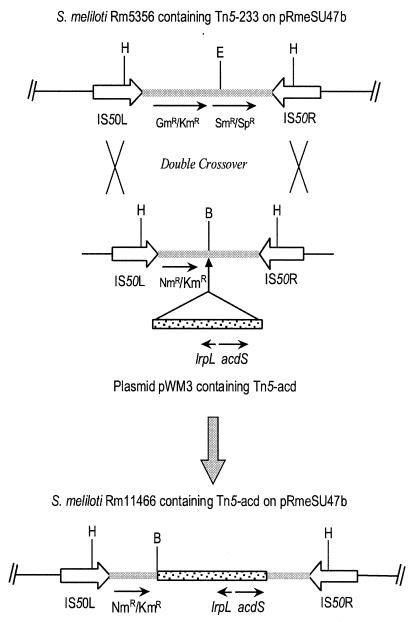
The same 4-kb DNA fragment containing the acdS and lrpL genes from R. leguminosarum bv. viciae 128C53K was inserted into transposon Tn5 inside plasmid pGS220 (10), which is unable to replicate in S. meliloti. The resulting plasmid, pWM3, was introduced into S. meliloti Rm5356 by triparental mating with E. coli MT616 (12) as a helper. After homologous recombination between the IS50 regions of the two transposons, the DNA fragment inside the flanking IS50 sequences of Tn5-233 in Rm5356 was replaced by that of Tn5-acd in pWM3 (Fig. 1). S. meliloti transconjugants were selected for neomycin resistance and then screened for loss of gentamicin resistance.
FIG. 1.
Construction of S. meliloti Rm11466 by transposon replacement. A 4-kb DNA fragment containing the lrpL and acdS genes from R. leguminosarum bv. viciae 128C53K was inserted into the BamHI site of transposon Tn5 in pGS220 to construct pWM3. pWM3 is suicidal after being introduced into S. meliloti Rm5356. After double crossover between the IS50 regions of the two transposons, Tn5-233 and Tn5-acd, the neomycin resistance gene, leucine-responsive regulatory-like gene (lrpL), and the ACC deaminase structural gene (acdS) were inserted into megaplasmid pRmeSU47b in S. meliloti Rm5356. The resulting variant was named Rm11466. Restriction sites: B, BamHI; E, EcoRI; H, HindIII. There are multiple HindIII sites in Tn5-233 and Tn5-acd, but only the locations of the sites inside the IS50 regions are shown on the restriction map.
Following the mating of E. coli DH5α(pWM3) with S. meliloti Rm5356, approximately 600 neomycin-resistant colonies were replica plated onto TY medium containing gentamicin to screen for loss of gentamicin resistance, and four of them were found to be gentamicin sensitive. The structures of these four transconjugants were confirmed by Southern hybridization with transposon Tn5-acd from pWM3, which was amplified by PCR (with primer 5′-CCTGAACGGAACCTTTCCCG-3′) and labeled with digoxigenin as a probe (data not shown). The hybridization results indicate that the fragments inside the IS50 sequences of Tn5-233 were replaced with those from Tn5-acd, whereas the external borders were maintained, confirming that the genomic DNA linked to the transposon remained intact. The resulting variant, S. meliloti Rm11466, was used in the subsequent nodulation and competition assays.
Detection of ACC deaminase activity in Rhizobia spp.
Rhizobial cells were grown in 5 ml of TY medium with appropriate antibiotics at 30°C for 2 days to the stationary phase. The cells were pelleted by centrifugation, washed twice with 0.1 M Tris-HCl (pH 7.5), suspended in 2 ml of M9 minimal medium supplemented with appropriate antibiotics as well as 5 mM ACC, and then incubated for 24 h at 25°C with shaking (100 rpm). ACC deaminase activity was determined by spectrophotometrically measuring the production of α-ketobutyrate (25).
Nodulation assay.
Wild-type and ACC deaminase-producing S. meliloti strains were grown overnight at 25°C with shaking in TY medium supplemented with appropriate antibiotics. The cells were washed with 0.85% NaCl to remove the antibiotics and diluted 1:50 (vol/vol) in 0.85% NaCl (approximately 108 CFU/ml); 5 ml of the diluted cell suspension was used to inoculate alfalfa seedlings in each Leonard's jar 2 days after the plants were transferred. Uninoculated plants were used as a control. Plants were harvested 4 weeks after inoculation, and the nodule numbers and shoot dry weights (after being baked at 105°C for 3 days) were determined. Eighteen plants were used for each treatment in one assay, and two independent assays were performed.
Competition assay.
S. meliloti Rm5356 and Rm11466 were grown at 25°C with shaking in TY medium for 2 days to reach the stationary phase. The cells were centrifuged and then washed with 0.85% NaCl to remove the antibiotics before they were diluted 1:50 with 0.85% NaCl. These two types of cells were mixed at different ratios, and 5 ml of each mixed cell suspension was used to inoculate 5-day-old alfalfa plants. A small portion of each inoculum was used to spread TY plates and grown at 25°C overnight, and 100 colonies from these TY plates were then randomly picked out and streaked on a TY-neomycin agar plate to identify the number of Rm11466 cells. The precise ratios of the two bacteria were determined by comparing the number of neomycin-sensitive colonies (Rm5356) to that of neomycin-resistant colonies (Rm11466).
Four replicate jars (three plants per jar) were used for each treatment. The plants were harvested 6 weeks after inoculation, and nodules were collected from the root systems. For each treatment, 25 mature pink nodules were randomly picked out and surface sterilized by soaking in 1% sodium hypochlorite for 15 min. Each nodule was macerated in 30 μl of TY medium containing 0.3 M sucrose and then spread onto TY plates. Five colonies from each nodule were tested for neomycin resistance for strain identification. In most instances, one nodule was only occupied with one of the two bacterial strains that were used to inoculate the plants, i.e., either all or none of the five colonies that were tested showed neomycin resistance. Approximately 2% of all the nodules identified were occupied with both strains, and we did not take the values for these nodules into the final analysis. The ratios of alfalfa nodules containing different S. meliloti strains were compared to the ratios of the number of two bacterial cells in the inocula to determine the competitiveness of the strains. The competition assay was repeated, and similar results were observed.
Statistics.
The data were analyzed by analysis of variance, and the treatment means were compared by Tukey's honestly significant difference multiple-comparison test. All hypotheses were tested at the 95% confidence interval (α = 0.05).
Molecular manipulations were performed according to Sambrook and Russell (44).
RESULTS
Characterization of the engineered ACC deaminase-producing mutants of S. meliloti.
Two strategies were used to generate ACC deaminase-producing variants of S. meliloti: one was the introduction of the gene and its regulatory region on a broad-host-range plasmid, and the other was the introduction of the genes into the genomic DNA as single copies by in situ transposon replacement. Tn5 insertion mutants of S. meliloti are easily obtained and stable, and secondary transposition has not been observed (10). In the present study, this method was used to introduce the ACC deaminase and its regulatory gene into the genome of S. meliloti Rm5356, which contains Tn5-233 in one of its megaplasmids, pRmeSU47b (9). Transposon replacement strategy is preferable to the use of plasmids or to random transposon mutation because it generates a mutant with a single copy of the target gene(s) integrated into the genome of the host strain, which is very stable and minimizes the extent of the metabolic burden on the host strain. In addition, the site of integration of the foreign gene(s) in the genomic DNA is known, which avoids the possibility of disrupting any important host genes. Finally, the extensive collection of various mapped transposon insertion mutants of S. meliloti provides a convenient starting point for this approach.
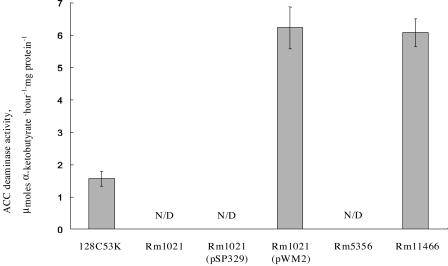
The ACC deaminase activities of S. meliloti Rm1021(pWM2) and Rm11466 as well as the parental strains were determined by ACC deaminase activity assays (Fig. 2). As expected, no activity was detected either in the parental strains, S. meliloti Rm1021 and Rm5356, or in Rm1021(pSP329), which carries the plasmid vector without an ACC deaminase gene. The level of ACC deaminase activity in S. meliloti Rm1021(pWM2) was similar to that of S. meliloti Rm11466 despite the fact that the former carries the ACC deaminase genes on a multicopy plasmid. The enzyme specific activity of S. meliloti Rm1021(pWM2) and Rm11466 was approximately four times greater than that of R. leguminosarum bv. viciae 128C53K, the source of the ACC deaminase genes (Fig. 2). In R. leguminosarum bv. viciae 128C53K, ACC is required for induction of the acdS gene, and l-leucine inhibits ACC deaminase expression, possibly by regulating the upstream LRP-like protein (32). It is possible that in S. meliloti, regulation of the LRP-like protein and/or uptake of ACC is different from that in R. leguminosarum bv. viciae 128C53K and therefore results in a higher expression level of ACC deaminase in the S. meliloti strains.
FIG. 2.
ACC deaminase activities of the S. meliloti strains induced by 5 mM ACC. R. leguminosarum bv. viciae 128C53K, which is the source of the lrpL and acdS genes, was used as a positive control. Rm1021 and Rm5356 are wild-type strains that were used as negative controls. Rm1021 containing the vector pSP329 was also used as a negative control for Rm1021(pWM2). N/D, no activity detected. The error bars represent the standard error of the mean of two independent assays.
ACC deaminase enhances the efficiency and competitiveness of S. meliloti in alfalfa nodulation.
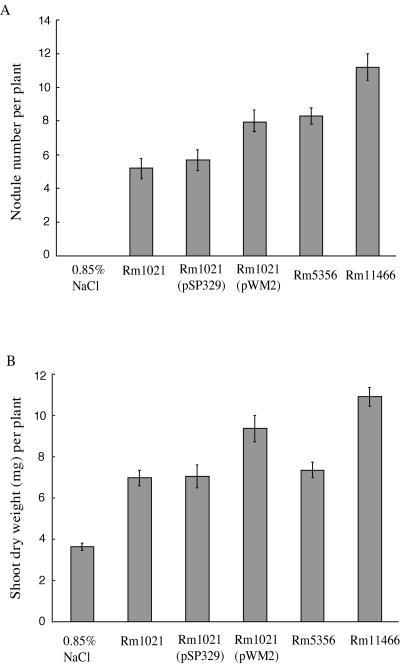
Figure 3 shows the results of nodulation assays with alfalfa inoculated with the wild-type as well as with the ACC deaminase-producing S. meliloti strains. Uninoculated plants were used as a control. No pink and well-developed nodules were detected on the roots of the control plants.
FIG. 3.
Nodule numbers (panel A) and shoot dry weights (panel B) of alfalfa inoculated with wild-type S. meliloti strains Rm1021 and Rm5356 and their ACC deaminase-producing variants, Rm1021(pWM2) and Rm11466. Plants not inoculated with a bacterium (0.85% NaCl only) were used as negative controls. Two independent nodulation assays were performed, and each had 18 plants per treatment. The error bars represent the standard error of the mean of a pool of 36 plants.
The nodule number and the shoot dry weight of plants inoculated with strain Rm1021(pSP329) were very similar to those of the wild-type strain Rm1021 (Fig. 3). Thus, any difference in nodule numbers or shoot dry weights of plants inoculated with the ACC deaminase-producing strain, Rm1021(pWM2), presumably resulted from the enzyme ACC deaminase produced by this strain. The data showed that approximately 40% more nodules formed on plants inoculated with S. meliloti Rm1021(pWM2) than on plants inoculated with Rm1021 or Rm1021(pSP329) (Fig. 3A). A significant increase (P < 0.05, approximately 33%) was also found in the shoot dry weights of the plants inoculated with S. meliloti Rm1021(pWM2) compared to the plants inoculated with Rm1021 or Rm1021(pSP329) (Fig. 3B). This higher biomass of the plants inoculated with Rm1021(pWM2) most probably resulted from the increased number of nodules formed by the bacterium.
Plants inoculated with S. meliloti Rm5356 had shoot dry weights similar to those of plants inoculated with S. meliloti Rm1021; however, the former strain formed an average of 8.3 nodules per plant, while S. meliloti Rm1021 formed approximately 5.5 nodules per plant (Fig. 3A). Whether this difference in nodule number is due to the interruption of the SMb21144 gene in Rm5356 is not clear at this time, and therefore, we only compared the ACC deaminase-producing strain Rm11466 to its parental strain, Rm5356. Similar to what was found with S. meliloti Rm1021 and Rm1021(pWM2), an approximately 35% increase in nodule number (Fig. 3A) and 45% increase in shoot dry weight (Fig. 3B), both statistically significant (P < 0.05), were found when comparing plants inoculated with S. meliloti Rm11466 to those inoculated with Rm5356. These data indicate that the ACC deaminase produced by the S. meliloti transformants Rm1021(pWM2) and Rm11466 significantly enhanced nodulation of alfalfa.
It was previously reported that the rhizobitoxine produced by Bradyrhizobium elkanii not only enhanced nodulation efficiency but also increased the competitiveness of the bacterium compared to a rhizobitoxine knockout mutant of the bacterium (52). To investigate the effect of ACC deaminase on the competitiveness of S. meliloti on nodulating alfalfa, the seedlings were coinoculated with S. meliloti Rm5356 and Rm11466. Table 2 shows that the majority of the nodules were occupied by Rm11466 even when the plants were inoculated with a mixed inoculum of Rm5356 and Rm11466 at a ratio of 5:1. These results suggest that S. meliloti Rm11466 is significantly more competitive than Rm5356 in nodulating alfalfa.
TABLE 2.
Competition assay of alfalfa inoculated with both S. melliloti Rm5356 (wild type) and Rm11466 (ACC deaminase-producing variant)a
| % of bacterial inocula
|
% of alfalfa nodules containing bacteria of strain:
|
||
|---|---|---|---|
| Rm5356 | Rm11466 | Rm5356 | Rm11466 |
| 100 | 0 | 100 | 0 |
| 0 | 100 | 0 | 100 |
| 30.4 | 69.6 | 2.5 | 97.5 |
| 82.8 | 17.2 | 15.8 | 84.2 |
Two independent assays were performed and similar results were obtained. Data from one representative experiment are presented.
DISCUSSION
Similar to what was found with pea plants and R. leguminosarum bv. viciae (32), ACC deaminase enhances nodulation of alfalfa by S. meliloti. Previously, ethylene production was found to be induced in the roots of alfalfa shortly after the plants were inoculated with S. meliloti (30). The ethylene evolution pattern showed three peaks after the addition of the bacterium, which was similar to the pattern with the uninoculated roots, but the amount of ethylene released was much higher (30). One sharp peak occurred shortly after the bacterium was added, which could be part of the defense reaction of the plant to the invasion by bacteria.
In alfalfa, a transient defense response including the induction of ethylene biosynthesis was induced by inoculation with a compatible Rhizobium strain (30). The ACC deaminase-producing variants might be able to reduce ethylene production by degrading some of the ACC at the infection sites. This localized reduction of ethylene might make it more likely that a bacterium enters the root cortex, and therefore the ACC deaminase-producing strains would be more competitive than those that do not possess this enzyme. As suggested by Ligero et al. (30), the second and the third peak of ethylene evolution in the ethylene release pattern inoculated with S. meliloti may be temporally coincident with nodule development and senescence.
A localized hypersensitive response in alfalfa, which is similar to that described in incompatible plant-pathogen interactions, is one of the mechanisms thought to be involved in the autoregulation of the infection threads (49). Ethylene is well known to be involved in pathogen-induced plant defenses (1). Rhizobia that produce ACC deaminase might be able to decrease ethylene levels in the plant cells in the immediate vicinity of the infection threads. Therefore, the infection threads containing ACC deaminase-producing bacterial cells can better suppress the defense signals in the plant cell and increase the persistence of the infection threads. This leads to higher nodule numbers formed on plants inoculated with S. meliloti Rm11466 as well as a higher portion of nodules formed by Rm11466 when the plants were coinoculated with both S. meliloti Rm5356 and Rm11466. In addition, since a high percentage of infection threads abort before a nodule is formed (49), infection threads containing Rm11466 could suppress the development of those containing Rm5356 by reaching nodule primordia and forming nodules faster. This also accounts for the higher competitiveness of Rm11466.
Gage et al. (15) observed that S. meliloti proliferates inside the infection threads of alfalfa. The population of bacteria inside the threads originates from the clonal expansion of a small number of founder cells which entered the threads early, and only the tipmost bacterial cells of the threads will eventually be released into the cytoplasm of the plant cell (14). This is consistent with our results that S. meliloti Rm11466 occupied the majority of the total nodules formed by plants coinoculated with a mixture of Rm11466 and Rm5356, since Rm11466 could enter the infection threads earlier than Rm5356 by suppressing the inhibitory effect of ethylene through ACC deaminase. It is also suggested that S. meliloti cells travel down the infection threads by proliferation instead of swimming, since the bacteria do not have flagella when inside the threads (16); however, what nutrient source(s) S. meliloti cells utilize to proliferate in the infection threads is not known.
It was previously shown that α-ketobutyrate and ammonia, produced from ACC by ACC deaminase, could be used by plant growth-promoting bacteria as carbon and nitrogen sources, respectively (19). Although the ACC concentrations in the infection threads are expected to be low, the possible ability of the ACC deaminase-producing cells to utilize ACC as an extra nutrient source could make the bacterium proliferate better in the infection threads than those that do not have the enzyme. Therefore, infecting cells that produce ACC deaminase are more likely to reach nodule primordia and form mature nodules, which results in the higher competitiveness of S. meliloti Rm11466 than of Rm5356.
In addition to the synthesis of an ACC synthase inhibitor, rhizobitoxine, ACC deaminase is the second strategy utilized by Rhizobium spp. to adjust ethylene levels in legumes during nodulation. The fate of rhizobial infection in the root hair of legumes has been proposed to be regulated by the levels of ethylene in the underlying plant cortex (1, 21, 51). A low level of ethylene is likely required for successful entry of the infection threads into the outermost layer of cortical cells; whereas higher levels of ethylene induce abortion of the threads. Through the action of ACC deaminase, on one hand, rhizobia could finely modulate ethylene production in legume roots to reach a level that allows the progression of the infection threads and the formation of functional nodules; on the other hand, the infecting rhizobial cells could use ACC as both a nitrogen and carbon source to facilitate the proliferation and thus increase the efficiency and competitiveness of the infection.
The results presented here suggest that by genetically engineering rhizobial strains to produce ACC deaminase, there is a real possibility of obtaining better strains for use as field inocula to increase the yield of leguminous crops. Furthermore, ethylene production can be induced during many environmental stresses, such as infection by pathogens, flooding, heavy metal poisoning, and mechanical wounding (1). In previous experiments with plant growth-promoting bacteria, ACC deaminase was found to be able to help the plants resist the inhibitory effects of stress ethylene on plant growth imposed by pathogens (50), heavy metals (5), high salt concentrations (33), drought (34), and flooding (20). When ACC deaminase is present in rhizobial strains, these bacteria may also help plants to overcome some of the detrimental effects of various environmental stresses.
REFERENCES
- 1.Abeles, F. B., P. W. Morgan, and M. E. Saltveit. 1992. Ethylene in plant biology, 2nd ed. Academic Press, New York, N.Y.
- 2.Barnett, M. J., R. F. Fisher, T. Jones, C. Komp, A. P. Abola, F. Barloy-Hubler, L. Bowser, D. Capela, F. Galibert, J. Gouzy, M. Gurjal, A. Hong, L. Huizar, R. W. Hyman, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, C. Palm, M. C. Peck, R. Surzycki, D. H. Wells, K.-C. Yeh, R. W. Davis, N. A. Federspiel, and S. R. Long. 2001. Nucleotide sequence and predicted functions of the entire Sinorhizobium meliloti pSymA. Proc. Natl. Acad. Sci. USA 98:9883-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beringer, J. E. 1974. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 84:188-198. [DOI] [PubMed] [Google Scholar]
- 4.Boncompagni, E., M. Østerås, M-C. Poggi, and D. Le Rudulier. 1999. Occurrence of choline and glycine betaine uptake and metabolism in the family Rhizobiaceae and their roles in osmoprotection. Appl. Environ. Microbiol. 65:2072-2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burd, G. I., D. G. Dixon, and B. R. Glick. 2000. Plant growth-promoting bacteria that decrease heavy metal toxicity in plants. Can. J. Microbiol. 46:237-245. [DOI] [PubMed] [Google Scholar]
- 6.Caba, J. M., L. Recalde, and F. Ligero. 1998. Nitrate-induced ethylene biosynthesis and the control of nodulation in alfalfa. Plant Cell Environ. 21:87-93. [Google Scholar]
- 7.Cangelosi, G. A., E. A. Best, G. Martinetti, and E. W. Nester. 1991. Genetic analysis of Agrobacterium. Methods Enzymol. 204:384-397. [DOI] [PubMed] [Google Scholar]
- 8.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dréano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Pühler, B. Purnelle, U. Ramsperger, C. Renard, P. Thébault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charles, T. C., and T. M. Finan. 1990. Genetic map of Rhizobium meliloti megaplasmid pRmeSU47b. J. Bacteriol. 172:2469-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Vos, G. F., G. C. Walker, and E. R. Signer. 1986. Genetic manipulations in Rhizobium meliloti utilizing two new transposon Tn5 derivatives. Mol. Gen. Genet. 204:485-491. [DOI] [PubMed] [Google Scholar]
- 11.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finan, T. M., B. Kundel, G. G. De Vos, and E. R. Signer. 1986. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J. Bacteriol. 167:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finan, T. M., S. Weidner, K. Wong, J. Buhrmester, P. Chain, F. J. Vorholter, I. Hernandez-Lucas, A. Becker, A. Cowie, J. Gouzy, B. Golding, and A. Pühler. 2001. The complete sequence of the 1,683-kb pSymB megaplasmid from the N2-fixing endosymbiont Sinorhizobium meliloti. Proc. Natl. Acad. Sci. USA 98:9889-9894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gage, D. J. 2002. Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J. Bacteriol. 184:7042-7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gage, D. J., T. Bobo, and S. R. Long. 1996. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J. Bacteriol. 178:7159-7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gage, D. J., and W. Margolin. 2000. Hanging by a thread: invasion of legume plants by rhizobia. Curr. Opin. Microbiol. 3:613-617. [DOI] [PubMed] [Google Scholar]
- 17.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, F. Barloy-Hubler, M. J. Barnett, A. Becker, P. Boistard, G. Bothe, M. Boutry, L. Bowser, J. Buhrmester, E. Cadieu, D. Capela, P. Chain, A. Cowie, R. W. Davis, S. Dreano, N. A. Federspiel, R. F. Fisher, S. Gloux, T. Godrie, A. Goffeau, B. Golding, J. Gouzy, M. Gurjal, I. Hernandez-Lucas, A. Hong, L. Huizar, R. W. Hyman, T. Jones, D. Kahn, M. L. Kahn, S. Kalman, D. H. Keating, E. Kiss, C. Komp, V. Lelaure, D. Masuy, C. Palm, M. C. Peck, T. M. Pohl, D. Portetelle, B. Purnelle, U. Ramsperger, R. Surzycki, P. Thebault, M. Vandenbol, F. J. Vorholter, S. Weidner, D. H. Wells, K. Wong, K. C. Yeh, and J. Batut. 2001. The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 18.Glick, B. R., D. M. Penrose, and J. Li. 1998. A model of the lowering of plant ethylene concentrations by plant growth-promoting bacteria. J. Theor. Biol. 190:63-68. [DOI] [PubMed] [Google Scholar]
- 19.Glick, B. R., C. L. Patten, G. Holguin, and D. M. Penrose. 1999. Biochemical and genetic mechanisms used by plant growth-promoting bacteria. Imperial College Press, London, United Kingdom.
- 20.Grichko, V. P., and B. R. Glick. 2001. Amelioration of flooding stress by ACC deaminase-containing plant growth-promoting bacteria. Plant Physiol. Biochem. 39:11-17. [Google Scholar]
- 21.Guinel, F. C., and R. D. Geil. 2002. A model for the development of the rhizobial and arbuscular mycorrhizal symbioses in legumes and its use to understand the roles of ethylene in the establishment of these two symbioses. Can. J. Bot. 80:695-720. [Google Scholar]
- 22.Guinel, F. C., and T. A. LaRue. 1992. Ethylene inhibitors partly restore nodulation to pea mutant E107 (brz). Plant Physiol. 99:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-575. [DOI] [PubMed] [Google Scholar]
- 24.Heidstra, R. W., W. C. Yang, Y. Yalcin, S. Peck, A. M. Emons, A. van Kammen, and T. Bisseling. 1997. Ethylene provides positional information on cortical cell division but is not involved in Nod factor-induced root hair tip growth in Rhizobium-legume interaction. Development 124:1781-1787. [DOI] [PubMed] [Google Scholar]
- 25.Honma, M., and T. Shimomura. 1978. Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 42:1825-1831. [Google Scholar]
- 26.Hosie, A. H. F., D. Allaway, M. A. Jones, D. L. Walshaw, A. W. B. Johnston, and P. S. Poole. 2001. Solute-binding protein-dependent ABC transporters are responsible for solute efflux in addition to solute uptake. Mol. Microbiol. 40:1449-1459. [DOI] [PubMed] [Google Scholar]
- 27.Jensen, H. L. 1942. Nitrogen fixation in leguminous plants. I. General characters of root-nodule bacteria isolated from species of Medicago and Trifolium in Australia. Proc. Linu. Soc. NSW 66:98-108. [Google Scholar]
- 28.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 29.Lee, K. H., and T. A. LaRue. 1992. Exogenous ethylene inhibits nodulation of Pisum sativum L. cv Sparkle. Plant Physiol. 100:1759-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ligero, F., C. Lluch, and J. Olivares. 1986. Evolution of ethylene from roots of Medicago sativa plants inoculated with Rhizobium meliloti. J. Plant Physiol. 125:361-365. [Google Scholar]
- 31.Ma, W., S. B. Sebestianova, J. Sebestian, G. I. Burd, F. C. Guinel, and B. R. Glick. 2003. Prevalence of 1-aminocyclopropane-1-carboxylate deaminase in Rhizobia spp. Antonie van Leeuwenhoek 83:285-291. [DOI] [PubMed] [Google Scholar]
- 32.Ma, W., F. C. Guinel, and B. R. Glick. 2003. Rhizobium leguminosarum biovar viciae 1-aminocyclopropane-1-carboxylate deaminase promotes nodulation of pea plants. Appl. Environ. Microbiol. 69:4396-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayak, S., T. Tirosh, and B. R. Glick. 2004. Plant growth-promoting bacteria that confer resistance in tomato to salt stress. Plant Physiol. Biochem. 42:565-572. [DOI] [PubMed]
- 34.Mayak, S., T. Tirosh, and B. R. Glick. 2004. Plant growth-promoting bacteria that confer resistance to water stress in tomato and pepper. Plant Sci. 166:525-530. [DOI] [PubMed] [Google Scholar]
- 35.Meade, H. M., S. R. Long, G. B. Ruvkum, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon mutagenesis. J. Bacteriol. 147:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Nukui, N., H. Ezura, K. Yuhashi, T. Yasuta, and K. Minamisawa. 2000. Effects of ethylene precursor and inhibitors for ethylene biosynthesis and perception on nodulation in Lotus japonicus and Macroptilium atropurpureum. Plant Cell Physiol. 41:893-897. [DOI] [PubMed] [Google Scholar]
- 38.Oldroyd, G. E. D., E. M. Engstrom, and S. R. Long. 2001. Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell 13:1835-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Penmetsa, R. V., and D. R. Cook. 1997. A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275:527-530. [DOI] [PubMed] [Google Scholar]
- 40.Penrose, D. M., and B. R. Glick. 2001. Levels of 1-aminocyclopropane-1-carboxylic acid (ACC) in exudates and extracts of canola seeds treated with plant growth-promoting bacteria. Can. J. Microbiol. 47:368-372. [DOI] [PubMed] [Google Scholar]
- 41.Peters, N. K., and D. Crist-Estes. 1989. Nodule formation is stimulated by ethylene inhibitor aminoethoxyvinylglycine. Plant Physiol. 91:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pocard, J.-A., T. Bernard, L. T. Smith, and D. Le Rudulier. 1989. Characterization of three choline transport activities in Rhizobium meliloti: modulation by choline and osmotic stress. J. Bacteriol. 171:531-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pocard, J.-A., N. Vincent, E. Boncompagni, L. T. Smith, M.-C. Poggi, and D. Le Rudulier. 1997. Molecular characterization of the bet genes encoding glycine betaine synthesis in Sinorhizobium meliloti 102F34. Microbiology 143:1369-1379. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:783-791. [Google Scholar]
- 46.Smith, L. T., J.-A. Pocard, T. Bernard, and D. Le Rudulier. 1988. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 170:3142-3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan, J. T., J. R. Trzebiatowski, R. W. Cruickshank, J. Gouzy, S. D. Brown, R. M. Elliot, D. J. Fleetwood, N. G. McCallum, U. Rossbach, G. S. Stuart, J. E. Weaver, R. J. Webby, F. J. Bruijn, and C. W. Ronson. 2002. Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J. Bacteriol. 184:3086-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Spronsen, P. C., A. A. N. van Brussel, and J. W. Kjine. 1995. Nod factors produced by Rhizobium leguminosarum biovar viciae induce ethylene-related changes in root cortical cells of Vicia sativa ssp. nigra. Euro. J. Cell Biol. 68:463-469. [PubMed] [Google Scholar]
- 49.Vasse, J., F. de Billy, and G. Truchet. 1993. Abortion of infection during the Rhizobium meliloti-alfalfa symbiotic interaction is accompanied by a hypersensitive reaction. Plant J. 4:555-566. [Google Scholar]
- 50.Wang, C., E. Knill, B. R. Glick, and G. Défago. 2000. Effect of transferring 1-aminocyclopopane-1-carboxylic acid (ACC) deaminase genes into Pseudomonas fluorescens strain CHA0 and its gacA derivative CHA96 on their growth-promoting and disease-suppressive capacities. Can. J. Microbiol. 46:898-907. [DOI] [PubMed] [Google Scholar]
- 51.Wisniewski, J. P., E. A. Rathbun, J. P. Knox, and N. J. Brewin. 2000. Involvement of diamine oxidase and peroxidase in insolubilization of the extracellular matrix: implications for pea nodule initiation by Rhizobium leguminosarum. Mol. Plant-Microbe Interact. 13:413-420. [DOI] [PubMed] [Google Scholar]
- 52.Yuhashi, K. I., N. Ichikawa, H. Ezura, S. Akao, Y. Minakawa, N. Nukui, T. Yasuta, and K. Minamisawa. 2000. Rhizobitoxine production by Bradyrhizobium elkanii enhances nodulation and competitiveness on Macroptilium atropurpureum. Appl. Environ. Microbiol. 66:2658-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]